Abstract
Clostridium perfringens is an important pathogen that provokes numerous different diseases. This bacterium is classified into five different types, each of which capable of causing a different disease. There are various methods for the bacterial identification, many are labor-intensive, time-consuming, expensive and also present low sensitivity and specificity. The aim of this research was to identify the different types of C. perfringens using PCR molecular method. In this study, 130 sheep-dung samples were randomly collected from areas around the city of Kerman, southeastern Iran. After processing and culturing of samples, the produced colonies were morphologically studied, gram stain test was also carried out and the genera of these bacteria were identified through biochemical tests. DNA extracted from isolated bacteria for genotyping was tested by multiplex PCR with specific primers. Based on length of synthesized fragments by PCR, toxin types and bacterial strains were detected. C. perfringens isolated types were divided as follows: 17.39% type A, 21.74% type B, 34.78% type C and 26.09% type D. It should be emphasized that, up to the present moment, C. perfringens type A has not been reported in Iran.
Clostridium perfringens; multiplex PCR; biochemical tests; genetic typing
ORIGINAL PAPER
Clostridium perfringens isolate typing by multiplex PCR
Ahsani MRI; Mohammadabadi MRI; Shamsaddini MBII
IDepartment of Animal Science, Shahid Bahonar University of Kerman, Kerman, Iran
IIRazi Vaccine and Serum Research Institute, Kerman, Iran
Correspondence to Correspondence to: MOHAMMAD REZA AHSANI Department of Animal Sciences, Shahid Bahonar University of Kerman 22 Bahman BLVD Kerman, Kerman, PO Box 76169133, Iran Email: Ahsani2001@gmail.com.
ABSTRACT
Clostridium perfringens is an important pathogen that provokes numerous different diseases. This bacterium is classified into five different types, each of which capable of causing a different disease. There are various methods for the bacterial identification, many are labor-intensive, time-consuming, expensive and also present low sensitivity and specificity. The aim of this research was to identify the different types of C. perfringens using PCR molecular method. In this study, 130 sheep-dung samples were randomly collected from areas around the city of Kerman, southeastern Iran. After processing and culturing of samples, the produced colonies were morphologically studied, gram stain test was also carried out and the genera of these bacteria were identified through biochemical tests. DNA extracted from isolated bacteria for genotyping was tested by multiplex PCR with specific primers. Based on length of synthesized fragments by PCR, toxin types and bacterial strains were detected. C. perfringens isolated types were divided as follows: 17.39% type A, 21.74% type B, 34.78% type C and 26.09% type D. It should be emphasized that, up to the present moment, C. perfringens type A has not been reported in Iran.
Key words:Clostridium perfringens, multiplex PCR, biochemical tests, genetic typing.
INTRODUCTION
C. perfringens has a notorious reputation for possessing an extremely wide toxin arsenal, with over 15 toxins and may be the most widely occurring pathogenic in the globe (1, 2). The organism is therefore capable of producing different pathological pictures and of causing numerous different histotoxic and enteric diseases in both humans and other animals (1). Alpha (α), beta (β), epsilon (µ) and iota (¹) comprise the four main C. perfringens toxins which form the basis of categorizing this bacterium into five different types, from A to E (3- 5). These different bacterial types provoke diverse diseases and even one strain may cause the infection in numerous hosts (6, 7).
In sheep, enterotoxemia is caused by different toxin types of C. perfringens (8). This infection causes considerable economic loss to the sheep industry due to high fatality rates, increased treatment costs, and decreased productivity (9). On the other hand, domestic animals are known to be sources of human food poisoning. In order to reduce or eliminate this risk, strategies must be developed to diagnose and prevent infected animals from entering the food chain (10).
In some laboratories, a serum neutralization test on mice or guinea pigs is employed to determine and diagnose bacterial toxins (11). This method is tedious, time-consuming, expensive and monovalent. Furthermore, it is improper and unethical to apply it at the expense of laboratory animals (10-12).
According to Timoney et al. (13), the antitoxin µ was measured in sheep blood sera by ELISA, which proven to be a specific, quick and economical method that may replace the serum neutralization test. ELISA utilizes polyclonal antibodies to identify C. perfringens toxins (14). However, its disadvantage is that the interaction reaction among the produced antibodies works against the toxins, which may difficult the identification of toxin types. Moreover, ELISA falls short of identifying β2 toxins and in order to identify toxins from spore-forming bacteria, it must be activated by means of special culture methods (15).
Biochemical tests are also incapable of distinguishing different types of C. perfringens (16). PCR is the most modern practical technology in diagnosing infectious diseases and compared with classical techniques, it has been shown to be more rapid, with results obtained in a few hours, and also more reliable (12, 14). PCR allows a faster bacterial identification directly from clinical samples (17). It should be noted that up to this moment, no research on this subject has been carried out in Iran.
MATERIALS AND METHODS
In the present study, sheep dung samples were obtained from 130 Kermani sheep, from nine locations in southeastern Iran. The sheep were from both genders and their ages ranged from 1 to 2.5 years. The samples were collected aseptically in sterile plastic bags and transferred to the laboratory within 1 to 2 hours, after which they were accordingly processed. Samples were diluted in PBS (1:10), the bath temperature was maintained at 80°C for ten minutes in order to eliminate the non-spore-forming bacteria, subsequently they were cultivated on 5% sheep blood agar and anaerobically incubated using Anoxomat® (Mart Microbiology, Netherlands) at 37°C for 48 hours.
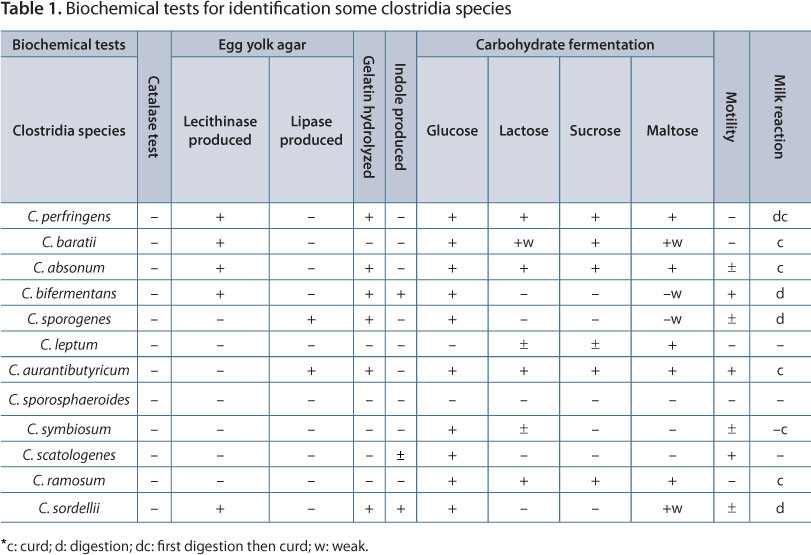
The suspected colonies were identified by characteristic colony morphology, gram staining and biochemical tests as described by MacFaddin (16). Biochemical identification consisted of gelatin and lecithin hydrolyses; and catalase, lipase and motility tests. Moreover, indole production, litmus milk reaction and carbohydrate fermentation (sucrose, glucose, lactose and maltose) were carried out in order to identify the bacterial genus according to MacFaddin (16) (Table 1).
DNA Extraction and PCR
Strains of C. perfringens types A, B, C, D and E were considered positive controls while C. septicum, C. sordellii, C. sporogenes, C. leptum and C. ramosum were the negative controls. Isolated strains were cultured on blood agar and total DNA was extracted as described by Sambrook and Russell (18). Two or three colonies that grown on blood agar were suspended in 350 μL of STET buffer (100 mM Tris-HCl, 10 mM EDTA, 100 mM NaCl, 5% Triton® X-100) in 1.5-mL microtubes. Then, 25 μL of lysozyme (10 mg/mL) was added to each solution and the content was mixed. Subsequently, the microtubes were placed in a boiling water bath for 40 seconds.
The bacterial lysate was centrifuged at 13,000 rpm for 15 minutes at room temperature in a Microfuge® refrigerated microcentrifuge (Beckman Coulter, USA). After that, the supernatant was poured into a fresh microcentrifuge tube and the nucleic acids were precipitated from it by adding 40 μL of 2.5 M sodium acetate (pH 5.2) and 420 μL of isopropanol. Precipitated nucleic acids were recovered by centrifugation at 13,000 rpm for ten minutes at 4°C. Next, the supernatant was removed and the pellet of nucleic acid was rinsed with 1 mL of 70% ethanol at 4°C. Finally, the pellet was dried and resuspended in 50 μL of TE buffer containing RNase.
Four specific primer sets corresponding to each toxin of C. perfringens were evaluated for their ability to detect C. perfringens and to discriminate between C. perfringens and other clostridia species (Table 2), according to Komoriya et al. (19) and van Asten et al. (20). The PCR was performed in a Thermal Cycler® (Bio-Rad, USA) in a total reaction volume of 50 μL containing: 5 μL of 10x PCR buffer (10 mM Tris-HCL, pH 9.0, 50 mM KCl), 2 μL 50 mM MgCl2, 250 μM of each deoxynucleotide triphosphate, 5 U of Taq DNA polymerase, 100 pmol of primers and 5 μL of template DNA.
Amplification was obtained with 35 cycles following an initial denaturating step at 95°C for ten minutes. Each cycle comprised denaturation at 94°C for 45 seconds, annealing at 55°C for 30 seconds, and synthesis at 72°C for 90 seconds. The final extension step occurred at 72°C for ten minutes. Then, 10 μL of the amplified product was electrophoresed in a 1.5% agarose gel and stained with ethidium bromide. Amplified bands were visualized and photographed under UV illumination.
RESULTS
The isolated C. perfringens comprised 30% of the samples, including C. perfringens, C. baratii, C. absonum, C. bifermentans, C. sporogenes, C. leptum, C. aurantibutyricum, C. sporosphaeroides, C. symbiosum, C. scatologenes, C. ramosum and C. sordellii. The DNA extracted from all bacterial strains was identified by biochemical tests and standard strains. The quality of the extracted DNA was examined through 1% agarose gel and confirmed.
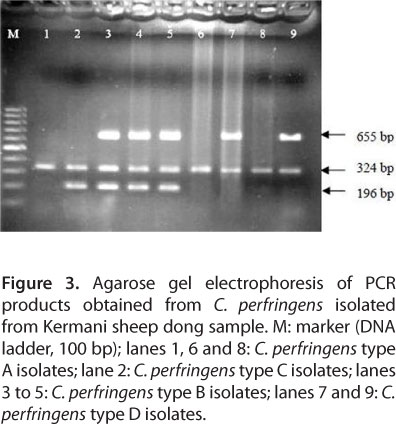
PCR was carried out with four sets of specific primers that were complementary to a fragment of α, β, µ and ¹ encoding genes of C. perfringens. Primer annealing of complementary genes and synthesization of the fragment of interest revealed a specified toxin gene. Thus, different types were identified in accordance with their toxins. The toxin encoding gene was amplified by means of single PCR, the fragment of 324 bp belonged to the α toxin gene, shared by all the types. The 196 bp fragment, on the other hand, was from the β toxin encoding gene, existing in types B and C. The 655 bp fragment was from the µ toxin encoding gene and is observable in type D. The ¹ toxin encoding gene found in type E generates the fragment of 446 bp (Figure 1). PCR results corresponding to positive and negative controls are displayed in Figure 2. Out of 23 C. perfringens types, isolated by biochemical tests, four (17.39%) were type A, five (21.74%) were type B, eight (34.78%) were type C and six (26.09%) were type D, with type C as the dominant one (Figure 3).
DISCUSSION
Toxins of C. perfringens are responsible for enterotoxaemia in sheep (21). Furthermore, it has been reported that the infection, which causes considerable economic losses to the sheep industry, may be provoked by all five types of C. perfringens (2, 11). It is well known that vaccination against C. perfringens comprises the main prophylactic measure to reduce losses or minimize the severity of enterotoxaemia (22). The vaccines should provide adequate protective immunity against all C. perfringens types of a certain area, since other diseases that affect humans and domestic animals originate in this bacterium (11, 23).
Biological gauge comprises one of the important methods of identifying C. perfringens; however, it presents several disadvantages including the long time the test takes (24 to 72 hours) (13). Another limitation consists of non-specific deaths that may lead to false diagnosis. Moreover, some C. perfringens strains are incapable of producing toxins in measurable amounts under laboratory conditions, which creates an obstacle for typing by classical methods (21).
Sensitivity and specificity are the two main characteristics of an efficient and practical technique, two qualities that PCR presents. Rapidity constitutes one of the major advantages of this method, so that bacterium identification and type determination lasts no more than four hours. Hence, the toxicogenic strain in the sample can be identified by means of a rapid evaluation by the PCR technique before it produces toxin.
In the current study, C. perfringens types A, B, C, and D were isolated from sheep-dong samples, with type C being the most frequent in Kerman. Therefore, it is recommended that vaccination against enterotoxemia in this province should provide adequate immunity, especially against C. perfringens types A and C. Furthermore, enterotoxemia in sheep is asymptomatic; thus, molecular detection of C. perfringens in farms may be useful for prophylaxis.
Up to our knowledge, no similar research has been conducted in Iran and the findings of the present study disagree with those from different countries (3, 7, 21, 24). The difference comprises the fact that the existence and emergence of some bacterium types is closely related to geographic features, consequently several regions may totally lack one or more types of a certain bacterium. For instance, C. perfringens type A has been reported as most prevalent in North America, while types C, D and E are uncommon. Moreover, type A constitutes 97% of isolates in Belgium (8). Although previous studies suggested that types A and E did not exist in Iran, the present study detected and identified type A for the first time in Kerman, southeastern Iran (13).
Primers used in this study were perfectly specific, only complementary to C. perfringens encoding toxin gene. As Figure 2 demonstrates, these primers do not identify other strains. In addition, PCR showed to be highly sensitive, so that the reaction was accomplished successfully with a small amount of DNA (less than 1 μL).
In general, molecular methods provide a new insight into bacterial classification. They offer essential genetic information about the organism of interest, which comprises one of the most practical and helpful aspects of PCR. As PCR employs specific primers, an individual species or strain may be traced among different species or types of organisms (17). Compared to other identification techniques that are based on large amounts of samples, PCR is safer for researchers; since sample preparation for PCR starts with cell lysis and DNA extraction. It is unnecessary to say that the cell cannot survive this stage and loses its pathogenic property (17). In conclusion, PCR has become an essential research and diagnostic tool, being a powerful technique with a vast and increasing range of applications.
ACKNOWLEDGEMENTS
The authors wish to thank Razi Vaccine and Serum Research of Kerman, Iran, for their help.
Submission status
Received: January 29, 2010.
Accepted: June 10, 2010.
Abstract published online: June 21, 2010.
Full paper published online: November 30, 2010.
FINANCIAL SOURCE
Razi Vaccine and Serum Research Institute of Kerman, Iran, provided the financial grant.
CONFLICTS OF INTEREST
There is no conflict.
- 1. Smedley JG 3rd, McClane BA. Fine mapping of the N-terminal cytotoxicity region of Clostridium perfringens enterotoxin by site-directed mutagenesis. Infect Immun. 2004;72(12):6914-23.
- 2. Songer JG. Clostridial enteric diseases of domestic animals. Clin Microbial Rev. 1996;9(2):216-34.
- 3. Yoo HS, Lee SU, Park KY, Park YH. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J Clin Microbiol. 1997;35(1):228-32.
- 4. Sawires YS, Songer JG. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe. 2006;12(1):23-43.
- 5. Gurjar AA, Hegde NV, Love BC, Jayarao BM. Real-time multiplex PCR assay for rapid detection and toxin typing of Clostridium perfringens toxin producing strains in feces of dairy cattle. Mol Cell Probes. 2008;22(2):90-5.
- 6. Wasinski B. Contribution of Clostridium perfringens type A with β2 toxin gene in aetiology of porcine enteric diseases. A case report. Bull Vet Inst Pulawy. 2007;51(1):509-13.
- 7. Wojdat E, Kwiatek K, Kozak M. Occurrence and characterization of some Clostridium species isolated from animal feeding stuffs. Bull Vet Inst Pulawy. 2006;50(1):63-7.
- 8. Songer JG. Bovine enteritis and enterotoxemia. In: Mainil J, editor. Genus Clostridium - clostridia in medical, veterinary and food microbiology: diagnosis and typing. Luxembourg: European Commission/EU Concerted Action; 2006. p. 43-80.
- 9. Greco G, Madio A, Buonavoglia D, Totaro M, Corrente M, Martella V, et al. Clostridium perfringens toxin-types in lambs and kids affected with gastroenteric pathologies in Italy. Vet J. 2005;170(3):346-50.
- 10. Piatti M, Ikuno AA, Baldassi L. Detection of bovine Clostridium perfringens by polymerase chain reaction. J Venom Anim Toxins incl Trop Dis. 2004;10(2):154-60.
- 11. Gokce HI, Genc O, Sozmen M, Gokce G. Determination of Clostridium perfringens toxin-types in sheep with suspected enterotoxemia in Kars Province, Turkey. Turk J Vet Anim Sci. 2007;31(5):355-60.
- 12. Miyashiro S, Nassar AFC, Del Fava C, Cabral AD, Silva M. Clostridium perfringens types A and D associated with enterotoxemia in an 18-month-old goat. J Venom Anim Toxins incl Trop Dis. 2007;13(4):885-93.
- 13. Timoney JF, Gillespie JH, Scott FW, Barlough JE. Hagan and Bruner's microbiology and infectious diseases of domestic animals. 8th ed. Ithaca: Comstock Publishing Associates; 1988. p. 214-40.
- 14. Baron EJ, Finegold SM, Martin WJ. Organisms encountered in the urinary tract. In: Baron EJ, Finegold SM, editors. Bailey and Scott's Diagnostic Microbiology. St. Louis: Mosby; 1990. p. 49-194.
- 15. Baums CG, Schotte U, Amtsberg G, Goethe R. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet Microbiol. 2004;100(1-2):11-6.
- 16. MacFaddin JF. Biochemical tests for identification of medical bacteria. Baltimore: Lippincott Williams & Wilkins; 2000. p. 1-450.
- 17. Bartlett JMS, Stirling D. PCR protocols (methods in molecular biology). 2th ed. Totowa: Human Press; 2003. p. 3-115 .
- 18. Sambrook J, Russell DW. Molecular cloning a laboratory manual. 3th ed. Woodbury: Cold Spring Harbor Laboratory Press; 2002. p. 18-96.
- 19. Komoriya T, Hashimoto A, Shinozaki A, Inoue M, Kohno H. Study on partial purification of α-toxin produced from obligate anaerobe Clostridium perfringens. Report of the Research Institute of Industrial Technology, Nihon University. 2007;88(1):1-11.
- 20. van Asten AJAM, van der Wiel CW, Nikolaou G, Houwers DJ, Grone A. A multiplex PCR for toxin typing of Clostridium perfringens Vet Microbiol. 2009;136(1):411-2.
- 21. Kalender H, Ertas HB, Cetinkaya B, Muz A, Arslan N, Kilic A. Typing of isolates of Clostridium perfringens from healthy and diseased sheep by multiplex PCR. Vet Med. 2005;50(10):439-42.
- 22. de la Rosa C, Hogue DE, Thonney ML. Vaccination schedules to raise antibody concentrations against epsilon-toxin of Clostridium perfringens in ewes and their triplet lambs. J Anim Sci. 1997;75(9):2328-34.
- 23. Juneja VK, Marks H, Thippareddi H. Predictive model for growth of Clostridium perfringens during cooling of cooked uncured beef. Food Microbiol. 2008;25(1):42-55.
- 24. Albini S, Brodard I, Jaussi A, Wollschlaeger N, Frey J, Miserez R, et al. Real-time multiplex PCR assays for reliable detection of Clostridium perfringens toxin genes in animal isolates. Vet Microbiol. 2008;127(1-2):179-85.
Publication Dates
-
Publication in this collection
30 Sept 2011 -
Date of issue
2010
History
-
Received
29 Jan 2010 -
Accepted
10 June 2010