Abstract
Mitochondrial DNA (mtDNA) has been widely employed in phylogeographic and phylogenetic studies. In the present study, the genetic identification of the scorpion Androctonus crassicauda (Olivier, 1807) was carried out by using the 16S mitochondrial gene, since this scorpion represents the most important species in Turkey regarding scorpionism and antivenom production. Two genetic groups were found according to the sequence analysis results, while five different loci at the nucleotide level presented genetic variation in the 16S region when compared to a known A. crassicauda sequence data (GenBank, AJ277598). Nucleotide variations found in the current work constitute the first descriptive report for A. crassicauda. Moreover, future studies may enlighten the genetic and venom composition variations for this scorpion species.
scorpion; Androctonus crassicauda; PCR; genotype; Turkey
ORIGINAL PAPER
A study on the genetic diversity of Androctonus crassicauda (Olivier, 1807; Scorpiones: Buthidae) from Turkey
Ozkan OI;II; Ahmet CIII; Zafer KII
IRefik Saydam Public Health Agency, Ankara, Turkey
IIDepartment of Entomology and Protozoology, Faculty of Veterinary Medicine, Ankara University, Ankara, Turkey
IIITurkish Accreditation Agency, Ankara, Turkey
Correspondence to Correspondence to: Ozcan Ozkan Refik Saydam Public Health Agency 06100 Ankara, Turkey Phone: 00 90 312 498 21 50. Email: ozcanozkan_62@hotmail.com.
ABSTRACT
Mitochondrial DNA (mtDNA) has been widely employed in phylogeographic and phylogenetic studies. In the present study, the genetic identification of the scorpion Androctonus crassicauda (Olivier, 1807) was carried out by using the 16S mitochondrial gene, since this scorpion represents the most important species in Turkey regarding scorpionism and antivenom production. Two genetic groups were found according to the sequence analysis results, while five different loci at the nucleotide level presented genetic variation in the 16S region when compared to a known A. crassicauda sequence data (GenBank, AJ277598). Nucleotide variations found in the current work constitute the first descriptive report for A. crassicauda. Moreover, future studies may enlighten the genetic and venom composition variations for this scorpion species.
Key words: scorpion, Androctonus crassicauda, PCR, genotype, Turkey.
INTRODUCTION
Scorpions can be regarded as "living fossils" because they have changed so little during the last 400 million years (1-3). They are distributed throughout the tropical, temperate, and cold zones of the Earth (4).
More than 1,500 species of scorpions have been described so far, but only 50 were stated as dangerous to humans (2). Scorpion sting is the most important type of arachnid envenomation resulting in adult morbidity and pediatric mortality (5, 6). Scorpionism remains a real health problem in developing countries, especially in tropical and subtropical regions including urban areas.
Lethal scorpions mostly belong to the family Buthidae, among which species of the genera Androctonus, Leiurus, and Mesobuthus are responsible for envenomation in Turkey (7, 8). Androctonus crassicauda is the most significant scorpion species in Turkey due to scorpionism and antivenom production (8, 9). This species can be found in Azerbaijan, Iran, Iraq, Syria, Jordan, Saudi Arabia and Yemen, besides Turkey (5, 10, 11).
The individual and geographical variability in venoms of one same species has become extremely important for the production of effective antivenoms and for the understanding of the clinical symptoms of patients. Intragenic differences can be a result of genetic and environmental factors (12).
Whittemore et al. (13) reported that the Turkish antivenom (antivenom produced against A. crassicauda venom) was more effective than homologous antivenom in neutralizing A. australis venom in Algeria. However, antivenom against A. crassicauda was found to have the same neutralization capacity as homologous antivenoms against B. occinatus venom in South Europe and North Africa, T. serrulatus and T. bahiensis venoms in South America, and L. quinquestriatus venom in Anatolia.
In this study, we aimed to use nucleotide sequences to obtain information about the taxonomic status of A. crassicauda scorpion, which is the most important scorpion species in Turkey considering envenomation cases and antivenom production.
MATERIALS AND METHODS
Specimens
The scorpions were originally collected from Sanliurfa province (36° 40'-38° 02' W, 37° 50'-42° 12' E) in the Southeast region of Turkey. The animals were captured at night by using a UV lamp. They were kept in plastic boxes at the Department of Entomology and Protozoology, Faculty of Veterinary Medicine, Ankara University.
Extraction of Genomic DNA (gDNA)
Genomic DNA was extracted from fresh or preserved (96% ethanol) muscle tissue (a leg) using the phenol/chloroform method (14). Tissues were rapidly frozen and ground in liquid nitrogen to produce readily digestible pieces. Samples were homogenized in homogenization buffer solution [10 mM Tris-Cl (pH 8.0); 0.1 M EDTA (pH 8.0); 20 g/mL RNAse A; 10% sodium dodecyl sulfate; 0.1 mg/mL proteinase K] by agitation for ten minutes under incubation at 50°C. After homogenization, DNA was purified in standard 25:24:1 phenol/chloroform/isoamyl alcohol series and precipitated by addition of 0.2 volumes of 10 M ammonium acetate and two volumes of 100% ethanol. The DNA pellet was centrifuged, washed twice in 70% ice-cold ethanol, dried at room temperature and resuspended in TE buffer (10 mM Tris-Cl, 1 mM EDTA) until dissolved. The quality of the genomic DNA was examined on 0.8% ethidium bromide-stained agarose gels and OD260 was determined by using UV spectrophotometer.
Amplification, Sequence Determination and Alignment
A part of the large-subunit mitochondrial ribosomal gene (16S rRNA) was amplified by using a pair of scorpion-specific 16S primers or LR-J-12887. The sequence of the forward primer was 5'-CGA TTT GAA CTC AGA TCA-3' and that of the reverse primer was 5'-GTG CAA AGG TAG CAT AAT CA-3', as reported by Gantenbein et al. (15). These primers corresponded to the positions 12.867-12.887 and 13.218-13.310 in the Drosophila yakuba mitochondrial genome (15). In a final volume of 25 μL, amplification mixtures contained 1.25U of Taq DNA polymerase (Promega, USA), 2.0 mM of MgCl2, 0.2 μM of dNTPs, 0.3 pmol of each primer (alpha DNA) and 100 ng of genomic DNA. The thermal cycling program started with an initial denaturing step at 94°C for five minutes, followed by 45 amplification cycles (94°C for one minute denaturation, 48°C for one minute annealing, 72°C for 1.5 minute extension), and a final extension at 72°C for ten minutes in a gradient PCR (Biometra T® Gradient, USA). Finally, the amplification products were verified on 1.5% agarose electrophoretic gel and purified by using the QIAquick PCR® Purification kit (Qiagen, Germany).
The sequencing reaction was carried out with the forward and reverse primers using the gel-purified PCR product as template. The purified templates were sequenced with BigDye®Terminator v.3.1 (Applied Biosystems, USA), run on an ABI Prism® 3100 Genetic Analyzer (RefGen, Turkey). The accuracy of data was confirmed by two-directional sequencing with the forward and reverse primers. Chromatograms and sequences were analyzed by using FinchTv program®, version 1.4.0 (Geospiza, Inc.; USA; http://www.geospiza.com). The obtained DNA sequences were edited and aligned by using Basic Local Alignment Search Tool software [BLAST, version 2.2.22 (http://blast.ncbi.nlm.nih.gov)] (16). All new DNA sequences reported in this paper were deposited in GenBank sequence database provided by the National Center for Biotechnology Information (accession numbers: FJ217732-FJ217739).
Phylogenetic Analyses
The neighbor-joining tree (17) was constructed from the distances by using MEGA4 software (http://www.megasoftware.net) (18, 19). As outgroup, we selected Pseudochactas ovchinnikovi Gromov, 1998 (20), the relict taxon of the monotypic family Pseudochactidae, which is considered the basal group for Buthidae (21).
RESULTS
All scorpions were examined under stereomicroscope. Most of them were reddish-brown, varying from brown to black. The patellae of pedipalps did not present ventral trichobothria. All scorpions were identified as A. crassicauda according to morphological data.
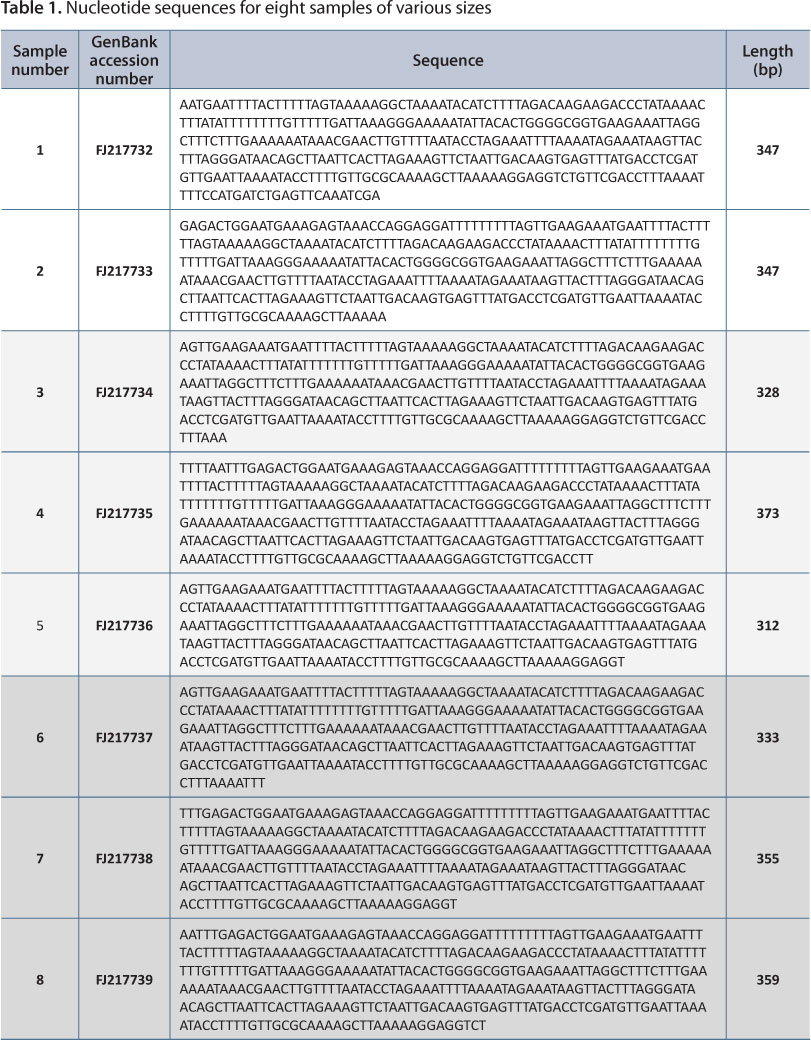
Eight samples were analyzed and amplified to ca. 400-base pair (bp) fragment of the 16S rRNA mitochondrial (mt) DNA by PCR. The DNA sequences of the mtDNA 16 S gene are listed in Table 1.
Eight 16S DNA sequences were blasted and aligned with interspecies. The sequences were divided into two genetic groups due to a T nucleotide Indel. Genetic group I (GI) was found to be 99.7% homologous with Genetic group II (GII), according to the alignment result. There are eight T nucleotides between positions 81 and 88 in the 315 base pairs in GI. However, GII has seven T nucleotides, as shown in Table 2. According to the alignment results, FJ217732, FJ217733 and FJ217737 make up GI, while FJ217734, FJ217735, FJ217736, FJ217738 and FJ217739 are GII. Genetic GI (Table 3) and GII (Table 4) were also blasted with a known A. crassicauda sequence data.
All results are based on the pairwise analysis of eight sequences. Analyses were conducted by using the maximum composite likelihood method in MEGA4 (18, 19). All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were 297 positions in the final data set.
The evolutionary history was inferred by using the neighbor-joining method (17). The percentage of replicate trees, in which the associated taxa clustered together in the bootstrap test (1000 replicates), are shown next to the branches (22). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the maximum composite likelihood method and are in the units of the number of base substitutions per site (18). All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were 286 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (19). Our sequence data are closely related (84%) to P. ovchinnikovi (GenBank accession number AY226167) (Figure 1).
DISCUSSION
Mitochondrial DNA (mtDNA) has been widely employed in phylogeographic and phylogenetic studies using conserved PCR primers (15). The order Scorpiones is under constant investigation regarding the taxonomic classification and phylogenetic relationships of its higher taxa (21, 23, 24). Recently, new scorpion species or subspecies have been recognized based on molecular techniques.
According to current reports, Androctonus crassicauda can be found in Sinai Peninsula (Egypt), across the entire Middle East (Southeastern Anatolia, Turkey), Arabian Peninsula and Armenia. In the present study, this species is reported as common in Sanl1urfa province near Syria and Iraqi borders in the Middle East. Androctonus crassicauda is one of the most medically important species in the Middle East and is also used for antivenom production.
The genotypes of A. crassicauda were determined according to the DNA sequence analysis of the 16S rRNA locus. The 16S rRNA locus in GI and GII was compared to the accession number AJ277598 (AcrTR from Urfa, Turkey).
GI was also blasted with AcrTR and was found to be 98.7% homologous with AcrTR. Differences in the first genetic group were due to a T for gap indel at position 81, a G for A substitution at positions 95 and 138, a T for C substitution at position 163, and a G for gap indel at position 312 (Table 3).
GII was blasted and was found to be 98.9% homologous with AcrTR. However, diversities in GII were due to a G for A substitution at positions 137 and 180, and a T for C substitution at position 205 (Table 4).
Comparing recent findings on A. crassicauda with the data from the present study, and according to the identification key, no morphological differences were detected between samples subjected to this study. However, this study determined two genetic groups according to the sequencing analysis. The genetic variation was determined in five different loci at the nucleotide level in the 16S region for the large-subunit mitochondrial ribosomal gene when compared to a known A. crassicauda sequence data (3). Nucleotide variations found in this study constitute the first descriptive report for A. crasscicauda.
Signs and symptoms of scorpionism depend on various factors including the genus, species, age, weight, feeding state and structure of scorpions, as well as the amount of venom injected, and the climate of the region. The genetic variation may also lead to diversity in A. crassicauda venom collected from one same locality.
In conclusion, the current study determined two genetic groups which have genetic variation (five different loci) in the 16S region when compared to a known A. crassicauda sequence (GenBank accession number AJ277598).
The sequence results obtained in this paper will increase the genetic information and benefit evolutionary taxonomists who work with scorpions.
In the future, the genetic variation and the composition of scorpion venom should be investigated for the development of more efficient antivenom to be used in scorpion envenomation. A combined morphological and molecular (DNA) phylogenetic analysis approach for the samples from the neighboring countries will also clarify the relationships among scorpion species. The investigation of diverse populations could determine the exact origin and possible ways of dispersal of A. crassicauda. In addition, the preliminary tree presented in this study may give an indication of the relationship among/within the species in the scorpion fauna of Turkey.
ACKNOWLEDGMENTS
We wish to thank Juliana Simionato for her valuable comments and high-quality editing on this manuscript.
Submission status
Received: April 15, 2010.
Accepted: July 12, 2010.
Abstract published online: August 13, 2010.
Full paper published online: November 30, 2010.
CONFLICTS OF INTEREST
There is no conflict.
- 1. Sissom WD. Systematics, biogeography and paleontology. In: Polis GA, editor. The biology of scorpions. Stanford, CA: Stanford University Press; 1990. p. 64-160.
- 2. Mullen G, Stockwell SA. Scorpions (Scorpiones). In: Mullen G, Durden L, editors. Medical and veterinary entomology. Amsterdam: Academic Press; 2002. p. 411-23.
- 3. Gantenbein B, Largiadèr CR. Mesobuthus gibbosus (Scorpiones: Buthidae) on the island of Rhodes: hybridization between Ulysses' stowaways and native scorpions? Mol Ecol. 2002;11(5):925-38.
- 4. Lourenço WR. Une nouvelle famille est nécessaire pour des microscorpions humicoles de Madagascar et d'afrique. C R Acad Paris Ser Sci Vie. 1998;321(1):845-8.
- 5. Radmanesh M. Androctonus crassicauda sting and its clinical study in Iran. J Trop Med Hyg. 1990;93(5):323-6.
- 6. Isbister GK, Graudins A, White J, Warrell D. Antivenom treatment in arachnidism. J Toxicol Clin Toxicol. 2003;41(3):291-300.
- 7. Demirsoy A, Durmu_ Y, Akbulut A. Türkiye scorpiones (akrep) faunas1n1n sistematik ve biyolojik yönden incelenmesi. Proje No: 1998 K 1001 40. Ankara: Çevre Bakanl11 Çevre Koruma Genel Müdürlüü Hayvanlar1 Koruma Dairesi Ba_kanl11; 2001. 118 p.
- 8. Ozkan O, Adiguzel S, Ates C, Bozyigit I, Filazi A. Optimization of antiscorpion venom production. J Venom Anim Toxins incl Trop Dis. 2006;12(3):390-9.
- 9. Trugut T. Türkiye akrepleri ve Türkiye'de haz1rlanm1_ anti-Androctonus crassicauda akrep serumunun paraspesifik etkisi. Türk Hijiyen ve Tercubei Biyoloji Dergisi. 1964;24(2):146-55.
- 10. Fet V, Braunwalder ME. The scorpions (Arachnida: Scorpiones) of the Aegean area: current problems in taxonomy and biogeography. Belg J Zool. 2000;130(Suppl 1):15-20.
- 11. Ozkan O, Adiguzel S, Kar S. Parametric values of Androctonus crassicauda (Olivier, 1807) (Scorpiones: Buthidae) from Turkey. J Venom Anim Toxins incl Trop Dis. 2006;12(4):549-59.
- 12. Abdel-Rahman MA, Omran MAA, Abdel-Nabi IM, Ueda H, McVean A. Intraspecific variation in the Egyptian scorpion Scorpio maurus palmatus venom collected from different biotopes. Toxicon. 2009;53(3):349-59.
- 13. Whittemore FW JR, Keegan HL, Borowitz JL. Studies of scorpion antivenins. 1. Paraspecificity. Bull World Health Organ. 1961;25(1):185-8.
- 14. Sambrook J, Fritschi EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1982. 545 p.
- 15. Gantenbein B, Fet V, Largiadèr C, Scholl A. First DNA phylogeny of the genus Euscorpius Thorell 1876 (Scorpiones, Euscorpiidae) and its bearing on the taxonomy and biogeography of this genus. Biogeographica (Paris). 1999;75(3):59-72.
- 16. Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1-2):203-14.
- 17. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-25.
- 18. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(1):1596-9.
- 19. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030-5.
- 20. Fet V, Gantenbein B, Gromov AV, Lowe G, Lourenço WR. The first molecular phylogeny of Buthidae (Scorpiones). Euscorpius. 2003;4(1):1-10.
- 21. Soleglad ME, Fet V. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius. 2003;11(1):1-75.
- 22. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-91.
- 23. Fet V, Soleglad ME. Contributions to scorpion systematics. I. On recent changes in high-level taxonomy. Euscorpius. 2005;31(1):1-13.
- 24. Prendini L, Wheeler WC. Scorpion higher phylogeny and classification, taxonomic anarchy, and standards for peer review in online publishing. Cladistics. 2005;21(5): 446-94.
Publication Dates
-
Publication in this collection
30 Sept 2011 -
Date of issue
2010
History
-
Accepted
12 July 2010 -
Received
15 Apr 2010