Abstract
Background:
Lachesis muta rhombeata is one of the venomous snakes of medical importance in Brazil whose envenoming is characterized by local and systemic effects which may produce even shock and death. Its venom is mainly comprised of serine and metalloproteinases, phospholipases A2 and bradykinin-potentiating peptides. Based on a previously reported fractionation of L. m. rhombeata venom (LmrV), we decided to perform a subproteome analysis of its major fraction and investigated a novel component present in this venom.
Methods:
LmrV was fractionated through molecular exclusion chromatography and the main fraction (S5) was submitted to fibrinogenolytic activity assay and fractionated by reversed-phase chromatography. The N-terminal sequences of the subfractions eluted from reversed-phase chromatography were determined by automated Edman degradation. Enzyme activity of LmrSP-4 was evaluated upon chromogenic substrates for thrombin (S-2238), plasma kallikrein (S-2302), plasmin and streptokinase-activated plasminogen (S-2251) and Factor Xa (S-2222) and upon fibrinogen. All assays were carried out in the presence or absence of possible inhibitors. The fluorescence resonance energy transfer substrate Abz-KLRSSKQ-EDDnp was used to determine the optimal conditions for LmrSP-4 activity. Molecular mass of LmrSP-4 was determined by MALDI-TOF and digested peptides after trypsin and Glu-C treatments were analyzed by high resolution MS/MS using different fragmentation modes.
Results:
Fraction S5 showed strong proteolytic activity upon fibrinogen. Its fractionation by reversed-phase chromatography gave rise to 6 main fractions (S5C1-S5C6). S5C1-S5C5 fractions correspond to serine proteinases whereas S5C6 represents a C-type lectin. S5C4 (named LmrSP-4) had its N-terminal determined by Edman degradation up to the 53rd amino acid residue and was chosen for characterization studies. LmrSP-4 is a fibrinogenolytic serine proteinase with high activity against S-2302, being inhibited by PMSF and benzamidine, but not by 1,10-phenantroline. In addition, this enzyme exhibited maximum activity within the pH range from neutral to basic and between 40 and 50 °C. About 68% of the LmrSP-4 primary structure was covered, and its molecular mass is 28,190 Da.
Conclusions:
Novel serine proteinase isoforms and a lectin were identified in LmrV. Additionally, a kallikrein-like serine proteinase that might be useful as molecular tool for investigating bradykinin-involving process was isolated and partially characterized.
Keywords:
bushmaster; snake venom; SVSP; kallikrein-like; plasminogen activator; kininogenase; lectin; protease; envenomation
Background
The genus Lachesis is represented only by the species L. muta in Brazil and the ophidian accidents caused by these snakes are the second most lethal (mortality/number of accidents) in this country [11. Brasil. Ministério da Saúde: Acidente por animais peçonhentos - Notificações registradas no sistema de informação de agravos de notificação - Brasil. Período 2013; 2017., 22. Málaque CMS, França FOS. Acidente laquético. In: Animais Peçonhentos no Brasil Biologia, Clínica e Terapêutica dos Acidentes. Cardoso JLC, França, F, Wen FH, Haddad Jr V, editors. São Paulo; 2003. pp. 87-90.]. This lethality index might be derived from the high amount of venom injected into the victim but also to the lacking (or delayed) treatment access in remote regions [33. Jorge MT, Sano-Martins IS, Tomy SC, Castro SC, Ferrari RA, Ribeiro LA, et al. Snakebite by the bushmaster (Lachesis muta) in Brazil: case report and review of the literature. Toxicon. 1997;35(4):545-54. -55. Chippaux JP. Epidemiology of envenomations by terrestrial venomous animals in Brazil based on case reporting: from obvious facts to contingencies. J Venom Anim Toxins incl Trop Dis. 2015;21:13. doi: 10.1186/s40409-015-0011-1.
https://doi.org/10.1186/s40409-015-0011-...
]. Ophidian accidents caused by Lachesis are very significant, especially due to the quantity of venom that this snake is able to inject into the victim. The envenoming caused by this snake is characterized by local pain, edema, hemorrhage, necrosis, nausea, vomiting, coagulopathies (e. g. hypofibrinogenemia), renal disturbs, bradycardia, hypotension and shock, followed by a fast and irreversible hypotension which leads to death [22. Málaque CMS, França FOS. Acidente laquético. In: Animais Peçonhentos no Brasil Biologia, Clínica e Terapêutica dos Acidentes. Cardoso JLC, França, F, Wen FH, Haddad Jr V, editors. São Paulo; 2003. pp. 87-90., 33. Jorge MT, Sano-Martins IS, Tomy SC, Castro SC, Ferrari RA, Ribeiro LA, et al. Snakebite by the bushmaster (Lachesis muta) in Brazil: case report and review of the literature. Toxicon. 1997;35(4):545-54. , 66. Torres JR, Torres MA, Arroyo-Parejo MA. Coagulation disorders in bushmaster envenomation. Lancet. 1995;346(8972):449-50.-1111. Ripa D. Ontogeny of the shock death in human beings. In: The bushmasters (Genus Lachesis Daudin, 1803) morphology in evolution and behavior, 5 edition. Wilmington: Cape Fear Serpentarium; 2007. ].
Lachesis muta rhombeata venom (LmrV) comprises mainly bradykinin potentiating peptides (BPPs), serine proteinases, metalloproteinases and phospholipases A2 (PLA2) besides lectins, cysteine-rich secretory proteins (CRISP), L-amino acid oxidase (LAAO), vascular endothelial growth factor (VEGF) and phospholipase B [1212. Pla D, Sanz L, Molina-Sánchez P, Zorita V, Madrigal M, Flores-Díaz M, et al. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J Proteomics. 2013;89:112-23., 1313. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.]. As described above, Lachesis venoms present local and systemic alterations. The serine proteinases from these venoms are usually involved in hydrolysis of coagulation factors and may act in blood pressure reduction [1414. Diniz MR, Oliveira EB. Purification and properties of a kininogenin from the venom of Lachesis muta (bushmaster). Toxicon. 1992;30(3):247-58.-1616. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta. Int J Biochem. 1989;21(8):863-71.] whereas metalloproteinases present hemorrhagic actions [1717. Rucavado A, Flores-Sánchez E, Franceschi A, Magalhaes A, Gutiérrez JM. Characterization of the local tissue damage induced by LHF-II, a metalloproteinase with weak hemorrhagic activity isolated from Lachesis muta muta snake venom. Toxicon. 1999;37(9):1297-312., 1818. Estêvão-Costa MI, Diniz CR, Magalhães A, Markland FS, Sanchez EF. Action of metalloproteinases mutalysin I and II on several components of the hemostatic and fibrinolytic systems. Thromb Res. 2000;99(4):363-76.]. On the other hand, PLA2s are myotoxic enzymes that cause extensive local damage in the bite site [1919. Cordeiro FA, Perini TG, Bregge-Silva C, Cremonez CM, Rodrigues RS, Boldrini-França J, et al. A new phospholipase A₂ from Lachesis muta rhombeata: purification, biochemical and comparative characterization with crotoxin B. Protein Pept Lett. 2015;22(9):816-27., 2020. Fuly AL, Calil-Elias S, Zingali RB, Guimarães JA, Melo PA. Myotoxic activity of an acidic phospholipase A2 isolated from Lachesis muta (Bushmaster) snake venom. Toxicon. 2000;38(7):961-72.] and LAAO present high cytotoxic action [2121. Bregge-Silva C, Nonato MC, de Albuquerque S, Ho PL, Junqueira de Azevedo IL, Vasconcelos Diniz MR, et al. Isolation and biochemical, functional and structural characterization of a novel L-amino acid oxidase from Lachesis muta snake venom. Toxicon. 2012;60(7):1263-76.].
However, there is a certain difficulty in maintain Lachesis snakes in captivity due to the peculiar characteristics of their natural habitat [2222. Melgarejo-Giménez AR. Criação e Manejo de Serpentes. In: Animais de Laboratório: criação e experimentação. Andrade A, Pinto SC, Oliveira RS editors. Fiocruz, Rio de Janeiro; 2002. pp. 175-99.] and this hinders the study of their venoms. Few components have been isolated from LmrV, including LAAO [2121. Bregge-Silva C, Nonato MC, de Albuquerque S, Ho PL, Junqueira de Azevedo IL, Vasconcelos Diniz MR, et al. Isolation and biochemical, functional and structural characterization of a novel L-amino acid oxidase from Lachesis muta snake venom. Toxicon. 2012;60(7):1263-76.], PLA2 [1919. Cordeiro FA, Perini TG, Bregge-Silva C, Cremonez CM, Rodrigues RS, Boldrini-França J, et al. A new phospholipase A₂ from Lachesis muta rhombeata: purification, biochemical and comparative characterization with crotoxin B. Protein Pept Lett. 2015;22(9):816-27.], serine proteinases [2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292.-2525. Aguiar AS, Alves CR, Melgarejo A, Giovanni-de-Simone S. Purification and partial characterization of a thrombin-like/gyroxin enzyme from bushmaster (Lachesis muta rhombeata) venom. Toxicon. 1996;34(5):555-65.] and hyaluronidase [1313. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.]. Hyaluronidase was the last isolated component to be reported about 3 years ago. Recently, the in vitro and in vivo effects of a synthetic version of a BPP identified in this venom was reported [2626. Pinheiro-Júnior EL, Boldrini-França J, de Campos Araújo LMP, Santos-Filho NA, Bendhack LM, Cilli EM, et al. LmrBPP9: A synthetic bradykinin-potentiating peptide from Lachesis muta rhombeata venom that inhibits the angiotensin-converting enzyme activity in vitro and reduces the blood pressure of hypertensive rats. Peptides. 2018;102:1-7.].
Studying animal venoms and their components have helped, for example, to understand the envenoming process [2727. Rucavado A, Escalante T, Gutiérrez JM. Effect of the metalloproteinase inhibitor batimastat in the systemic toxicity induced by Bothrops asper snake venom: understanding the role of metalloproteinases in envenomation. Toxicon. 2004;43(4):417-24.], improve the effectiveness of antivenoms [2828. Boldrini-França J, Corrêa-Netto C, Silva MM, Rodrigues RS, De La Torre P, Pérez A, et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: assessment of geographic variation and its implication on snakebite management. J Proteomics. 2010;73(9):1758-76.], search for targets in the antivenom therapy [2929. Xiao H, Pan H, Liao K, Yang M, Huang C. Snake venom PLA2, a promising target for broad-spectrum antivenom drug development. Biomed Res Int. 2017;2017:6592820.] and to develop diagnostic reagents [3030. Karapetian H. Reptilase time (RT). Methods Mol Biol. 2013;992:273-7.] and more specific therapeutic agents to fight against different diseases [3131. Scarborough RM, Rose JW, Hsu MA, Phillips DR, Fried VA, Campbell AM, et al. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m. barbouri. J Biol Chem. 1991;266(15):9359-62., 3232. Dardevet L, Rani D, Aziz TA, Bazin I, Sabatier JM, Fadl M, et al. Chlorotoxin: a helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins (Basel). 2015;7(4):1079-101.]. Therefore, based on a previous fractionation of LmrV [1313. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.], we decided to investigate the subproteome of its major fraction (S5) and search for compounds which may be further investigated as a tool in the development of novel drugs or diagnostic reagents.
Methods
Venom
L. m. rhombeata (IBAMA registration number 647.998) was maintained by the Serpentarium “Bosque da Saúde”, city of Americana (São Paulo state, Brazil, 22º 44' 21" S, 47º 19' 53" W). The venom provided was desiccated and stored at -20 ºC until used.
Venom fractionation protocol
LmrV (23 mg) was dissolved in 500 μL of 50 mM sodium acetate buffer with 0.15 M NaCl (pH 6) and centrifuged (13,400 xg, 4 ºC, 10 min). The supernatant was applied on a HiPrep Sephacryl S-100 HR column (1.6 x 60 cm; GE Healthcare, Sweden) previously equilibrated with the same buffer, and fractions of 1.5 mL were collected at a flow rate of 0.5 mL/min [1313. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.]. The S5 fraction was pooled and submitted to a reversed-phase fast protein liquid chromatography (RP-FPLC) on a 214MS® C4 column (250 x 4.6 mm, 5 µm, 300 Å, Grace™ Vydac™, USA) previously equilibrated with 0.1% trifluoroacetic acid (TFA). Samples were eluted following a discontinuous gradient of 60% acetonitrile (ACN) in 0.1% TFA at 0.7 mL/min. Absorbance was monitored at 280 nm by FPLC Äkta Purifier UPC-10 system (GE Healthcare) in both steps. Protein recovery of the eluted fractions was calculated by the software Unicorn® 5.20 (GE Healthcare).
N-terminal sequencing
The N-terminal sequences of the fractions eluted from the RP-FPLC were determined by Edman degradation [3333. Edman P, Begg G. A protein sequenator. Eur J Biochem. 1967;1(1):80-91.]. About 200 pmol of each fraction S5C1-S5C5 and 400 pmol of fraction S5C6 were sequenced by an automated protein sequencer model PPSQ-33A (Shimadzu Co., Japan) according to the manufacturer’s instructions. The obtained sequences were compared with non-redundant protein sequences (nr) from snakes (taxid: 8570) available at BLAST database using the online blastp suite (protein-protein blast) (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Internal peptides analysis
The fraction S5C4 (20 µg) dissolved in NuPAGE LDS sample buffer (Life Technologies, USA) was reduced with 0.2 M 1,4-dithiothreitol (DTT) and alkylated with 0.5 M iodoacetamide. Reduced and alkylated sample was divided into two wells to be loaded in a NuPAGE 4-12% Bis-Tris Gel (Novex, USA) and gel run was performed at 200 V for 45 min using NuPAGE® MOPS-SDS Running Buffer (Invitrogen). Gel was stained with GelCode® Blue Stain Reagent (Thermo Scientific, USA) and SeeBlue® Plus2 Pre-stained Protein Standard (Invitrogen, USA) was used as molecular weight marker (4-250 kDa).
Protein band from each well was excised from the gel, destained and digested with 200 ng of modified trypsin (Promega, USA) or 400 ng of endoproteinase Glu-C (Roche, Germany) at 25 °C, for 15 h and under shaking. Peptides were extracted using C18 ZipTip ZTC18S960 (Merck Millipore, USA) [3434. Cotto-Rios XM, Békés M, Chapman J, Ueberheide B, Huang TT. Deubiquitinases as a signaling target of oxidative stress. Cell Rep. 2012;2(6):1475-84.].
1/5 of each digested sample was loaded onto an EASY-Spray PepSwift Monolithic Capillary column (Thermo Scientific) using an Easy-nLC 1000 (Thermo Scientific) coupled to an Orbitrap Elite™ Mass Spectrometer (Thermo Scientific). Peptides were eluted for 65 min using a gradient from 2 to 90% of ACN in 0.5% acetic acid. High-resolution full MS spectra were acquired with resolution of 60,000 (at m/z 400) and automatic gain control (AGC) target of 1e6. The twenty most intense ions were subsequently fragmented by higher-energy collisional dissociation (HCD) in a data-dependent mode. The HCD MS/MS spectra were acquired with a resolution of 15,000 (at m/z 400), AGC target of 5e4, normalized collision energy of 27, and isolation window of ±2 Da. Additionally, another run was performed and here the 20 most intense ions were fragmented by electron transfer dissociation (ETD) also in a data-dependent mode and the ETD MS/MS spectra were acquired with a resolution of 15,000 (at m/z 400), AGC target of 5e4, activation time of 60 ms, and isolation window of ±2 Da after each full MS scan.
Data were searched against a database downloaded from UniProt [3535. UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204-12.] using the keywords “serine proteinase” and “Lachesis”. This database was downloaded in July 13th, 2015 and contains the 5 sequences of serine proteinases available for this snake genus. The search was performed by the error tolerant search engine Byonic™ v2.3.5 (Protein Metrics, USA) [3636. Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;13.], setting the protein false discovery rate (FDR) cutoff as 1%, the precursor tolerance as 10 ppm and the fragment tolerance as 20 ppm and the ‘wildcard’ feature (which adds customizable mass windows to specific or all amino acids to search amino acid substitutions/modifications) as ±150 Da. Cysteine residues are carbamidomethylated and methionine oxidation, pyro-Glu at N-terminal, amidated C-terminal and HexNAc addition were set as variable modifications. Results were manually confirmed by de novo sequencing to exclude false positives.
Carbohydrate content analysis
MS/MS data were searched as described in the section above and both the ‘wildcard’ feature (adding a customizable mass window of +5000 Da to specific or all amino acids) and HexNAc (+203.079373 Da) at N-Glycan set as variable modification were enabled in Byonic to detect possible glycosylation sites in LmrSP-4 amino acid sequence in comparison to the sequences available in the database.
In addition, LmrSP-4 (~50 μg) diluted in 50 mM ammonium bicarbonate buffer (AMBIC) was added to 5% SDS (0.36%, final concentration) and 1 M DTT (71 mM, final concentration) and heated at 95 °C for 10 min. Reduced LmrSP-4 was let at room temperature for 5 min and added of 10% Triton™ X-100 (1.25%, final concentration). Deglycosylation reaction occurred with 10 units of PNGase F (G5166, Sigma-Aldrich, USA) at 37 °C for 5 h. Reaction was stopped by heat denaturation (100 °C, 10 min) and sample was stored at -20 °C until used. Positive control for glycosylation was the reduced LmrSP-4 with 5% SDS, 1 M DTT and 10% Triton™ X-100 as described above. Reduced and deglycosylated LmrSP-4 were analyzed by 13.5% SDS-PAGE [3737. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680-5.] and gel was stained with periodic acid-Schiff to the detection of glycoproteins [3838. Doerner KC, White BA. Detection of glycoproteins separated by nondenaturing polyacrylamide gel electrophoresis using the periodic acid-Schiff stain. Anal Biochem. 1990;187(1):147-50.] and Coomassie Brilliant Blue G-250 ® (Sigma-Aldrich).
In silico analysis of the amino acid sequences
Multiple sequence alignments were generated by Clustal Omega [3939. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539.] and edited by the free software BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and the ESPript server [4040. Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320-3.].
Molecular mass
LmrSP-4 sample was dissolved in water and diluted in the proportion 1:1 with sinapinic acid (5 mg/mL) in 50% ACN and 0.1% TFA, spotted onto a sample plate and allowed to dry at room temperature. LmrSP-4 molecular mass was determined by matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry (MALDI-TOF MS) using the system Ultraflex II MALDI TOF/TOF (Bruker Daltonics, USA). The equipment was calibrated in the masse range 5.7-66.4 kDa using the kit ProteoMass™ Protein MALDI-MS Calibration Kit (Sigma-Aldrich), and it was operated in linear positive mode with 4400 shots per spectrum. Data was analyzed by the software flexAnalysis version 3.3 (Bruker Daltonics).
Fibrinogenolytic activity assay
Fibrinogenolytic activity assay was performed according to modifications on Edgar and Prentice method [4141. W Edgar, Prentice CRM. The proteolytic action of ancrod on human fibrinogen and its polypeptide chains. Thromb Res. 1973;2(1):85-95.]. Bovine fibrinogen (0.5 mg/mL; Sigma Chemical Co.) in 0.1 M Tris-HCl buffer (pH 8) was incubated with S5 or LmrSP-4 at 37 ºC for 3 h. Reaction was stopped with 50 mM Tris-HCl buffer (pH 6.8) containing 10% glycerol, 10% β-mercaptoethanol, 2% SDS and 0.05% bromophenol blue. Sample was taken under boiling for 5 min and analyzed by 12% SDS-PAGE [3737. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680-5.]. Gel was stained with Coomassie Brilliant Blue G-250 ®. The action of possible inhibitors was evaluated pre-incubating the enzyme with ethylenediamine tetraacetic acid (EDTA, 20 mM final concentration) and phenylmethylsulfonyl fluoride (PMSF, 10 mM final concentration) at 37 °C for 15 minutes.
Enzyme activity evaluation upon chromogenic substrates
Amidolytic activities upon chromogenic substrates (S-2238: substrate for thrombin; S-2302: substrate for plasma kallikrein; S-2251: substrate for plasmin and streptokinase-activated plasminogen; and S-2222: substrate for Factor Xa) were performed as described in the manufacturer’s protocols (Chromogenix, Italy). Each substrate was dispersed in ultrapure water (18.2 MΩ cm, Milli-Q water, Millipore, USA) and diluted in 50 mM Tris-HCl buffer with 5 mM CaCl2 (pH 7.5). The substrate solution and LmrSP-4 (1 μg dispersed in the same buffer) were added to a 96-well microplate and incubated at 37 ºC for 40 min. After that, the samples absorbance was measured at 405 nm. The assay was performed in triplicate and the negative control consisted of chromogenic substrate solution added to the previous buffer. Enzyme activity was also evaluated in the presence of some proteinase inhibitors (benzamidine, 1,10-phenantroline and PMSF). For this purpose, the enzyme (1 μg) was previously incubated with each inhibitor (20 mM) at 37 ºC for 60 min.
Optimal conditions
Optimal conditions (pH and temperature) for LmrSP-4 were determined with the fluorescence resonance energy transfer (FRET) Abz-KLRSSKQ-EDDnp substrate as previously reported [4242. da Silva RR, de Oliveira LCG, Juliano MA, Juliano L, Rosa JC, Cabral H. Activity of a peptidase secreted by Phanerochaete chrysosporium depends on lysine to subsite S'1. Int J Biol Macromol. 2017;94(Pt A):474-83.]. Proteolytic activity was monitored using excitation and emission wavelengths of 320 and 420 nm, respectively, in a Lumina Fluorescence Spectrometer (Thermo Scientific). Effects of pH were evaluated using the following 0.1 M buffer solutions: Mes (pH 6.0 and 6.5), Hepes (pH 7.0, 7.5 and 8.0) and Bicine (pH 8.5 and 9.0) at 40 °C. Temperature effects were evaluated in 0.1 M Hepes buffer (pH 7) in the range 40-60 °C.
Statistical analysis
Statistical analyses for the LmrSP-4 activity upon chromogenic substrates were performed by GraphPad Prism 6.01 (GraphPad Software Inc., USA) using unpaired t-test and comparing the activity in each tested substrate with its own negative control. The data (mean ± standard error of the mean) obtained in the activity assays against chromogenic and FRET substrates were analyzed by one-way ANOVA followed by Dunnett’s multiple comparisons test and considered statistically significant when p < 0.05.
Results
Venom fractionation and N-terminal sequencing
LmrV was fractionated by molecular exclusion chromatography (Fig. 1a) and the majoritarian fraction (S5), which corresponds to approximately 26% of the crude soluble LmrV (Table 1), presented proteolytic activity upon fibrinogen except in the presence of PMSF (Fig. 1a). S5 fraction was then fractionated by RP-FPLC, giving rise to 6 main subfractions named S5C1-S5C6 (Fig. 1b). These fractions were submitted to N-terminal sequencing and results are shown in Table 2. Fractions S5C1-S5C5 were identified as belonging to the snake venom serine proteinase (SVSP) family and represent together more than 50% of the protein families identified in the fraction S5. However, S5C5 fraction also contains lectin. The S5C6 belongs to the lectins family and accounts to about 42% of the S5 proteins (Table 2). A complete list containing the blast results for each fraction is available in Additional file 1. Additional File 1: Each spreadsheet contains the first 100 results from the blastp search performed for the N-terminal sequence from each reversed-phase chromatography fraction (S5C1-S5C6).
Lachesis muta rhombeata venom fractionation. A: LmrV was applied on a HiPrep Sephacryl S-100 HR previously equilibrated with 50 mM sodium acetate buffer with 0.15 M NaCl (pH 6) and fractions were eluted in the same buffer. Insert: 13.5% SDS-PAGE of S5 submitted to fibrinogenolytic activity. Gel run was carried out at 90 V. Lanes: kDa: molecular weight marker; 1: S5; 2: S5 plus fibrinogen; 3: S5 plus fibrinogen and EDTA; 4: S5 plus fibrinogen and PMSF; 5: fibrinogen (negative control). B: Fraction S5 was submitted to RP-FPLC on a C4 column previously equilibrated with 0.1% TFA (solution A). Fractions S5C1-S5C6 were eluted using a gradient of 60% ACN in 0.1% TFA (solution B). Insert: 13.5% SDS-PAGE of S5C4 submitted to fibrinogenolytic activity. Gel run was carried out at 90 V. Lanes: 1: fibrinogen (negative control); 2: S5; 3: S5 plus fibrinogen; 4: S5 plus fibrinogen and EDTA; 5: S5 plus fibrinogen and PMSF; kDa: molecular mass marker. C: Densitometry analysis of the protein bands from the SDS-PAGE of S5C4 submitted to fibrinogenolytic activity. After dried, the gel was scanned and analyzed by the system Gel Doc™ EZ Gel Documentation and the software Image Lab™ (Bio-Rad, USA). The intensities of the main bands are in parentheses.
LmrSP-4 structural characterization
LmrSP-4 had its N-terminal determined by Edman degradation up to the 53rd amino acid residue as previously shown (Table 2). We also investigated the internal peptides of this novel serine proteinase to obtain the highest sequence coverage. The primary sequence of LmrSP-4 was verified by digesting the protein with trypsin and Glu-C and peptides were analyzed by MS/MS using two different fragmentation modes (HCD and ETD). Additional file 2 Additional File 2: Table containing all MS/MS-derived sequences after the digestion of LmrSP-4 with trypsin and Glu-C. contains all peptides identified in the MS/MS database search (including the false positive ones) and Table 3 shows the correct peptides after manual investigation of the MS/MS search. Multiple sequence alignment of LmrSP-4 with other SVSPs is shown in Fig. 2. In addition, we investigated the occurrence of N-linked sugars. HCD MS/MS spectrum of the ion [M + 4H] 4+ = 4971.2538 shows characteristic oxonium ions for N-acetylhexoseamine (HexNAc), N-acetylneuraminic acid (Neu5Ac) and the disaccharide Hex-HexNAc (hexose galactose/mannose linked to a HexNAc), although the peptide containing the glycosylation site could not be identified (Fig. 3). The digestion with PNGase F confirmed that LmrSP-4 is N-linked with a carbohydrate content estimated in 12% and no glycoprotein band was stained after PNGase F treatment (Fig. 4). The molecular mass of LmrSP-4 was determined through MALDI-TOF as 28,190 Da (Fig. 5).
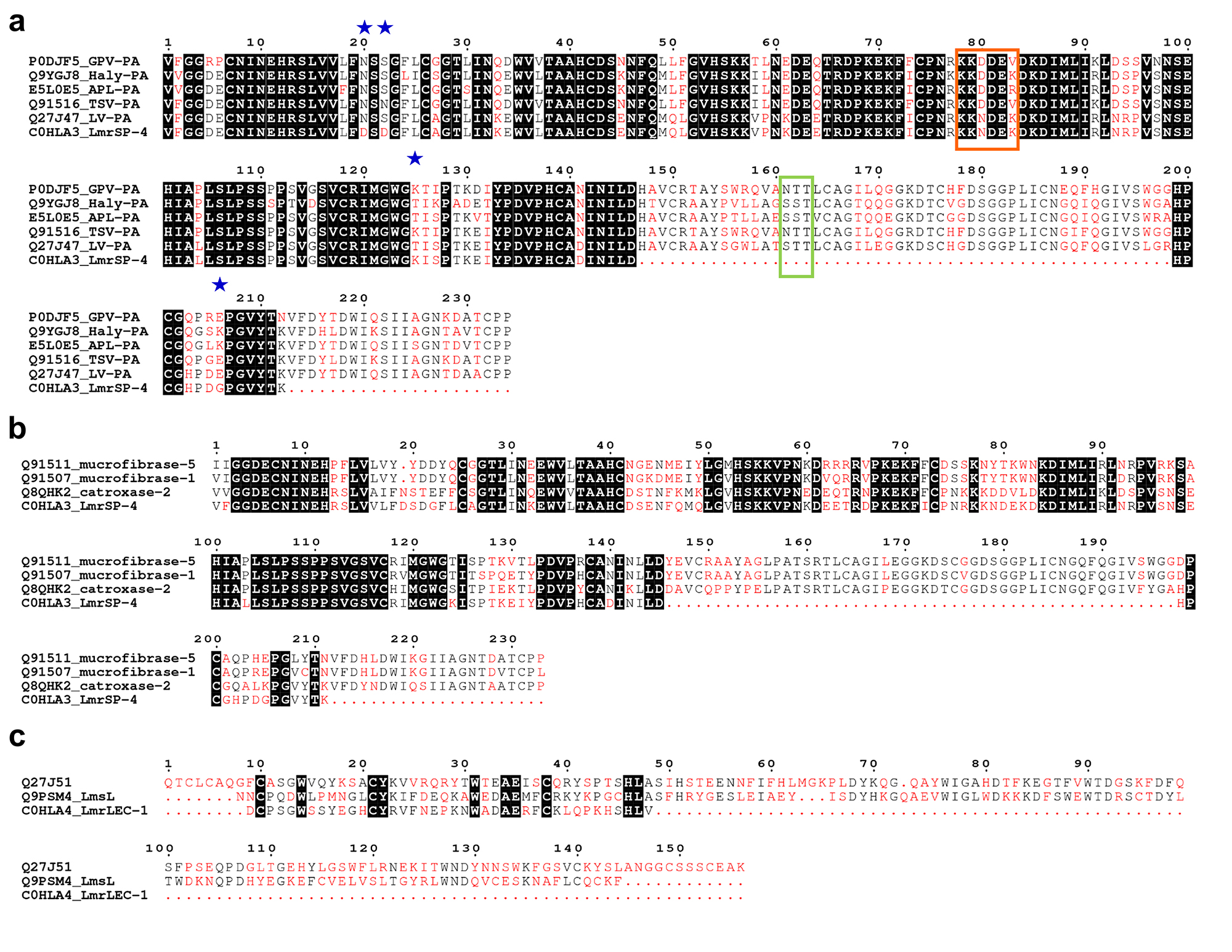
Multiple sequence alignments of SVSPs and snake venom lectins. Multiple sequence alignment among (a) LmrSP-4 and plasminogen activators SVSPs, (b) LmrSP-4 and kallikrein-like SVSPs and (c) LmrLEC-1 and lectins from Lachesis. The highly conserved residues are highlighted in black while low consensus residues are shown in red. Main differences between LmrSP-4 and LV-PA discussed in the text are indicated by blue stars. The green box shows a region with a putative N-glycosylation site in LmrSP-4 and the orange box represents the peptide important for the interaction between TSV-PA and its substrate plasminogen. P0DJF5: venom plasminogen activator GPV-PA from Trimeresurus albolabris; Q9YGJ8: venom plasminogen activator Haly-PA from Gloydius brevicaudus; E5L0E5: venom plasminogen activator APL-PA from Agkistrodon piscivorus leucostoma; Q91516: venom plasminogen activator TSV-PA from Trimeresurus stejnegeri; Q27J47: venom plasminogen activator LV-PA from L. m. muta; C0HLA3: serine proteinase LmrSP-4 from L. m. rhombeata; Q91511: beta-fibrinogenase mucrofibrase-5 from Protobothrops mucrosquamatus; Q91507: beta-fibrinogenase mucrofibrase-1 from Protobothrops mucrosquamatus; Q8QHK2: SVSP catroxase-2 from Crotalus atrox; Q27J51: C-type lectin from L. muta; Q9PSM4: C-type lectin from L. stenophrys.
Carbohydrate content analysis by MS/MS. HCD MS/MS spectrum of the ion [M+4H]4+ = 4971.2538 from a LmrSP-4 glycopeptide acquired by an Orbitrap Elite™ Mass Spectrometer with resolution of 60,000 (at m/z 400). Abbreviations: HexNAc: N-acetylhexoseamine; Hex-HexNAc (hexose galactose/mannose-N-acetylhexoseamine); Neu5Ac: N-acetylneuraminic acid.
Deglycosylation of LmrSP-4 by PNGase F and visualization of the digested product by 13.5% SDS-PAGE. Gel run was performed at 90 V and gel was divided into two pieces for staining protocol. One of them was stained with Coomassie Brilliant Blue G-250 ® (blue) and the other one was stained with periodic acid-Schiff (pink for glycoproteins). Picture was acquired in black and white by the Gel Doc™ EZ Gel Documentation System (Bio-Rad, USA). The red arrow indicates PNGase F (36 kDa). Lanes: kDa: molecular weight marker; rLmrSP-4: reduced LmrSP-4; dLmrSP-4: deglycosylated LmrSP-4.
Molecular mass determination of LmrSP-4 through MALDI-TOF mass spectrometry. Sample was diluted in a sinapinic acid matrix and data were acquired in a positive linear mode using the mass spectrometer Ultraflex II MALDI TOF/TOF (Bruker).
Enzyme activities of LmrSP-4
LmrSP-4 degrades the fibrinogen Aα-chain as shown in Fig. 1b and 1c, even in the presence of EDTA, but not in the presence of PMSF, evidencing the presence of serine proteinase activity. A densitometry analysis reveals the decreasing of Aα-chain proportion in comparison to the Bβ and γ-chains of fibrinogen. Besides fibrinogen, LmrSP-4 is also active upon the substrate for plasma kallikrein (S-2302) and, in a minor extension, upon the substrate for plasmin and streptokinase-activated plasminogen (S-2251). However, LmrSP-4 has no activity upon the substrate for thrombin (S-2238) nor the substrate for factor Xa (S-2222) (Fig. 6a). Enzyme activity upon S-2302 was also evaluated in the presence of different enzyme inhibitors. The proteolytic activity was decreased by PMSF and benzamidine, but in the presence of 1,10-phenantroline the enzyme remained active (Fig. 6b).
Enzyme activity upon chromogenic substrates. A: Enzyme activity was evaluated upon 0.4 mM chromogenic substrates for Factor Xa (S-2222), substrate for thrombin (S-2238), plasmin and streptokinase-activated plasminogen (S-2251) and plasma kallikrein (S-2302), according to the manufacturer’s protocol. Data were analyzed using unpaired t-test and comparing the activity on each substrate tested with its own negative control (** for p < 0.01 and **** for p < 0.0001). B: Enzyme activity of LmrSP-4 was evaluated upon substrate S-2302 in the presence of possible inhibitors (PMSF, benzamidine and 1,10-phenantroline). Data were analyzed by one-way ANOVA followed by Dunnett’s multiple comparisons test (****p < 0.0001 comparing results to LmrSP-4 without inhibitor and #### p < 0.0001 compared to the control). Abbreviations: C: control.
Determination of the optimal conditions of LmrSP-4
The optimal conditions for LmrSP-4 activity was evaluated at several pHs (6.0-9.0) and temperatures (40-60 °C) using the FRET substrate Abz-KLRSSKQ-EDDnp. LmrSP-4 showed the highest activities within the range from neutral to basic pH values (Fig. 7a) and was very active at high temperatures, starting to denaturate at 60 °C (Fig. 7b).
Determination of serine proteinase activity of LmrSP-4 on FRET substrate Abz-KLRSSKQEDDnp. A: pH-profile. B: temperature-profile. Data were analyzed by one-way ANOVA followed by Dunnett’s multiple comparisons test (**p < 0.01 and ****p < 0.0001). Results were compared to the pH/temperature with the highest activity.
Discussion
A transcriptomic study of L. muta venom gland [4343. Junqueira-de-Azevedo IL, Ching AT, Carvalho E, Faria F, Nishiyama MY Jr, Ho PL, et al. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics. 2006;173(2):877-89.] revealed major toxin transcripts encode BPPs (73.2%), SVMPs (5.9%), C-type lectin (5.8%), PLA2 (4.7%), LAAO (3.7%) and SVSP (3.5%). However, several factors are involved in protein production and proteomic studies from the genus Lachesis have shown these venoms are mainly comprised by serine and metalloproteinases (21-35% and 18-38%, respectively), PLA2s (2-13%), BPPs (14-28%) and LAAO (0.5-10%) [1212. Pla D, Sanz L, Molina-Sánchez P, Zorita V, Madrigal M, Flores-Díaz M, et al. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J Proteomics. 2013;89:112-23., 4444. Madrigal M, Sanz L, Flores-Díaz M, Sasa M, Núñez V, Alape-Girón A, et al. Snake venomics across genus Lachesis. Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J Proteomics. 2012;77:280-97., 4545. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.]. Despite this, there is a long road ahead to unveil novel components from these venoms. Based on a previous fractionation of LmrV [1313. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.], we decided to perform the subproteome of the major fraction from this venom and investigate some of its components.
The S5 fraction mainly presents components of about 30 kDa (with minor components of around 15 kDa) comprising about 26% of the crude soluble LmrV (Fig. 1a-insert; Table 1). This fraction presented proteolytic activity upon fibrinogen (except in the presence of PMSF), indicating the presence of serine proteinases (Fig. 1a-insert). S5 was then submitted to a RP-FPLC using a C4 column and the 6 subfractions (S5C1-S5C6) obtained were analyzed, showing a diverse array of serine proteinases, but also a C-type lectin (Fig. 1b, Table 2).
SVSPs are trypsin-like enzymes since the mammalian trypsin cysteine pattern is conserved in their structure [4646. Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477(1-2):146-56.]. These proteins are usually related to hemostasis disturbances in snake envenoming and have demonstrated interesting biological activities, including (anti)coagulant action, fibrinolysis, blood pressure reduction and (in)activation of platelet aggregation [4747. Kini RM. Serine proteases affecting blood coagulation and fibrinolysis from snake venoms. Pathophysiol Haemost Thromb. 2005;34(4-5):200-4.-5151. Markland FS, Kettner C, Schiffman S, Shaw E, Bajwa SS, Reddy KN, et al. Kallikrein-like activity of crotalase, a snake venom enzyme that clots fibrinogen. Proc Natl Acad Sci U S A. 1982;79(6):1688-92.].
Concerning the lectin class, there are usually two types of lectins in snake venoms: the sugar binding lectins (or C-type lectins) and the C-type lectin-like proteins (CLPs). The C-type lectins usually show weak toxicity, being the erythrocytes agglutination one of its key roles. In addition, these lectins may contribute to the venom’s antibacterial action during the digestion of preys by recognizing some specific pathogen carbohydrates. On the other hand, CLPs present the carbohydrate recognition domain but lack this activity due to the loss of the calcium binding site, and are usually implicated in anticoagulation effects, platelet activation, as well as antithrombotic action. C-type-lectins and CLPs are reviewed elsewhere [5252. Du XY, Clemetson KJ. Reptile C-type lectins. In: Handbook of Venoms and Toxins of Reptiles. Mackessy SP editor. Boca Raton: CRC Press; 2010. pp. 359-75.-5555. Clemetson KJ. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon. 2010;56(7):1236-46.].
S5C6 fraction contains the first lectin described in LmrV, which was named LmrLEC-1. It is only 43% and 38% identical (but 55% and 51% similar) to a C-type lectin from L. stenophrys (sp|Q9PSM4) [5656. Aragón-Ortíz F, Brenes-Brenes JR, Gubensek F. Characterization of a lectin-like protein isolated from Lachesis muta snake venom. Rev Biol Trop. 1989;37(1):79-83.] and a C-type lectin precursor from L. muta (sp|Q27J51) [4343. Junqueira-de-Azevedo IL, Ching AT, Carvalho E, Faria F, Nishiyama MY Jr, Ho PL, et al. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics. 2006;173(2):877-89.], respectively (Fig. 2c). Lectins already isolated from Lachesis venoms are generally dimers of around 28 kDa which are separated under reducing conditions [5656. Aragón-Ortíz F, Brenes-Brenes JR, Gubensek F. Characterization of a lectin-like protein isolated from Lachesis muta snake venom. Rev Biol Trop. 1989;37(1):79-83., 5757. Ogilvie ML, Dockter ME, Wenz L, Gartner TK. Isolation and characterization of lactose-binding lectins from the venoms of the snakes Lachesis muta and Dendroaspis jamesonii. J Biochem. 1986;100(6):1425-31.]. Therefore, LmrLEC-1 and possibly other lectins in LmrV are likely represented by the low molecular weight band (~14 kDa) in the SDS-PAGE carried out with S5 fraction under reducing conditions (Fig. 1a-insert). The study of snake venom lectins is of utmost importance. It has enabled the discovery of biochemical pathways in homeostasis helping to understand the mechanisms involved in platelet aggregation. Furthermore, they are potential leads for the design of antithrombotic and anticoagulation drugs [5353. Arlinghaus FT, Eble JA. C-type lectin-like proteins from snake venoms. Toxicon. 2012;60(4):512-9.].
Regarding the serine proteinases present in S5 fraction, most of them (LmrSP-2, LmrSP-3 and LmrSP-5) were identified as belonging to the thrombin-like enzymes (TLEs) class. In fact, this class is mainly reported in snakes from Crotalinae subfamily, which comprises the genus Lachesis among others [5858. Castro HC, Zingali RB, Albuquerque MG, Pujol-Luz M, Rodrigues CR. Snake venom thrombin-like enzymes: from reptilase to now. Cell Mol Life Sci. 2004;61(7-8):843-56.]. TLEs have been already isolated from L. muta venoms [1616. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta. Int J Biochem. 1989;21(8):863-71., 5959. Aragon-Ortiz F, Gubensek F. A thrombin-like enzyme from bushmaster (Lachesis muta stenophyrs) venom. Toxicon. 1993;31(11):1435-43.-6161. Yarleque A, Campos S, Escobar E, Lazo F, Sanchez N, Hyslop S, et al. Isolation and characterization of a fibrinogen-clotting enzyme from venom of the snake, Lachesis muta muta (Peruvian bushmaster). Toxicon. 1989;27(11):1189-97.], and two are TLEs from LmrV [2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292., 6060. Magalhães A, de Oliveira GJ, Diniz CR. Purification and partial characterization of a thrombin-like enzyme from the venom of the bushmaster snake, Lachesis muta noctivaga. Toxicon. 1981;19(2):279-94.]. The first one was isolated in 1981 when L. m. rhombeata was designated as L. m. noctivaga [6060. Magalhães A, de Oliveira GJ, Diniz CR. Purification and partial characterization of a thrombin-like enzyme from the venom of the bushmaster snake, Lachesis muta noctivaga. Toxicon. 1981;19(2):279-94., 6262. Roge AR, Alma S, Romano RWDL. Lachesis muta rhombeata (Serpentes : Viperidae, Crotalinae). Mem Inst Butantan. 1976;40:53-4.] and LMR-47 (sp|Q9PRP4) [2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292.] was the only serine proteinase from LmrV whose sequence was available at UniProt so far. A third and last SVSP from LmrV was published in 1985, but it is not a TLE [6363. Silva LM, Diniz CR, Magalhães A. Purification and partial characterization of an arginine ester hydrolase from the venom of the bushmaster snake, Lachesis muta noctivaga. Toxicon. 1985;23(4):707-18.].
Furthermore, a vast variety of serine proteinases have been reported in venom proteomes across the genus Lachesis [1212. Pla D, Sanz L, Molina-Sánchez P, Zorita V, Madrigal M, Flores-Díaz M, et al. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J Proteomics. 2013;89:112-23., 4444. Madrigal M, Sanz L, Flores-Díaz M, Sasa M, Núñez V, Alape-Girón A, et al. Snake venomics across genus Lachesis. Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J Proteomics. 2012;77:280-97., 4545. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.] but the sequenced amino acid residues presented 100% of identity and unable to identify different isoforms of SVSPs [6464. Doley R, Mackessy SP, Kini RM. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol Biol. 2009;9:146., 6565. Vaiyapuri S, Wagstaff SC, Harrison RA, Gibbins JM, Hutchinson EG. Evolutionary analysis of novel serine proteases in the venom gland transcriptome of Bitis gabonica rhinoceros. PLoS One. 2011;6(6):e21532.]. For example, serine proteinases containing the N-terminal sequence IVGGDECNINEHRFL were identified in the venom proteomes of L. muta [4545. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.], L. stenophrys [4545. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.] and L. melanocephala [4444. Madrigal M, Sanz L, Flores-Díaz M, Sasa M, Núñez V, Alape-Girón A, et al. Snake venomics across genus Lachesis. Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J Proteomics. 2012;77:280-97.], and the same sequence was determined in fractions S5C2 and S5C3 not allowing differentiating the present enzymes. These enzymes may be result of amino acid mutations, post-translational modifications (e. g. glycosylation) or other modifications (e. g. metoxilation) that lead to different interactions with the chromatographic resin and, consequently, different elution times.
In this study, the SVSPs identified were denominated from the number 2 (e. g. LmrSP-2) since they will be the second, third, fourth and fifth sequences from LmrV available at UniProt database. The accession numbers are listed on Table 2. Sequences of LmrSP-2 and LmrSP-3, represented by fractions S5C1 and S5C2, respectively, resemble those of other serine proteinases from L. muta subspecies. The presence of distinct serine proteinases within snake venoms may lead to different substrate specificities and functional diversity in these venoms. The first 20 amino acid residues of LmrSP-2 are 100% identical to a TLE isolated from L. m. muta [6666. Magalhaes A, Da Fonseca BC, Diniz CR, Gilroy J, Richardson M. The complete amino acid sequence of a thrombin-like enzyme/gyroxin analogue from venom of the bushmaster snake (Lachesis muta muta). FEBS Lett. 1993;329(1-2):116-20.] while LmrSP-3 is similar to a kallikrein-like enzyme from L. m. rhombeata [2424. Giovanni-De-Simone S, Aguiar AS, Gimenez AR, Novellino K, de Moura RS. Purification, properties, and N-terminal amino acid sequence of a kallikrein-like enzyme from the venom of Lachesis muta rhombeata (Bushmaster). J Protein Chem. 1997;16(8):809-18.]. However, they are not the same enzymes as those already published and represent a novelty in LmrV. The TLE isolated from L. m. muta has more than 40 kDa [6666. Magalhaes A, Da Fonseca BC, Diniz CR, Gilroy J, Richardson M. The complete amino acid sequence of a thrombin-like enzyme/gyroxin analogue from venom of the bushmaster snake (Lachesis muta muta). FEBS Lett. 1993;329(1-2):116-20.] and this molecular mass was not detected in our analysis (Fig. 1a-insert) although it is ~10% of S5 fraction, and LmrSP-3 has a Leu residue in the 10th position while the kallikrein-like enzyme from L. m. rhombeata presents a Leu [2424. Giovanni-De-Simone S, Aguiar AS, Gimenez AR, Novellino K, de Moura RS. Purification, properties, and N-terminal amino acid sequence of a kallikrein-like enzyme from the venom of Lachesis muta rhombeata (Bushmaster). J Protein Chem. 1997;16(8):809-18.].
Concerning LmrSP-5 present in the S5C5 fraction, the first 25 amino acid residues are identical to LmrSP-4. The enzymes are assumed to be different since they are eluting at different points in the gradient (Fig. 1b), but it was not possible to differentiate them solely based on the N-terminal sequencing. LmrSP-4 sequence was chosen for database submission since it is the longest one. The first 13 N-terminal amino acid residues (VFGGDECNINEHR) from LmrSP-4 have been already detected in proteomic analyses of L. muta [4545. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.], L. m. rhombeata [1212. Pla D, Sanz L, Molina-Sánchez P, Zorita V, Madrigal M, Flores-Díaz M, et al. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J Proteomics. 2013;89:112-23.] and L. stenophrys venoms [4444. Madrigal M, Sanz L, Flores-Díaz M, Sasa M, Núñez V, Alape-Girón A, et al. Snake venomics across genus Lachesis. Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda. J Proteomics. 2012;77:280-97.], but this sequence obtained in these studies was too short that the identification of specific isoforms was impossible. The N-terminal from LmrSP-4 shared the highest identity with LV-PA (EC 3.4.21), a plasminogen activator from L. m. muta (sp|Q27J47), whose complete amino acid sequence was obtained through molecular cloning [6767. Sanchez EF, Felicori LF, Chavez-Olortegui C, Magalhaes HBP, Hermogenes AL, Diniz MV, et al. Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from bushmaster snake venom. Biochim Biophys Acta. 2006;1760(12):1762-71.]. The N-terminal fragment determined by Edman degradation revealed differences in the positions 20 and 22 in comparison to LV-PA. LV-PA presents Asn20 and Ser22 whilst both positions are represented by Asp residue in LmrSP-4. Asn20 is the unique N-glycosylation in LV-PA [6767. Sanchez EF, Felicori LF, Chavez-Olortegui C, Magalhaes HBP, Hermogenes AL, Diniz MV, et al. Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from bushmaster snake venom. Biochim Biophys Acta. 2006;1760(12):1762-71.], therefore its change by an Asp residue leads to the loss of a potential N-glycosylation site in LmrSP-4. Other differences between them are at positions 125 (T-K) and 206 (E-G). LV-PA and LmrSP-4 share 97.5% of identity and 98.1% of similarity taking into account only the aligned residues between them.
LmrSP-4 is also very similar to TSV-PA from Trimeresurus stejnegeri [6868. Zhang Y, Wisner A, Maroun RC, Choumet V, Xiong Y, Bon C. Trimeresurus stejnegeri snake venom plasminogen activator. Site-directed mutagenesis and molecular modeling. J Biol Chem. 1997;272(33):20531-7.], sharing 87.5% of identity in the aligned amino acid residues. Interesting modification is found in the region 80-82 (numbering in this study), specifically in the position 80, where there is a replacement of an Asp by an Asn. Implications of this modification will be discussed further. Additionally, LmrSP-4 is structurally very similar (64 to 74%) to snake venom kallikrein-like enzymes (Fig. 2b), which are intrinsically responsible for causing hypotension in preys [1414. Diniz MR, Oliveira EB. Purification and properties of a kininogenin from the venom of Lachesis muta (bushmaster). Toxicon. 1992;30(3):247-58., 4949. Felicori LF, Souza CT, Velarde DT, Magalhaes A, Almeida AP, Figueiredo S, et al. Kallikrein-like proteinase from bushmaster snake venom. Protein Expr Purif. 2003;30(1):32-42.].
LmrSP-4 showed reduction of about 4 kDa, corresponding to 12% (m/m) of its molecular mass estimated by SDS-PAGE, after PNGase F treatment. LmrSP-4 was stained by Schiff reagent only before PNGase F treatment, indicating all carbohydrate content is N-linked and was removed by PNGase F. While plasminogen activators usually present one N-glycosylation site in the N-terminal region (position 20th, Fig. 2a), LmrSP-4 present a substitution in this position leading to the loss of this N-glycan site. MS/MS analysis revealed the presence of glycopeptides, but the glycan site could not be determined. Fig 3 shows the carbohydrate marker ions HexNAc, Hex-HexNAc and Neu5Ac. Considering that N-linked carbohydrate are linked to an Asn residues in the consensus sequence Asn-X-Ser/Thr, where X can be any amino acid except Pro [6969. Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3(5):433-42.], the LmrSP-4 N-glycan site is likely present in the region 161-163 for which there was no peptide sequence coverage by MS/MS (Fig. 2a).
The degree of glycosylation varies among SVSPs and this post-translational modification seems to be related to the macromolecular selectivity and unusual thermal stability that these enzymes may present [7070. Mackessy SP. Thrombin-like enzymes in nnake venoms. In: Toxins and Hemostasis - from bench to bedside. Kini MR, Markland FS, Morita T, Clemetson K, McLane MA editors. Dordrecht, Springer; 2010. pp. 519-57., 7171. Swenson S, Markland FS Jr. Snake venom fibrin(ogen)olytic enzymes. Toxicon. 2005;45(8):1021-39.]. TLEs are usually glycosylated in a major extent than plasminogen activators or kallikrein-like enzymes. SVSPs with plasminogen-activating or kallikrein-like activity from the genus Lachesis present molecular mass within the range of 27.9 to 33 kDa estimated by SDS-PAGE [1414. Diniz MR, Oliveira EB. Purification and properties of a kininogenin from the venom of Lachesis muta (bushmaster). Toxicon. 1992;30(3):247-58., 2424. Giovanni-De-Simone S, Aguiar AS, Gimenez AR, Novellino K, de Moura RS. Purification, properties, and N-terminal amino acid sequence of a kallikrein-like enzyme from the venom of Lachesis muta rhombeata (Bushmaster). J Protein Chem. 1997;16(8):809-18., 6767. Sanchez EF, Felicori LF, Chavez-Olortegui C, Magalhaes HBP, Hermogenes AL, Diniz MV, et al. Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from bushmaster snake venom. Biochim Biophys Acta. 2006;1760(12):1762-71., 7272. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42.]. On the other hand, Lachesis TLEs are usually larger proteins of more than 40 kDa [1616. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta. Int J Biochem. 1989;21(8):863-71., 2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292., 7373 Magalhaes A, Ferreira RN, Richardson M, Gontijo S, Yarleque A, Magalhaes HP, et al. Coagulant thrombin-like enzymes from the venoms of Brazilian and Peruvian bushmaster (Lachesis muta muta) snakes. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(2):255-66.]. However, after deglycosylation reaction, all of them present 27-28 kDa [6767. Sanchez EF, Felicori LF, Chavez-Olortegui C, Magalhaes HBP, Hermogenes AL, Diniz MV, et al. Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from bushmaster snake venom. Biochim Biophys Acta. 2006;1760(12):1762-71., 7272. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42., 7373 Magalhaes A, Ferreira RN, Richardson M, Gontijo S, Yarleque A, Magalhaes HP, et al. Coagulant thrombin-like enzymes from the venoms of Brazilian and Peruvian bushmaster (Lachesis muta muta) snakes. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(2):255-66.]. Glycosylation is also responsible for heterogeneity among SVSPs. TLE-B and TLE-P from L. m. muta present very similar structural, functional and immunological properties but different glycosylation degrees [7373 Magalhaes A, Ferreira RN, Richardson M, Gontijo S, Yarleque A, Magalhaes HP, et al. Coagulant thrombin-like enzymes from the venoms of Brazilian and Peruvian bushmaster (Lachesis muta muta) snakes. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(2):255-66.]. In addition, stenoxobin from L. stenophrys [5959. Aragon-Ortiz F, Gubensek F. A thrombin-like enzyme from bushmaster (Lachesis muta stenophyrs) venom. Toxicon. 1993;31(11):1435-43.] and a kininogenin from Vipera ammodytes ammodytes [7474. Bailey GS, Shipolini RA. Purification and properties of a kininogenin from the venom of Vipera ammodytes ammodytes. Biochem J. 1976;153(2):409-14.] has heterogeneity due to sialic acid. Therefore, glycosylation is a post-translational modification intrinsically related to the presence of serine proteinases isoforms in snake venoms and might explain the same N-terminal sequences for S5C2 and S5C3 (Table 2). However, other assays are necessary to further characterize these proteins. The molecular mass of LmrSP-4, determined by MALDI-TOF as 28,190 Da, is in accordance with the mass estimated by SDS-PAGE and with the mass of other SVSPs as discussed above.
Although SVSPs are very similar in terms of primary structure, post-translational modifications and/or surface residues may exert important role in substrate specificity. Partially deglycosylated proteinases from Russell’s viper venom have shown lower fibrinogenolytic activity than the native enzymes [7575. Mukherjee AK. The pro-coagulant fibrinogenolytic serine protease isoenzymes purified from Daboia russelii russelii venom coagulate the blood through factor V activation: role of glycosylation on enzymatic activity. PLoS One. 2014;9(2):e86823.]. Another example includes the key residue Asp80 (our numbering) in TSV-PA. The peptide KKDDEV (amino acid residues 78-83) is close to the catalytic site in the three-dimensional structure and the residue Asp80 takes part in electrostatically interactions between TSV-PA and plasminogen. The replacement of Asp80 by an Asn residue resulted in lower activity upon plasminogen [6868. Zhang Y, Wisner A, Maroun RC, Choumet V, Xiong Y, Bon C. Trimeresurus stejnegeri snake venom plasminogen activator. Site-directed mutagenesis and molecular modeling. J Biol Chem. 1997;272(33):20531-7.]. LmrSP-4 naturally presents the residue Asn80, which explains its lower activity upon the substrate S-2251.
In comparison to TLEs, plasminogen activators comprehend a small number of proteins among the snake venom serine proteinases [4646. Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477(1-2):146-56.]. LV-PA from L. m. muta, is an acid glycoprotein active upon S-2251 and Tos-Gly-Pro-Lys-pNA (another substrate for plasmin) and cleaves fibrinogen but does not induce blood coagulation [1515. Sanchez EF, Santos CI, Magalhaes A, Diniz CR, Figueiredo S, Gilroy J, et al. Isolation of a proteinase with plasminogen-activating activity from Lachesis muta muta (bushmaster) snake venom. Arch Biochem Biophys. 2000;378(1):131-41.].
LmrSP-4 has an outstanding activity upon the substrate for plasma kallikrein (S-2302) but was not active upon S-2222 and S-2238, substrates for factor Xa and thrombin, respectively. Kallikreins are trypsin-like proteins and thus are able to hydrolyze different substrates, although their specificities are not as broad as trypsin [7676. Raspi G. Kallikrein and kallikrein-like proteinases: purification and determination by chromatographic and electrophoretic methods. J Chromatogr B Biomed Appl. 1996;684(1-2):265-87.]. The first kallikrein-like protein from Lachesis was isolated from its venom more than 20 years ago and released bradykinin (BK) from bovine kininogen [1414. Diniz MR, Oliveira EB. Purification and properties of a kininogenin from the venom of Lachesis muta (bushmaster). Toxicon. 1992;30(3):247-58.]. LV-Ka is another kallikrein-like serine proteinase isolated from Lachesis muta whose structure and function was extensively studied [4949. Felicori LF, Souza CT, Velarde DT, Magalhaes A, Almeida AP, Figueiredo S, et al. Kallikrein-like proteinase from bushmaster snake venom. Protein Expr Purif. 2003;30(1):32-42., 7272. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42.]. This enzyme is active against substrates for plasma and glandular kallikrein and plasmin [4949. Felicori LF, Souza CT, Velarde DT, Magalhaes A, Almeida AP, Figueiredo S, et al. Kallikrein-like proteinase from bushmaster snake venom. Protein Expr Purif. 2003;30(1):32-42., 7272. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42.], releases BK from bovine fibrinogen [7272. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42.] and decreases blood pressure in rats [4949. Felicori LF, Souza CT, Velarde DT, Magalhaes A, Almeida AP, Figueiredo S, et al. Kallikrein-like proteinase from bushmaster snake venom. Protein Expr Purif. 2003;30(1):32-42.].
Enzyme activity upon substrate S-2302 in the presence of possible inhibitors confirmed that LmrSP-4 is a serine proteinase. Proteolytic activity decreased in the presence of PMSF and benzamidine. Both are serine proteinase inhibitors but differ in their inhibition mechanism. While PMSF reacts with catalytic Ser residues in these proteins [7777. Gold AM. Sulfonyl fluorides as inhibitors of esterases. 3. Identification of serine as the site of sulfonylation in phenylmethanesulfonyl alpha-chymotrypsin. Biochemistry. 1965;4:897-901.], benzamidine is a reversible competitive inhibitor [7878. Markwardt F, Landmann H, Walsmann P. Comparative studies on the inhibition of trypsin, plasmin, and thrombin by derivatives of benzylamine and benzamidine. Eur J Biochem. 1968;6(4):502-6.]. On the other hand, 1,10-phenantroline is a metalloproteinase inhibitor which competes by the zinc ions needed for the proteinase activity [7979. Felber JP, Coombs TL, Vallee BL. The mechanism of inhibition of carboxypeptidase A by 1,10-phenanthroline. Biochemistry. 1962;1:231-8.]. Herein, this inhibitor did not affect the LmrSP-4 proteolytic activity, indicating that zinc ions are not essential for the catalytic activity. However, it is interesting that zinc ions are structurally relevant for ABUSV-PA, a plasminogen activator from Agkistrodon blomhoffii Ussurensis venom, although they do not influence its activity [8080. Liu SQ, Sun MZ, Greenaway FT. A novel plasminogen activator from Agkistrodon blomhoffii Ussurensis venom (ABUSV-PA): purification and characterization. Biochem Biophys Res Commun. 2006;348(4):1279-87.].
LmrSP-4 consumes fibrinogen Aα-chain (Fig. 1b and 1c) except in the presence of PMSF, a serine proteinase inhibitor. The proportion among the Aα, Bβ and γ chains remains the same comparing fibrinogen chains after its treatment with LmrSP-4 pre-incubated with PMSF (Fig. 1c). Besides the Aα consumption, there is a difference in the migration pattern close to the Aα chains (Fig. 1c), which might indicate the release of fibrinopeptide A from the Aα chain. However, a more specific assay is needed to confirm this hypothesis. TLEs from Lachesis usually release fibrinopeptide A but some of them also slowly split off fibrinopeptide B from fibrinogen [1616. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta. Int J Biochem. 1989;21(8):863-71., 2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292., 6161. Yarleque A, Campos S, Escobar E, Lazo F, Sanchez N, Hyslop S, et al. Isolation and characterization of a fibrinogen-clotting enzyme from venom of the snake, Lachesis muta muta (Peruvian bushmaster). Toxicon. 1989;27(11):1189-97., 7373 Magalhaes A, Ferreira RN, Richardson M, Gontijo S, Yarleque A, Magalhaes HP, et al. Coagulant thrombin-like enzymes from the venoms of Brazilian and Peruvian bushmaster (Lachesis muta muta) snakes. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(2):255-66.]. During the envenoming process, thrombin-like serine proteinases act on fibrinogen converting that in a different form of fibrin which does not form a solid fibrin clot. As time goes by, the fibrinogen of prey/victim is all consumed, and blood becomes uncoagulable [8181. Kini RM. Anticoagulant proteins from snake venoms: structure, function and mechanism. Biochem J. 2006;397(Pt 3):377-87.]. On the other hand, the action of TLEs upon fibrinogen has enable the production of the fibrin sealant: a mixture of fibrinogen-rich cryoprecipitate from Bubalus bubalis blood and a TLE from Crotalus durissus terrificus snake venom, which is in clinical trial for the treatment of chronic venous ulcers and has shown promising results as adjuvant in the peripheral nerve injury treatment [8282. Ferreira RS Jr, de Barros LC, Abbade LPF, Barraviera SRCS, Silvares MRC, de Pontes LG, et al. Heterologous fibrin sealant derived from snake venom: from bench to bedside - an overview. J Venom Anim Toxins incl Trop Dis. 2017;23:21. doi: 10.1186/s40409-017-0109-8.
https://doi.org/10.1186/s40409-017-0109-...
-8484. Biscola NP, Cartarozzi LP, Ulian-Benitez S, Barbizan R, Castro MV, Spejo AB, et al. Multiple uses of fibrin sealant for nervous system treatment following injury and disease. J Venom Anim Toxins incl Trop Dis. 2017;23:13. doi: 10.1186/s40409-017-0103-1.
https://doi.org/10.1186/s40409-017-0103-...
].
Regarding optimal conditions for LmrSP-4 enzyme activity, we firstly evaluated the pH condition incubating enzyme and FRET substrate in different pH values at 40 °C, a medium temperature for enzymes that act in physiological conditions. LmrSP-4 showed highest substrate hydrolysis in pH 7 although there was no statistically significant difference in activity within the pH range from 7 to 8 (Fig. 7a). The optimum pH determined for LmrSP-4 is in accordance to other SVSPs, especially those from Lachesis venoms [1616. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta. Int J Biochem. 1989;21(8):863-71., 2323. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292., 6060. Magalhães A, de Oliveira GJ, Diniz CR. Purification and partial characterization of a thrombin-like enzyme from the venom of the bushmaster snake, Lachesis muta noctivaga. Toxicon. 1981;19(2):279-94., 6161. Yarleque A, Campos S, Escobar E, Lazo F, Sanchez N, Hyslop S, et al. Isolation and characterization of a fibrinogen-clotting enzyme from venom of the snake, Lachesis muta muta (Peruvian bushmaster). Toxicon. 1989;27(11):1189-97.]. Temperature influence was evaluated at pH 7 and the enzyme showed the highest activity at 50 °C but there was no statistically significant difference in activity at 40, 45 and 55 ºC (Fig. 7b). Substrate hydrolysis decreased at 60 °C showing the beginning of a denaturation process and loss of activity. SVSPs are usually resistant to high temperatures [8585. Yonamine CM, Kondo MY, Juliano MA, Icimoto MY, Baptista GR, Yamane T, et al. Kinetic characterization of gyroxin, a serine protease from Crotalus durissus terrificus venom. Biochimie. 2012;94(12):2791-3.-8787. Boldrini-França J, Santos Rodrigues R, Santos-Silva LK, de Souza DL, Gomes MS, Cologna CT, et al. Expression of a new serine protease from Crotalus durissus collilineatus venom in Pichia pastoris and functional comparison with the native enzyme. Appl Microbiol Biotechnol. 2015;99(23):9971-86.] and this may be due to their sugar content and three-dimensional structure stabilized by the six conserved disulfide bonds [7070. Mackessy SP. Thrombin-like enzymes in nnake venoms. In: Toxins and Hemostasis - from bench to bedside. Kini MR, Markland FS, Morita T, Clemetson K, McLane MA editors. Dordrecht, Springer; 2010. pp. 519-57.].
Conclusions
Analysis of the dominant fraction of LmrV after molecular exclusion chromatography revealed the presence of novel and different serine proteinase isoforms in this venom as well as the first lectin from LmrV. A kallikrein-like serine proteinase (LmrSP-4) that might be useful as molecular tool for investigating bradykinin-involving process was isolated and biochemically characterized.
Abbreviations
ACN: acetonitrile; AGC: automatic gain control; AMBIC: ammonium bicarbonate; BK: bradykinin; BPPs: bradykinin potentiating peptides; CLP: C-type lectin-like protein; CRISP: cysteine-rich secretory protein; DTT: 1,4-dithiothreitol; EDTA: ethylenediamine tetraacetic acid; ETD: electron transfer dissociation; FRET: fluorescence resonance energy transfer; HCD: higher-energy collisional dissociation; HexNAc: N-acetylhexoseamine; LAAO: L-amino acid oxidase; Lmr: Lachesis muta rhombeata; LmrV: Lachesis muta rhombeata venom; MALDI-TOF MS: matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry; Neu5Ac: N-acetylneuraminic acid; PAGE: polyacrylamide gel electrophoresis; PLA2: phospholipase A2; PMSF: phenylmethylsulfonyl fluoride; RP-FPLC: reversed-phase fast protein liquid chromatography; SDS: sodium dodecylsulphate; SVSP: snake venom serine proteinase; TFA: trifluoroacetic acid; TLEs: thrombin-like enzymes; VEGF: vascular endothelial growth factor.
Acknowledgements
The authors are thankful to Iara Aimê Cardoso for technical assistance as well as to the Center for the Study of Venoms and Venomous Animals (CEVAP) of UNESP for enabling the publication of this special collection.
References
- 1. Brasil. Ministério da Saúde: Acidente por animais peçonhentos - Notificações registradas no sistema de informação de agravos de notificação - Brasil. Período 2013; 2017.
- 2. Málaque CMS, França FOS. Acidente laquético. In: Animais Peçonhentos no Brasil Biologia, Clínica e Terapêutica dos Acidentes. Cardoso JLC, França, F, Wen FH, Haddad Jr V, editors. São Paulo; 2003. pp. 87-90.
- 3. Jorge MT, Sano-Martins IS, Tomy SC, Castro SC, Ferrari RA, Ribeiro LA, et al. Snakebite by the bushmaster (Lachesis muta) in Brazil: case report and review of the literature. Toxicon. 1997;35(4):545-54.
- 4. Chippaux JP. Incidence and mortality due to snakebite in the Americas. PLoS Negl Trop Dis. 2017;11(6):e0005662.
- 5. Chippaux JP. Epidemiology of envenomations by terrestrial venomous animals in Brazil based on case reporting: from obvious facts to contingencies. J Venom Anim Toxins incl Trop Dis. 2015;21:13. doi: 10.1186/s40409-015-0011-1.
» https://doi.org/10.1186/s40409-015-0011-1 - 6. Torres JR, Torres MA, Arroyo-Parejo MA. Coagulation disorders in bushmaster envenomation. Lancet. 1995;346(8972):449-50.
- 7. Otero R, Furtado MF, Gonçalves C, Núñez V, García ME, Osorio RG, et al. Comparative study of the venoms of three subspecies of Lachesis muta (bushmaster) from Brazil, Colombia and Costa Rica. Toxicon. 1998;36(12):2021-7.
- 8. Pardal PP, Souza SM, Monteiro MR, Fan HW, Cardoso JL, França FO, et al. Clinical trial of two antivenoms for the treatment of Bothrops and Lachesis bites in the north eastern Amazon region of Brazil. Trans R Soc Trop Med Hyg. 2004;98(1):28-42.
- 9. Stephano MA, Guidolin R, Higashi HG, Tambourgi DV, Sant'Anna OA. The improvement of the therapeutic anti-Lachesis muta serum production in horses. Toxicon. 2005;45(4):467-73.
- 10. Bolaños R, Rojas O, Ulloa Flores CE. [Biomedical aspects of 4 cases of snake bites by Lachesis muta (Ophidia: Viperidae) in Costa Rica]. Rev Biol Trop. 1982;30(1):53-8. [Article in Spanish].
- 11. Ripa D. Ontogeny of the shock death in human beings. In: The bushmasters (Genus Lachesis Daudin, 1803) morphology in evolution and behavior, 5 edition. Wilmington: Cape Fear Serpentarium; 2007.
- 12. Pla D, Sanz L, Molina-Sánchez P, Zorita V, Madrigal M, Flores-Díaz M, et al. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis J Proteomics. 2013;89:112-23.
- 13. Wiezel GA, dos Santos PK, Cordeiro FA, Bordon KCF, Selistre-de-Araujo HS, Ueberheide B, et al. Identification of hyaluronidase and phospholipase B in Lachesis muta rhombeata venom. Toxicon. 2015;107(Pt B):359-68.
- 14. Diniz MR, Oliveira EB. Purification and properties of a kininogenin from the venom of Lachesis muta (bushmaster). Toxicon. 1992;30(3):247-58.
- 15. Sanchez EF, Santos CI, Magalhaes A, Diniz CR, Figueiredo S, Gilroy J, et al. Isolation of a proteinase with plasminogen-activating activity from Lachesis muta muta (bushmaster) snake venom. Arch Biochem Biophys. 2000;378(1):131-41.
- 16. Silveira AM, Magalhães A, Diniz CR, de Oliveira EB. Purification and properties of the thrombin-like enzyme from the venom of Lachesis muta muta Int J Biochem. 1989;21(8):863-71.
- 17. Rucavado A, Flores-Sánchez E, Franceschi A, Magalhaes A, Gutiérrez JM. Characterization of the local tissue damage induced by LHF-II, a metalloproteinase with weak hemorrhagic activity isolated from Lachesis muta muta snake venom. Toxicon. 1999;37(9):1297-312.
- 18. Estêvão-Costa MI, Diniz CR, Magalhães A, Markland FS, Sanchez EF. Action of metalloproteinases mutalysin I and II on several components of the hemostatic and fibrinolytic systems. Thromb Res. 2000;99(4):363-76.
- 19. Cordeiro FA, Perini TG, Bregge-Silva C, Cremonez CM, Rodrigues RS, Boldrini-França J, et al. A new phospholipase A₂ from Lachesis muta rhombeata: purification, biochemical and comparative characterization with crotoxin B. Protein Pept Lett. 2015;22(9):816-27.
- 20. Fuly AL, Calil-Elias S, Zingali RB, Guimarães JA, Melo PA. Myotoxic activity of an acidic phospholipase A2 isolated from Lachesis muta (Bushmaster) snake venom. Toxicon. 2000;38(7):961-72.
- 21. Bregge-Silva C, Nonato MC, de Albuquerque S, Ho PL, Junqueira de Azevedo IL, Vasconcelos Diniz MR, et al. Isolation and biochemical, functional and structural characterization of a novel L-amino acid oxidase from Lachesis muta snake venom. Toxicon. 2012;60(7):1263-76.
- 22. Melgarejo-Giménez AR. Criação e Manejo de Serpentes. In: Animais de Laboratório: criação e experimentação. Andrade A, Pinto SC, Oliveira RS editors. Fiocruz, Rio de Janeiro; 2002. pp. 175-99.
- 23. Torres-Huaco FD, Werneck CC, Vicente CP, Vassequi-Silva T, Nery-Diez AC, Mendes CB, et al. Rapid purification and procoagulant and platelet aggregating activities of Rhombeobin: a thrombin-like/gyroxin-like enzyme from Lachesis muta rhombeata snake venom. Biomed Res Int. 2013;2013:903292.
- 24. Giovanni-De-Simone S, Aguiar AS, Gimenez AR, Novellino K, de Moura RS. Purification, properties, and N-terminal amino acid sequence of a kallikrein-like enzyme from the venom of Lachesis muta rhombeata (Bushmaster). J Protein Chem. 1997;16(8):809-18.
- 25. Aguiar AS, Alves CR, Melgarejo A, Giovanni-de-Simone S. Purification and partial characterization of a thrombin-like/gyroxin enzyme from bushmaster (Lachesis muta rhombeata) venom. Toxicon. 1996;34(5):555-65.
- 26. Pinheiro-Júnior EL, Boldrini-França J, de Campos Araújo LMP, Santos-Filho NA, Bendhack LM, Cilli EM, et al. LmrBPP9: A synthetic bradykinin-potentiating peptide from Lachesis muta rhombeata venom that inhibits the angiotensin-converting enzyme activity in vitro and reduces the blood pressure of hypertensive rats. Peptides. 2018;102:1-7.
- 27. Rucavado A, Escalante T, Gutiérrez JM. Effect of the metalloproteinase inhibitor batimastat in the systemic toxicity induced by Bothrops asper snake venom: understanding the role of metalloproteinases in envenomation. Toxicon. 2004;43(4):417-24.
- 28. Boldrini-França J, Corrêa-Netto C, Silva MM, Rodrigues RS, De La Torre P, Pérez A, et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: assessment of geographic variation and its implication on snakebite management. J Proteomics. 2010;73(9):1758-76.
- 29. Xiao H, Pan H, Liao K, Yang M, Huang C. Snake venom PLA2, a promising target for broad-spectrum antivenom drug development. Biomed Res Int. 2017;2017:6592820.
- 30. Karapetian H. Reptilase time (RT). Methods Mol Biol. 2013;992:273-7.
- 31. Scarborough RM, Rose JW, Hsu MA, Phillips DR, Fried VA, Campbell AM, et al. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m. barbouri J Biol Chem. 1991;266(15):9359-62.
- 32. Dardevet L, Rani D, Aziz TA, Bazin I, Sabatier JM, Fadl M, et al. Chlorotoxin: a helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins (Basel). 2015;7(4):1079-101.
- 33. Edman P, Begg G. A protein sequenator. Eur J Biochem. 1967;1(1):80-91.
- 34. Cotto-Rios XM, Békés M, Chapman J, Ueberheide B, Huang TT. Deubiquitinases as a signaling target of oxidative stress. Cell Rep. 2012;2(6):1475-84.
- 35. UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204-12.
- 36. Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;13.
- 37. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680-5.
- 38. Doerner KC, White BA. Detection of glycoproteins separated by nondenaturing polyacrylamide gel electrophoresis using the periodic acid-Schiff stain. Anal Biochem. 1990;187(1):147-50.
- 39. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539.
- 40. Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320-3.
- 41. W Edgar, Prentice CRM. The proteolytic action of ancrod on human fibrinogen and its polypeptide chains. Thromb Res. 1973;2(1):85-95.
- 42. da Silva RR, de Oliveira LCG, Juliano MA, Juliano L, Rosa JC, Cabral H. Activity of a peptidase secreted by Phanerochaete chrysosporium depends on lysine to subsite S'1. Int J Biol Macromol. 2017;94(Pt A):474-83.
- 43. Junqueira-de-Azevedo IL, Ching AT, Carvalho E, Faria F, Nishiyama MY Jr, Ho PL, et al. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics. 2006;173(2):877-89.
- 44. Madrigal M, Sanz L, Flores-Díaz M, Sasa M, Núñez V, Alape-Girón A, et al. Snake venomics across genus Lachesis Ontogenetic changes in the venom composition of Lachesis stenophrys and comparative proteomics of the venoms of adult Lachesis melanocephala and Lachesis acrochorda J Proteomics. 2012;77:280-97.
- 45. Sanz L, Escolano J, Ferretti M, Biscoglio MJ, Rivera E, Crescenti EJ, et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J Proteomics, 2008;71(1):46-60.
- 46. Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477(1-2):146-56.
- 47. Kini RM. Serine proteases affecting blood coagulation and fibrinolysis from snake venoms. Pathophysiol Haemost Thromb. 2005;34(4-5):200-4.
- 48. White J. Snake venoms and coagulopathy. Toxicon. 2005;45(8):951-67.
- 49. Felicori LF, Souza CT, Velarde DT, Magalhaes A, Almeida AP, Figueiredo S, et al. Kallikrein-like proteinase from bushmaster snake venom. Protein Expr Purif. 2003;30(1):32-42.
- 50. Serrano SM, Maroun RC. Snake venom serine proteinases: sequence homology vs. substrate specificity, a paradox to be solved. Toxicon. 2005;45(8):1115-32.
- 51. Markland FS, Kettner C, Schiffman S, Shaw E, Bajwa SS, Reddy KN, et al. Kallikrein-like activity of crotalase, a snake venom enzyme that clots fibrinogen. Proc Natl Acad Sci U S A. 1982;79(6):1688-92.
- 52. Du XY, Clemetson KJ. Reptile C-type lectins. In: Handbook of Venoms and Toxins of Reptiles. Mackessy SP editor. Boca Raton: CRC Press; 2010. pp. 359-75.
- 53. Arlinghaus FT, Eble JA. C-type lectin-like proteins from snake venoms. Toxicon. 2012;60(4):512-9.
- 54. Sartim MA, Sampaio SV. Snake venom galactoside-binding lectins: a structural and functional overview. J Venom Anim Toxins incl Trop Dis. 2015;21:35. doi: 10.1186/s40409-015-0038-3.
» https://doi.org/10.1186/s40409-015-0038-3 - 55. Clemetson KJ. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon. 2010;56(7):1236-46.
- 56. Aragón-Ortíz F, Brenes-Brenes JR, Gubensek F. Characterization of a lectin-like protein isolated from Lachesis muta snake venom. Rev Biol Trop. 1989;37(1):79-83.
- 57. Ogilvie ML, Dockter ME, Wenz L, Gartner TK. Isolation and characterization of lactose-binding lectins from the venoms of the snakes Lachesis muta and Dendroaspis jamesonii J Biochem. 1986;100(6):1425-31.
- 58. Castro HC, Zingali RB, Albuquerque MG, Pujol-Luz M, Rodrigues CR. Snake venom thrombin-like enzymes: from reptilase to now. Cell Mol Life Sci. 2004;61(7-8):843-56.
- 59. Aragon-Ortiz F, Gubensek F. A thrombin-like enzyme from bushmaster (Lachesis muta stenophyrs) venom. Toxicon. 1993;31(11):1435-43.
- 60. Magalhães A, de Oliveira GJ, Diniz CR. Purification and partial characterization of a thrombin-like enzyme from the venom of the bushmaster snake, Lachesis muta noctivaga Toxicon. 1981;19(2):279-94.
- 61. Yarleque A, Campos S, Escobar E, Lazo F, Sanchez N, Hyslop S, et al. Isolation and characterization of a fibrinogen-clotting enzyme from venom of the snake, Lachesis muta muta (Peruvian bushmaster). Toxicon. 1989;27(11):1189-97.
- 62. Roge AR, Alma S, Romano RWDL. Lachesis muta rhombeata (Serpentes : Viperidae, Crotalinae). Mem Inst Butantan. 1976;40:53-4.
- 63. Silva LM, Diniz CR, Magalhães A. Purification and partial characterization of an arginine ester hydrolase from the venom of the bushmaster snake, Lachesis muta noctivaga Toxicon. 1985;23(4):707-18.
- 64. Doley R, Mackessy SP, Kini RM. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol Biol. 2009;9:146.
- 65. Vaiyapuri S, Wagstaff SC, Harrison RA, Gibbins JM, Hutchinson EG. Evolutionary analysis of novel serine proteases in the venom gland transcriptome of Bitis gabonica rhinoceros PLoS One. 2011;6(6):e21532.
- 66. Magalhaes A, Da Fonseca BC, Diniz CR, Gilroy J, Richardson M. The complete amino acid sequence of a thrombin-like enzyme/gyroxin analogue from venom of the bushmaster snake (Lachesis muta muta). FEBS Lett. 1993;329(1-2):116-20.
- 67. Sanchez EF, Felicori LF, Chavez-Olortegui C, Magalhaes HBP, Hermogenes AL, Diniz MV, et al. Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from bushmaster snake venom. Biochim Biophys Acta. 2006;1760(12):1762-71.
- 68. Zhang Y, Wisner A, Maroun RC, Choumet V, Xiong Y, Bon C. Trimeresurus stejnegeri snake venom plasminogen activator. Site-directed mutagenesis and molecular modeling. J Biol Chem. 1997;272(33):20531-7.
- 69. Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3(5):433-42.
- 70. Mackessy SP. Thrombin-like enzymes in nnake venoms. In: Toxins and Hemostasis - from bench to bedside. Kini MR, Markland FS, Morita T, Clemetson K, McLane MA editors. Dordrecht, Springer; 2010. pp. 519-57.
- 71. Swenson S, Markland FS Jr. Snake venom fibrin(ogen)olytic enzymes. Toxicon. 2005;45(8):1021-39.
- 72. Weinberg MLD, Felicori LF, Bello CA, Magalhaes HPB, Almeida AP, Magalhaes A, et al. Biochemical properties of a bushmaster snake venom serine proteinase (LV-Ka), and its kinin releasing activity evaluated in rat mesenteric arterial rings. J Pharmacol Sci. 2004;96(3):333-42.
- 73 Magalhaes A, Ferreira RN, Richardson M, Gontijo S, Yarleque A, Magalhaes HP, et al. Coagulant thrombin-like enzymes from the venoms of Brazilian and Peruvian bushmaster (Lachesis muta muta) snakes. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(2):255-66.
- 74. Bailey GS, Shipolini RA. Purification and properties of a kininogenin from the venom of Vipera ammodytes ammodytes Biochem J. 1976;153(2):409-14.
- 75. Mukherjee AK. The pro-coagulant fibrinogenolytic serine protease isoenzymes purified from Daboia russelii russelii venom coagulate the blood through factor V activation: role of glycosylation on enzymatic activity. PLoS One. 2014;9(2):e86823.
- 76. Raspi G. Kallikrein and kallikrein-like proteinases: purification and determination by chromatographic and electrophoretic methods. J Chromatogr B Biomed Appl. 1996;684(1-2):265-87.
- 77. Gold AM. Sulfonyl fluorides as inhibitors of esterases. 3. Identification of serine as the site of sulfonylation in phenylmethanesulfonyl alpha-chymotrypsin. Biochemistry. 1965;4:897-901.
- 78. Markwardt F, Landmann H, Walsmann P. Comparative studies on the inhibition of trypsin, plasmin, and thrombin by derivatives of benzylamine and benzamidine. Eur J Biochem. 1968;6(4):502-6.
- 79. Felber JP, Coombs TL, Vallee BL. The mechanism of inhibition of carboxypeptidase A by 1,10-phenanthroline. Biochemistry. 1962;1:231-8.
- 80. Liu SQ, Sun MZ, Greenaway FT. A novel plasminogen activator from Agkistrodon blomhoffii Ussurensis venom (ABUSV-PA): purification and characterization. Biochem Biophys Res Commun. 2006;348(4):1279-87.
- 81. Kini RM. Anticoagulant proteins from snake venoms: structure, function and mechanism. Biochem J. 2006;397(Pt 3):377-87.
- 82. Ferreira RS Jr, de Barros LC, Abbade LPF, Barraviera SRCS, Silvares MRC, de Pontes LG, et al. Heterologous fibrin sealant derived from snake venom: from bench to bedside - an overview. J Venom Anim Toxins incl Trop Dis. 2017;23:21. doi: 10.1186/s40409-017-0109-8.
» https://doi.org/10.1186/s40409-017-0109-8 - 83. Mozafari R, Kyrylenko S, Castro MV, Ferreira RS, Barraviera B, Oliveira ALR. Combination of heterologous fibrin sealant and bioengineered human embryonic stem cells to improve regeneration following autogenous sciatic nerve grafting repair. J Venom Anim Toxins incl Trop Dis. 2018;24:11. doi: 10.1186/s40409-018-0147-x.
» https://doi.org/10.1186/s40409-018-0147-x - 84. Biscola NP, Cartarozzi LP, Ulian-Benitez S, Barbizan R, Castro MV, Spejo AB, et al. Multiple uses of fibrin sealant for nervous system treatment following injury and disease. J Venom Anim Toxins incl Trop Dis. 2017;23:13. doi: 10.1186/s40409-017-0103-1.
» https://doi.org/10.1186/s40409-017-0103-1 - 85. Yonamine CM, Kondo MY, Juliano MA, Icimoto MY, Baptista GR, Yamane T, et al. Kinetic characterization of gyroxin, a serine protease from Crotalus durissus terrificus venom. Biochimie. 2012;94(12):2791-3.
- 86. Menaldo DL, Bernardes CP, Santos-Filho NA, Moura LeA, Fuly AL, Arantes EC, et al. Biochemical characterization and comparative analysis of two distinct serine proteases from Bothrops pirajai snake venom. Biochimie. 2012;94(12):2545-58.
- 87. Boldrini-França J, Santos Rodrigues R, Santos-Silva LK, de Souza DL, Gomes MS, Cologna CT, et al. Expression of a new serine protease from Crotalus durissus collilineatus venom in Pichia pastoris and functional comparison with the native enzyme. Appl Microbiol Biotechnol. 2015;99(23):9971-86.
-
Availability of data and material
All data generated or analyzed during this study are included in this published article. -
Funding
This study was supported by the National Institute of Health (NIH, USA) grant R41 GM103362 and the Brazilian funding agencies São Paulo Research Foundation (FAPESP, grants 2011/23236-4 and 2015/18432-0, and scholarships to GAW 2010/06199-5 and 2014/23285-3), Minas Gerais Research Foundation (FAPEMIG, CBB-APQ-01637-15), Bahia Research Foundation (FAPESB), the National Council for Scientific and Technological Development (CNPq, grants 303689/2013-7; 449960/2014-5 and 440954/2017-7) and the Coordination for the Improvement of Higher Education Personnel (CAPES), Programa Editoração CAPES grant n. 88881.142062/2017-01). The authors are also thankful to the Federal University of Uberlândia (UFU, Brazil) and the National Institute of Science and Technology in Theranostics and Nanobiotechnology INCT-TeraNano-CNPq/CAPES/FAPEMIG (grant number CNPq-465669/2014-0). -
Ethics approval and consent to participate
Not applicable. -
Consent for publication
Not applicable.
Supplementary material
TThe following online material is available for this article
Publication Dates
-
Publication in this collection
15 Apr 2019 -
Date of issue
2019
History
-
Received
29 June 2018 -
Accepted
02 Oct 2018