Abstract
Background:
Bothrops atrox is known to be the pit viper responsible for most snakebites and human fatalities in the Amazon region. It can be found in a wide geographical area including northern South America, the east of Andes and the Amazon basin. Possibly, due to its wide distribution and generalist feeding, intraspecific venom variation was reported by previous proteomics studies. Sex-based and ontogenetic variations on venom compositions of Bothrops snakes were also subject of proteomic and peptidomic analysis. However, the venom peptidome of B. atrox remains unknown.
Methods:
We conducted a mass spectrometry-based analysis of the venom peptides of individual male and female specimens combining bottom-up and top-down approaches.
Results:
We identified in B. atrox a total of 105 native peptides in the mass range of 0.4 to 13.9 kDa. Quantitative analysis showed that phospholipase A2 and bradykinin potentiating peptides were the most abundant peptide families in both genders, whereas disintegrin levels were significantly increased in the venoms of females. Known peptides processed at non-canonical sites and new peptides as the Ba1a, which contains the SVMP BATXSVMPII1 catalytic site, were also revealed in this work.
Conclusion:
The venom peptidomes of male and female specimens of B. atrox were analyzed by mass spectrometry-based approaches in this work. The study points to differences in disintegrin levels in the venoms of females that may result in distinct pathophysiology of envenomation. Further research is required to explore the potential biological implications of this finding.
Keywords
Bothrops atrox; Venom; Peptidomics; Sexual dimorphism; Disintegrin
Background
Snake venoms are toxic glandular secretions containing high concentrations of proteins and peptides. Their biologically active components were elaborated and refined over millions of years of evolution through an arms race with its preys [11. Calvete JJ. Venomics: integrative venom proteomics and beyond. Biochem J. 2017 Feb 20;474(5):611-34. doi: 10.1042/BCJ20160577.
https://doi.org/10.1042/BCJ20160577...
,22. Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013 Apr;28(4):219-29. doi:10.1016/j.tree.2012.10.020.
https://doi.org/10.1016/j.tree.2012.10.0...
]. Particularly, Bothrops atrox is a highly adapted and widely distributed species found in many countries of northern South America [33. Núñez V, Cid P, Sanz L, de la Torre P, Angulo Y, Lomonte B, et al. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics. 2009 Aug 5;73(1):57-78. doi: 10.1016/j.jprot.2009.07.013.
https://doi.org/10.1016/j.jprot.2009.07....
,44. Calvete JJ, Sanz L, Pérez A, Borges A, Vargas AM, Lomonte B, et al. Snake population venomics and antivenomics of Bothrops atrox: paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J Proteomics. 2011 Apr 1;74(4):510-27. doi: 10.1016/j.jprot.2011.01.003.
https://doi.org/10.1016/j.jprot.2011.01....
]. The snake is responsible for most snakebites in Northern Brazil [55. Santos Barreto GN, Oliveira SS, Anjos IV, Chalkidis HM, Mourão RH, Moura-da-Silva AM, et al. Experimental Bothrops atrox envenomation: efficacy of antivenom therapy and the combination of Bothrops antivenom with dexamethasone. PLoS Negl Trop Dis. 2017 Mar 17;11(3):e0005458. doi: 10.1371/journal.pntd.0005458.
https://doi.org/10.1371/journal.pntd.000...
] and its venom is characterized by three main pathophysiological activities: coagulant, hemorrhagic, and acute inflammatory effects [66. Oliveira SS, Sampaio VS, Sachett JA, Alves EC, Silva VC, Lima JA, et al. Snakebites in the Brazilian Amazon: current knowledge and perspectives. In: Gopalakrishnakone P, editor. Clinical toxinology in Australia, Europe, and Americas. Dordrecht: Springer; 2018. p. 73-99. doi: 10.1007/978-94-017-7438-3_61.
https://doi.org/10.1007/978-94-017-7438-...
]. Previous proteomics studies revealed intraspecific variation in B. atrox venom composition associated to its wide geographical distribution range [33. Núñez V, Cid P, Sanz L, de la Torre P, Angulo Y, Lomonte B, et al. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics. 2009 Aug 5;73(1):57-78. doi: 10.1016/j.jprot.2009.07.013.
https://doi.org/10.1016/j.jprot.2009.07....
,44. Calvete JJ, Sanz L, Pérez A, Borges A, Vargas AM, Lomonte B, et al. Snake population venomics and antivenomics of Bothrops atrox: paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J Proteomics. 2011 Apr 1;74(4):510-27. doi: 10.1016/j.jprot.2011.01.003.
https://doi.org/10.1016/j.jprot.2011.01....
,77. Sousa LF, Portes JA Jr, Nicolau CA, Bernardoni JL, Nishiyama MY Jr, Amazonas DR, et al. Functional proteomic analyses of Bothrops atrox venom reveals phenotypes associated with habitat variation in the Amazon. J Proteomics. 2017 Apr 21;159:32-46. doi: 10.1016/j.jprot.2017.03.003.
https://doi.org/10.1016/j.jprot.2017.03....
]. Differences and similarities in venom compositions were found in these studies, suggesting that venom phenotypes may be classified according to specific regions [44. Calvete JJ, Sanz L, Pérez A, Borges A, Vargas AM, Lomonte B, et al. Snake population venomics and antivenomics of Bothrops atrox: paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J Proteomics. 2011 Apr 1;74(4):510-27. doi: 10.1016/j.jprot.2011.01.003.
https://doi.org/10.1016/j.jprot.2011.01....
]. Other intraspecific venom variations related to sex [88. Zelanis A, Menezes MC, Kitano ES, Liberato T, Tashima AK, Pinto AF, et al. Proteomic identification of gender molecular markers in Bothrops jararaca venom. J Proteomics. 2016 Apr 29;139:26-37. doi: 10.1016/j.jprot.2016.02.030.
https://doi.org/10.1016/j.jprot.2016.02....
-1010. Amorim FG, Costa TR, Baiwir D, Pauw E, Quinton L, Sampaio SV. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: influence of snake gender on venom composition. Toxins (Basel). 2018 Apr 26;10(5):177. doi: 10.3390/toxins10050177.
https://doi.org/10.3390/toxins10050177...
], diet [1111. Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996 Feb 8;379(6565):537-40. doi: 10.1038/379537a0.
https://doi.org/10.1038/379537a0...
,1212. Barlow A, Pook CE, Harrison RA, Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci. 2009 Jul 7;276(1666):2443-9. doi: 10.1098/rspb.2009.0048.
https://doi.org/10.1098/rspb.2009.0048...
] and ontogeny [1313. Zelanis A, Tashima AK, Rocha MM, Furtado MF, Camargo AC, Ho PL, et al. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J Proteome Res. 2010 May 7;9(5):2278-91. doi: 10.1021/pr901027r.
https://doi.org/10.1021/pr901027r...
] are well-documented phenomena in snake species. A remarkable sexual dimorphism in B. atrox is the size difference between males and females [1414. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.]. Males are significantly smaller than females and as a result may present higher motilities [1414. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.].
On the other side of venom research, toxins have been sources of inspiration for drug research and also significant in elucidating major biochemical and physiological mechanisms in vertebrates [1515. Escoubas P, Quinton L, Nicholson GM. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J Mass Spectrom. 2008 Mar;43(3):279-95. doi: 10.1002/jms.1389.
https://doi.org/10.1002/jms.1389...
-1717. Tashima AK, Zelanis A. Snake venom peptidomics. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Dordrecht: Springer Netherlands; 2016. p. 317-31. doi: 10.1007/978-94-007-6416-3_49.
https://doi.org/10.1007/978-94-007-6416-...
]. Biochemical approaches of isolation and analysis of purified venom components revealed important biologically active peptides including the bradykinin potentiating peptides (BPPs) [1818. Ferreira SH, Greene LH, Alabaster VA, Bakhle YS, Vane JR. Activity of various fractions of bradykinin potentiating factor against angiotensin I converting enzyme. Nature. 1970 Jan 1;225:379-80. doi: 10.1038/225379a0.
https://doi.org/10.1038/225379a0...
,1919. Ondetti MA, Williams NJ, Sabo E, Pluscec J, Weaver ER, Kocy O. Angiotensin-converting enzyme inhibitors from the venom of Bothrops jararaca. Isolation, elucidation of structure, and synthesis. Biochemistry. 1971 Oct 26;10(22):4033-9. doi: 10.1021/bi00798a004.
https://doi.org/10.1021/bi00798a004...
], sarafotoxins [2020. Kloog Y, Ambar I, Sokolovsky M, Kochva E, Wollberg Z, Bdolah A. Sarafotoxin, a novel vasoconstrictor peptide: phosphoinositide hydrolysis in rat heart and brain. Science. 1988 Oct 14;242(4876):268-70. doi: 10.1126/science.2845579.
https://doi.org/10.1126/science.2845579...
], disintegrins [2121. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
https://doi.org/10.1016/0167-4889(90)900...
] and analgesic peptides as crotalphine [2222. Konno K, Picolo G, Gutierrez VP, Brigatte P, Zambelli VO, Camargo AC, et al. Crotalphine, a novel potent analgesic peptide from the venom of the South American rattlesnake Crotalus durissus terrificus. Peptides. 2008 Aug;29(8):1293-304. doi: 10.1016/j.peptides.2008.04.003.
https://doi.org/10.1016/j.peptides.2008....
], for instance. However, despite the discovery of important venom toxins and the maturity achieved by the snake venomics [2323. Lomonte B, Calvete JJ. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J Venom Anim Toxins Incl Trop Dis. 2017;23(1):26. doi: 10.1186/s40409-017-0117-8.
https://doi.org/10.1186/s40409-017-0117-...
] and other venom proteomics approaches [2424. Galizio NC, Serino-Silva C, Stuginski DR, Abreu PA, Sant’Anna SS, Grego KF, et al. Compositional and functional investigation of individual and pooled venoms from long-term captive and recently wild-caught Bothrops jararaca snakes. J Proteomics. 2018 Aug 30;186:56-70. doi: 10.1016/j.jprot.2018.07.007.
https://doi.org/10.1016/j.jprot.2018.07....
,2525. Morais-Zani K, Serino-Silva C, Galizio NC, Tasima LJ, Pagotto JF, Rocha MM, et al. Does the administration of pilocarpine prior to venom milking influence the composition of Micrurus corallinus venom? J Proteomics. 2018 Mar 1;174:17-27. doi: 10.1016/j.jprot.2017.12.010.
https://doi.org/10.1016/j.jprot.2017.12....
], venom peptidomics is still an emerging research field [1717. Tashima AK, Zelanis A. Snake venom peptidomics. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Dordrecht: Springer Netherlands; 2016. p. 317-31. doi: 10.1007/978-94-007-6416-3_49.
https://doi.org/10.1007/978-94-007-6416-...
,2626. Zelanis A, Keiji Tashima A. Unraveling snake venom complexity with ‘omics’ approaches: challenges and perspectives. Toxicon. 2014 Sep;87:131-4. doi: 10.1016/j.toxicon.2014.05.011.
https://doi.org/10.1016/j.toxicon.2014.0...
]. Only a few peptides have been characterized in the venom of B. atrox, as the BPP-12a, BPP-BAX12 [2727. Menin L, Perchuć A, Favreau P, Perret F, Michalet S, Schöni R, et al. High throughput screening of bradykinin-potentiating peptides in Bothrops moojeni snake venom using precursor ion mass spectrometry. Toxicon. 2008 Jun 1;51(7):1288-302. doi: 10.1016/j.toxicon.2008.02.019.
https://doi.org/10.1016/j.toxicon.2008.0...
,2828. Coutinho-Neto A, Caldeira CA, Souza GH, Zaqueo KD, Kayano AM, Silva RS, et al. ESI-MS/MS identification of a bradykinin-potentiating peptide from Amazon Bothrops atrox snake venom using a hybrid Qq-oaTOF mass spectrometer. Toxins (Basel). 2013 Feb 18;5(2):327-35. doi: 10.3390/toxins5020327.
https://doi.org/10.3390/toxins5020327...
] and the disintegrin batroxostatin [2121. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
https://doi.org/10.1016/0167-4889(90)900...
].
Peptidomics analysis of other Bothrops snake venoms revealed new BPPs, poly-His-poly-Gly peptides and other protein fragments [1313. Zelanis A, Tashima AK, Rocha MM, Furtado MF, Camargo AC, Ho PL, et al. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J Proteome Res. 2010 May 7;9(5):2278-91. doi: 10.1021/pr901027r.
https://doi.org/10.1021/pr901027r...
,2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,3030. Nicolau CA, Carvalho PC, Junqueira-de-Azevedo IL, Teixeira-Ferreira A, Junqueira M, Perales J, et al. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca. J Proteomics. 2017 Jan 16;151:214-31. doi: 10.1016/j.jprot.2016.06.029.
https://doi.org/10.1016/j.jprot.2016.06....
]. Biological assays indicated that few amino acid mutations have significant effects on the activities of peptides within the same class [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
]. Thus, considering the richness of the yet unexplored peptidome and the sexual dimorphism of B. atrox, we used in this work a combination of mass spectrometry-based analysis and bioinformatics to compare the male and female Bothrops atrox venom peptidomes.
Methods
Reagents
Proteolytic enzymes (Asp-N, Glu-C and trypsin) were purchased from Promega. Dithiothreitol (DTT) and iodoacetamide were obtained from GE Healthcare. Acetonitrile was purchased from Avantor Pierce. Unless otherwise stated, all other reagents were acquired from Sigma-Aldrich.
Animals
Adult Bothrops atrox specimens from Northeastern of Brazil (Viana, MA) were maintained in the biotherium of the Laboratório de Herpetologia, Instituto Butantan (SP, Brazil). Experiments were approved by the Ethical Committee of Instituto Butantan (number 6303280220), the Ethical Committee of Universidade Federal de São Paulo (number 3437250719) and performed in accordance to the Brazilian laws for the use of experimental animals and with the ethical principles adopted by the Brazilian College of Animal Experimentation (COBEA).
Venom extraction and fractionation
Venom samples were extracted from four female and four male specimens of B. atrox. The animals were previously anesthetized with carbon dioxide. Venoms were individually extracted into beakers kept in ice bath and immediately mixed with the proteinase inhibitors EDTA and PMSF to final concentrations of 5 mM and 2 mM, respectively [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,3131. Serrano SMT, Zelanis A, Kitano ES, Tashima AK. Analysis of the snake venom peptidome. Methods Mol Biol. 2018;1719:349-58. doi: 10.1007/978-1-4939-7537-2_23.
https://doi.org/10.1007/978-1-4939-7537-...
]. The venom solutions were centrifuged at 16,000 g and 4 °C for 5 min to remove debris, lyophilized and stored at -20 °C for further fractionation.
Venom peptidomic fractions were obtained from 50 µg aliquots of crude venoms subjected to solid-phase extraction with C18 stage tips and eluted with 40% ACN [3232. Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896-906. doi: 10.1038/nprot.2007.261.
https://doi.org/10.1038/nprot.2007.261...
]. Stage tips were assembled with InertSep RP-C18 resin (GL Sciences) and SDB-XC membrane (Empore, 3M) inside P200 pipette tips. The eluates were dried in a vacuum concentrator (Concentrator Plus, Eppendorf) and the dried venom eluates were stored at -20 °C until MS analysis or digestion prior to analysis.
Enzyme digestion
Pools of venoms (males and females distinctly) containing 50 µg of crude venom each were separately digested with three different enzymes. Venoms were dissolved in specific buffer solutions for each enzyme and digested as previously described [3333. Abreu TF, Sumitomo BN, Nishiyama MY Jr, Oliveira UC, Souza GH, Kitano ES, et al. Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J Proteomics. 2017 Jan 16;151:232-42. doi: 10.1016/j.jprot.2016.07.012.
https://doi.org/10.1016/j.jprot.2016.07....
,3434. Lomazi RL, Nishiduka ES, Silva PI Jr, Tashima AK. Identification of peptides in spider venom using mass spectrometry. Methods Mol Biol. 2018;1719:359-67. doi: 10.1007/978-1-4939-7537-2_24.
https://doi.org/10.1007/978-1-4939-7537-...
]. Briefly, for trypsin and Asp-N, samples were dissolved in 50 mM NH4HCO3 and in 50 mM sodium phosphate for digestion with Glu-C. Additionally, two other pools of each gender containing 100 µg of crude venom were digested only with trypsin. The enzyme to protein ratio of 1:100 was used for all digestions. Samples were incubated with 0.2% RapiGest surfactant (Waters) at 80 °C for 15 min, followed by centrifugation at 2000 g for 3 min. All samples were reduced with 5 mM DTT for 30 min at 60 °C and alkylated with 10 mM iodoacetamide for 30 min in the dark at room temperature. Incubations with the enzymes were conducted for 30 minutes at 37 °C. TFA at final concentration of 0.5% was added to the samples to stop the digestions and to cleave the RapiGest surfactant. Samples were cleaned in stage tips, as described in the fractionation section, before LC-MS/MS analysis.
Mass spectrometry acquisition
LC-MS/MS analysis of native and digested toxins were performed on a Synapt G2 HDMS mass spectrometer (Waters) coupled to a nanoAcquity UPLC (Waters) chromatographic system. Samples were injected into a trap column (nanoAquity C18 trap column Symmetry 180 µm x 20 mm, Waters) and transferred by an elution gradient to an analytical column (nanoAcquity C18 BEH 75 µm x 150 mm, 1.7 mm, Waters). Mobile phase A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) were used to generate a 7-35% B elution gradient run over 60 min at a flow rate of 275 nl/min. Data were acquired in the data-independent acquisition modes MSE and UDMSE with ion mobility separation [3333. Abreu TF, Sumitomo BN, Nishiyama MY Jr, Oliveira UC, Souza GH, Kitano ES, et al. Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J Proteomics. 2017 Jan 16;151:232-42. doi: 10.1016/j.jprot.2016.07.012.
https://doi.org/10.1016/j.jprot.2016.07....
,3535. Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014 Feb;11(2):167-70. doi: 10.1038/nmeth.2767.
https://doi.org/10.1038/nmeth.2767...
], in the m/z range of 50-2000 and operating in resolution mode. Peptide ions were fragmented by collision induced dissociation (CID) switching from low (4 eV) to high (ramped from 19 to 45 eV) collision energy, for accurate measurement of both precursor and fragment ions. Scan times were set to 1.25 s. The ESI source was operated in the positive mode with a capillary voltage of 3.0 kV, block temperature of 100 °C and cone voltage of 40 V. Glu-fibrinopeptide B (Peptide 2.0) was infused through the nanoLockSpray source and sampled for 1 s every 60 s for external calibration. Native venom peptides and digested samples were analyzed in technical duplicates, totalizing 46 LC-MS/MS runs.
Bioinformatics analysis
Quantitative analysis of native peptides
Raw data of native peptides were processed and analyzed in Progenesis QI for Proteomics (Nonlinear Dynamics). Relative quantification and retention time alignment were based on peptide ion data of a reference run automatically selected. Only native peptide ions with normalized abundance above 200 counts and detected in at least 2 biological replicates of male or female groups were considered for further analysis. Entries with differences in monoisotopic mass and retention time below 30 ppm and 2 min, respectively, were regarded as redundant and only the entry with higher average abundance was considered.
Peptide identification
MS/MS spectra peak lists were generated in the software ProteinLynx Global Server 3.0.3 (Waters) as .mzML files. Spectra were processed by the Apex3D module using low energy threshold of 750 counts and high energy threshold of 50 counts. The peak lists of native peptide samples were submitted to searches using MASCOT 2.2.04 (Matrix Science) and PEAKS Studio 7.5 (Bioinformatics Solution Inc.) against the following Uniprot databases: Bothrops atrox with 202 entries (date of fasta file: June 21, 2018), Bothrops with 1,120 entries (date of fasta file: June 21, 2018) and Serpentes with 156,483 entries (date of fasta file: May 28, 2020). The search parameters set in PEAKS Studio were: no enzyme specificity, pyroglutamic acid from N-terminal Gln or Glu and methionine oxidation as variable modifications, mass tolerances of 10 ppm for precursor ions and 0.025 Da for fragments ions and FDR of 1% at the peptide level. De novo (ALC ≥ 50%), post-translational modifications (PEAKS PTM) and homology (SPIDER module) searches were also performed in PEAKS Studio. The same database and variable modifications were set on MASCOT engine. Peptide and fragment mass tolerances were set to 0.1 Da and ion identifications were considered for expectation values lower than 0.05 (p < 0.05). The expectation cut-off value of 0.05 was applied in the MASCOT ion score to avoid peptide identifications out of the 95% confidence interval to be selected.
The MS/MS spectra of digested samples were submitted to database search in PEAKS Studio using the same databases and mass tolerances. Enzyme specificity was defined for each sample and up to one non-specific and three missed cleavages were allowed per peptide. Carbamidomethylation of Cys was set as fixed modification and Met oxidation, N-terminus acetylation and Asn/Gln deamidation were set as variable modifications.
Native peptidome analysis
Identified peptides from digested samples on PEAKS Studio were manually reviewed and N- and C-terminii from native peptides were determined by consensus of non-specific cleavages and overlapping peptides. To validate the native sequences of heavy peptides (> 5 kDa), identified by overlapping cleaved peptides in PEAKS Studio, the experimental mass of each ion was compared to its theoretical mass calculated in ProteinProspector v 5.22.1 (http://prospector.ucsf.edu/prospector/mshome.htm). For peptides < 8 kDa, the monoisotopic masses were used in the comparisons. The sequences were validated if the relative mass difference in ppm was equal or less than 30 and a minimum of 4 fragments matching b+ or y+ ion series were found [3636. Morano C, Zhang X, Fricker LD. Multiple isotopic labels for quantitative mass spectrometry. Anal Chem. 2008 Dec 1;80(23):9298-309. doi: 10.1021/ac801654h.
https://doi.org/10.1021/ac801654h...
,3737. Tashima AK, Fricker LD. Quantitative peptidomics with five-plex reductive methylation labels. J Am Soc Mass Spectrom. 2018;29(5):866-78. doi: 10.1007/s13361-017-1852-3.
https://doi.org/10.1007/s13361-017-1852-...
]. For peptides ( 8 kDa, the average masses were used in the comparisons and the sequences were validated if the relative mass difference in ppm was equal or less than 200 ppm.
Peptide alignment
Primary structures of selected peptides were analyzed by homology searches using protein BLAST (https://www.uniprot.org/blast/) and aligned in TCoffee [3838. Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000 Sep 8;302(1):205-17. doi: 10.1006/jmbi.2000.4042.
https://doi.org/10.1006/jmbi.2000.4042...
] or in PEAKS Studio 7.5.
Peptide folding and visualization
The three-dimensional structure of peptide sequences were predicted by PEP-FOLD3 [3939. Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016 Jul 8;44(W1):W449-54. doi: 10.1093/nar/gkw329.
https://doi.org/10.1093/nar/gkw329...
] using default parameters. Structure visualization and comparison with other proteins were performed in PyMOL Molecular Graphics System, Version 2.3.4 (Schrödinger, LLC).
Results
Identification of B. atrox venom peptides
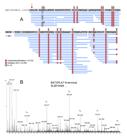
The biometric data of the specimens used for venom extraction are shown in Table 1. Native and digested B. atrox venom peptidome samples were analyzed by LC-MS/MS in the data-independent acquisition mode. The native samples of female and male groups were analyzed individually, totalizing 16 runs (4 individual samples for each group and analyzed in technical duplicates). Processing of the raw data in Progenesis QI for Proteomics resulted in 4112 features detected, that after application of the inclusion criteria were reduced to 878 precursor ions (Additional file 1 Additional file 1. Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens. ). Automated de novo analysis, followed by database search resulted in 375 peptide-spectrum matches (PSM) from 88 unique peptides and 31 precursor proteins (Table 2 and Additional file 2 Additional file 2. Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins. ). Three additional peptides were sequenced by de novo analysis, summing 91 (Table 2). Most of the peptides were from the SVMP family, 46 from SVMPI, 24 from SVMPII and 7 from SVMPIII. The other 14 were BPPs (Table 2). The metalloprotease BATXSVMPI1 contributed with the majority of the SVMPI peptides, covering 39 of the identified peptides, followed by the BATXSVMPI3, BATXSVMPI4 and BATXSVMPI5 with 37 peptides (Additional file 2 Additional file 2. Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins. ). Several of the peptides are shared among these homologous toxins (Figure 1 and Additional file 2 Additional file 2. Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins. ). The same is true for the SVMPII and SVMPIII peptides (Additional file 2 Additional file 2. Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins. ).
Biometric data of the B. atrox specimens used for venom extraction. Size 1 is the length from the head to the cloaca and Size 2 is the total length of the animal.
Alignment of the SVMPI peptides identified in the venoms of B. atrox with the precursor proteins BATXSVMPI1, BATXSVMPI4, BATXSVMPI5 and BATXSVMPI3. The peptides are shared among several of the homologous proteins. Letters in bold blue represent the amino acids covered by MS/MS spectra. Capital letters represent complete identity among the aligned sequences. Alignment performed in PEAKS Studio 7.5.
Additional searches with the Bothrops and Serpentes databases resulted in 119 and 63 unique peptides, respectively (Venn diagram [4040. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. JVENN: an interactive Venn diagram viewer. BMC Bioinformatics. 2014 Aug 29;15(1):293. doi: 10.1186/1471-2105-15-293.
https://doi.org/10.1186/1471-2105-15-293...
] in Additional file 3
Additional file 3.
Venn diagram of unique peptides identified by database search of the B. atrox peptidome LC-MS/MS data. Green: B. atrox database; blue: Bothrops database; pink: Serpentes database. Venn diagram plot in Jvenn.
). The Bothrops database resulted in more peptide identifications. However, opposed to the expected, the Serpentes database (that contains all Bothrops sequences) resulted in less identifications. The decrease can be explained by the exponential expansion of the search space for peptidomics searches [4141. Borges D, Perez-Riverol Y, Nogueira FC, Domont GB, Noda J, da Veiga Leprevost F, et al. Effectively addressing complex proteomic search spaces with peptide spectrum matching. Bioinformatics. 2013 May 15;29(10):1343-4. doi: 10.1093/bioinformatics/btt106.
https://doi.org/10.1093/bioinformatics/b...
], as the sensitivity of a peptide-spectrum match search tool varies inversely with the size of the sequence database [4242. Yen CY, Russell S, Mendoza AM, Meyer-Arendt K, Sun S, Cios KJ, et al. Improving sensitivity in shotgun proteomics using a peptide-centric database with reduced complexity: protease cleavage and SCX elution rules from data mining of MS/MS spectra. Anal Chem. 2006 Feb 15;78(4):1071-84. doi: 10.1021/ac051127f.
https://doi.org/10.1021/ac051127f...
]. As a result, the number of identifications decreases. Furthermore, when we applied the inclusion criteria of ion intensity and presence in biological replicates, the list of relevant peptides of the B. atrox and Bothrops databases did not differ.
The SVMP peptides found in the venoms of B. atrox are homologous to venom peptides previously identified in the venom of B. jararaca [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
]. For instance, the peptide EVWSKKDLIKVEKDSSKTLTSFGEWR (Pep #182, Table 2 and Additional file 1
Additional file 1.
Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens.
) and its fragments from BATXSVMPII1, BATXSVMPII2 and BATXSVMPII3 (Table 2, Additional file 2
Additional file 2.
Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins.
) are identical to the corresponding region of the SVMPII insularinase-A [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,4343. Modesto JC, Junqueira-de-Azevedo IL, Neves-Ferreira AG, Fritzen M, Oliva ML, Ho PL, et al. Insularinase A, a prothrombin activator from Bothrops insularis venom, is a metalloprotease derived from a gene encoding protease and disintegrin domains. Biol Chem. 2005 Jun;386(6):589-600. doi: 10.1515/BC.2005.069.
https://doi.org/10.1515/BC.2005.069...
], leucurolysin-A [4444. Ferreira RN, Rates B, Richardson M, Guimarães BG, Sanchez EO, Pimenta AM, et al. Complete amino-acid sequence, crystallization and preliminary X-ray diffraction studies of leucurolysin-a, a nonhaemorrhagic metalloproteinase from Bothrops leucurus snake venom. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Aug 1;65(Pt 8):798-801. doi: 10.1107/S1744309109025767.
https://doi.org/10.1107/S174430910902576...
], neuwiedase [4545. Rodrigues VM, Soares AM, Guerra-Sá R, Rodrigues V, Fontes MRM, Giglio JR. Structural and functional characterization of neuwiedase, a nonhemorrhagic fibrin(ogen)olytic metalloprotease from Bothrops neuwiedi snake venom. Arch Biochem Biophys. 2000 Sep 15;381(2):213-24. doi: 10.1006/abbi.2000.1958.
https://doi.org/10.1006/abbi.2000.1958...
], and homologous to several other SVMPs. The peptide EVVYP is the most conserved sequence found, shared among 19 SVMPs of the three classes (Additional file 2
Additional file 2.
Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins.
). The sequence SFGEWR from the metalloprotease domain is present in 32 of the SVMP peptides (Table 2), suggesting that this region may be exposed to frequent proteolytic processing. The peptides #182 and #32 (ZTLDSFGEWRKTDLLNRKSHDNAQ, Table 2 and Additional file 1) cover a significant homologous region of the SVMPI leucurolysin-A sequence, comprising the amino acids 57 to 96. These peptides were aligned and highlighted in yellow in leucurolysin-A crystallographic structure (4Q1L [4444. Ferreira RN, Rates B, Richardson M, Guimarães BG, Sanchez EO, Pimenta AM, et al. Complete amino-acid sequence, crystallization and preliminary X-ray diffraction studies of leucurolysin-a, a nonhaemorrhagic metalloproteinase from Bothrops leucurus snake venom. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Aug 1;65(Pt 8):798-801. doi: 10.1107/S1744309109025767.
https://doi.org/10.1107/S174430910902576...
]), as shown in Figure 2A, covering a random coil and an α-helix of the protein not constrained by disulfide bonds. Interestingly, the native 15-aa peptide AHELGHNLGMRHDGN covers the three histidines of the SVMP BATXSVMPII1 catalytic site. This peptide, named Ba1a (Table 2), was also aligned in the 3D structure of leucurolysin-A (in cyan, Figure 2A). The Ba1a sequence contains the consensus motif HEXXHXXGXXH, characteristic of the “metzincin” superfamily of Zn-dependent metalloproteases [4646. Bode W, Gomis-Rüth FX, Stöckler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993 Sep 27;331(1-2):134-40. doi: 10.1016/0014-5793(93)80312-i.
https://doi.org/10.1016/0014-5793(93)803...
] with three histidines residues (in red) involved in the catalytic Zn-binding region. The Ba1a fold detached from leucurolysin-A structure was simulated and the conformations of the histidines in the peptide differed from the positions in leucurolysin-A (Figure 2B). Although the transcripts of B. atrox precursor proteins have been previously reported [4747. Freitas-de-Sousa LA, Amazonas DR, Sousa LF, Sant’Anna SS, Nishiyama MY Jr, Serrano SM, et al. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie. 2015 Nov;118:60-70. doi: 10.1016/j.biochi.2015.08.006.
https://doi.org/10.1016/j.biochi.2015.08...
], to our knowledge these native SVMP peptides had not been previously found at the protein level in the venoms.
(A) Ribbon diagram of the SVMPI leucurolysin-A 3D structure (4Q1L [44]). The region in yellow (1) corresponds to the native homologous peptides Pep #182 and Pep #32 (Additional file 1) identified in Bothrops atrox venoms. The sequence highlighted in cyan (2) corresponds to the homologous peptide Ba1a, which aligns to the catalytic site containing the three Zn-binding histidines (red). (B) Ba1a fold predicted by PEP-FOLD3 (in green) compared to the original crystallographic structure of leucurolysin-A (in red).
In regard to bradykinin potentiating peptides, B. atrox venoms contain the well-known peptide BPP-5a (ZKWAP, in which Z stands for the N-terminal pyroglutamic acid), that provided the basis for the development of important antihypertensive drugs [1616. McCleary RJ, Kini RM. Non-enzymatic proteins from snake venoms: a gold mine of pharmacological tools and drug leads. Toxicon. 2013 Feb;62:56-74. doi: 10.1016/j.toxicon.2012.09.008.
https://doi.org/10.1016/j.toxicon.2012.0...
,1818. Ferreira SH, Greene LH, Alabaster VA, Bakhle YS, Vane JR. Activity of various fractions of bradykinin potentiating factor against angiotensin I converting enzyme. Nature. 1970 Jan 1;225:379-80. doi: 10.1038/225379a0.
https://doi.org/10.1038/225379a0...
,4848. Ferreira SH, Bartelt DC, Greene LJ. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry. 1970 Jun 23;9(13):2583-93. doi: 10.1021/bi00815a005.
https://doi.org/10.1021/bi00815a005...
], and its fragment ZKW (Table 2). The peptide ZSWPGPNIPP (BPP-10a) was previously reported in the venom of B. jararaca [1919. Ondetti MA, Williams NJ, Sabo E, Pluscec J, Weaver ER, Kocy O. Angiotensin-converting enzyme inhibitors from the venom of Bothrops jararaca. Isolation, elucidation of structure, and synthesis. Biochemistry. 1971 Oct 26;10(22):4033-9. doi: 10.1021/bi00798a004.
https://doi.org/10.1021/bi00798a004...
] and the ZKWPRPGPEIPP and its fragment ZKWP, in the venoms of B. atrox [2828. Coutinho-Neto A, Caldeira CA, Souza GH, Zaqueo KD, Kayano AM, Silva RS, et al. ESI-MS/MS identification of a bradykinin-potentiating peptide from Amazon Bothrops atrox snake venom using a hybrid Qq-oaTOF mass spectrometer. Toxins (Basel). 2013 Feb 18;5(2):327-35. doi: 10.3390/toxins5020327.
https://doi.org/10.3390/toxins5020327...
] and B. moojeni [2727. Menin L, Perchuć A, Favreau P, Perret F, Michalet S, Schöni R, et al. High throughput screening of bradykinin-potentiating peptides in Bothrops moojeni snake venom using precursor ion mass spectrometry. Toxicon. 2008 Jun 1;51(7):1288-302. doi: 10.1016/j.toxicon.2008.02.019.
https://doi.org/10.1016/j.toxicon.2008.0...
]. But we also observed new isoforms of these peptides processed in non-canonical sites, as the sequences ZSWPGPNIP, ZKWPRPGPEIP, ZKWPRPGPEIPPLT and ZQWAQKWPRPGPEIPP. Similar processing was also observed in the venoms of B. jararaca [1313. Zelanis A, Tashima AK, Rocha MM, Furtado MF, Camargo AC, Ho PL, et al. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J Proteome Res. 2010 May 7;9(5):2278-91. doi: 10.1021/pr901027r.
https://doi.org/10.1021/pr901027r...
,2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,3030. Nicolau CA, Carvalho PC, Junqueira-de-Azevedo IL, Teixeira-Ferreira A, Junqueira M, Perales J, et al. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca. J Proteomics. 2017 Jan 16;151:214-31. doi: 10.1016/j.jprot.2016.06.029.
https://doi.org/10.1016/j.jprot.2016.06....
,4949. Ianzer D, Konno K, Marques-Porto R, Vieira Portaro FC, Stöcklin R, Martins de Camargo AC, et al. Identification of five new bradykinin potentiating peptides (BPPs) from Bothrops jararaca crude venom by using electrospray ionization tandem mass spectrometry after a two-step liquid chromatography. Peptides. 2004 Jul;25(7):1085-92. doi: 10.1016/j.peptides.2004.04.006.
https://doi.org/10.1016/j.peptides.2004....
] and B. moojeni [2727. Menin L, Perchuć A, Favreau P, Perret F, Michalet S, Schöni R, et al. High throughput screening of bradykinin-potentiating peptides in Bothrops moojeni snake venom using precursor ion mass spectrometry. Toxicon. 2008 Jun 1;51(7):1288-302. doi: 10.1016/j.toxicon.2008.02.019.
https://doi.org/10.1016/j.toxicon.2008.0...
]. The sequences ZKWPSPKVP and ZKWPSPKVPP are novel BPPs. The ZKWPSPKVPP differs only in the second amino acid from the B. cotiara’s ZNWPSPKVPP (BPP-10e) and B. fonsecai’s ZRWPSPKVPP (BPP-10f) [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
]. The 14 BPPs mapped to 7 protein precursors (Figure 3 and Additional file 2
Additional file 2.
Significant peptides identified in the venoms of Bothrops atrox and the originating proteins. Homologous peptides are shared by several proteins.
).
Alignment of the BPP sequences identified in the venoms of B. atrox with the precursor proteins. The peptides are shared among seven homologous proteins. Bold blue letters represent the amino acids covered by MS/MS spectra. Capital letters represent complete identity among the aligned sequences. Alignment performed in PEAKS Studio 7.5.
The pooled crude venoms of the two groups (females and males) were split in three separated aliquots and digested with trypsin, Asp-N and Glu-C. The analysis of all digested samples resulted in the identification of additional 1,152 unique cleaved peptides (Additional file 4
Additional file 4.
Peptides identified in the crude venoms of Bothrops atrox digested with the enzymes Asp-N, Clu-C and trypsin. Database search performed in PEAKS Studio 7.5 against B. atrox venom proteins. Peptide scores are presented in the columns.
). The multiple enzyme approach provides a deeper venom proteome coverage. In addition, protein N- and C-terminii consensus can be found by the overlapping of peptides cleaved in different sites and with unexpected amino acids for the enzyme at one of the peptide ends. However, the proteomic analysis was not in the scope of this work and only peptides that assisted in the identification of heavier native peptides were considered. The experimental data of native peptide ions matched the primary structures of 12 proteoforms of the disintegrin derived from BATXPII1 (Figure 4 and Additional file 5
Additional file 5.
Isotopic patterns of six of the most intense B. atrox disintegrins with multiple charges.
). These disintegrins contain the RGD motif and differs from the batroxostatin sequence [2121. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
https://doi.org/10.1016/0167-4889(90)900...
] only in the C-terminal amino acids (FH or FHA instead of FY in the batroxostatin, Figure 4). The disintegrin cotiarin [5050. Scarborough RM, Rose JW, Naughton MA, Phillips DR, Nannizzi L, Arfsten A, et al. Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J Biol Chem. 1993 Jan 15;268(2):1058-65. ] only lacks the C-terminal Ala in comparison to the new BATXDIS1. Consensus analysis of the N- and C-terminal by verification of enzyme cleavages, overlapping peptides, formation of disulfide bonds and comparison of the theoretical monoisotopic masses with the experimental values were used to confirm the identity of these native heavier peptides (Additional file 1
Additional file 1.
Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens.
and Figure 4). All disintegrins are medium-sized and form 6 disulfide bonds [5151. Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, et al. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005 Jun 15;45(8):1063-74. doi: 10.1016/j.toxicon.2005.02.024.
https://doi.org/10.1016/j.toxicon.2005.0...
].
Alignment of 12 disintegrins identified in the venoms of B. atrox with BATXPII1 (A0A1L8D600) and batroxostatin (P18618). The sequences were derived from the disintegrin domain of the BATXPII1 precursor. Alignment performed in Tcoffee.
This combination of bottom-up and top-down data analysis also revealed the presence of two PLA2 sequences, a mutated form of the BATXPLA6, (with the K23N mutation, Figure 5), the D49 PLA2 that we denominated BATXPLA7. Top-down fragmentation of multiple charged peaks (Additional file 6 Additional file 6. ESI-MS spectra of multiple charged peaks of B. atrox PLA2 toxins showing average m/z values. (A) BATXPLA7; (B) BATXPLA8. ) and de novo analysis of the MS/MS spectra confirmed the first 8 N-terminal amino acids of the toxin, SLIEFANM (Figure 5). Intact mass analysis shows that it forms 7 disulfide bonds (Additional file 1 Additional file 1. Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens. and Additional file 6 Additional file 6. ESI-MS spectra of multiple charged peaks of B. atrox PLA2 toxins showing average m/z values. (A) BATXPLA7; (B) BATXPLA8. ). The presence of BATXPLA6 was not confirmed in the native peptidome data. We identified the peptide GSLIEFANMILEETKK, showing an additional glycine to the BATXPLA7 N-terminus (Additional file 7 Additional file 7. ESI-MS/MS spectrum of the B. atrox PLA2 peptide SLIEFANMILEETKK. The N-terminal GS may also be the carbamidomethylated S, as both have the same mass. Peptide ion observed at m/z 608.33+3. ). However, the peptide ion corresponding to the native extended sequence was not found. As this BATXPLA7 N-terminal peptide was identified by the bottom-up approach, another possibility is that the first Ser was just carbamidomethylated during sample preparation, which results in the same mass difference of a glycine extension (+57.02 Da). The other new PLA2, BATXPLA8, also had its first 7 N-terminal amino acids determined by top-down fragmentation of the multiple charged precursor ions (Additional file 6 Additional file 6. ESI-MS spectra of multiple charged peaks of B. atrox PLA2 toxins showing average m/z values. (A) BATXPLA7; (B) BATXPLA8. ) and de novo analysis (Figure 6). Automated de novo analysis of the digested peptide ion at m/z 589.0+3 revealed the first 15 N-terminal amino acids of BATXPLA8: HLVQFEKLLQLLAGR (Figure 6).
(A) Sequence coverage of the BATXPLA7 showing the consensus of the N-terminal amino acid (S17, indicated by the red arrow) by overlapping of peptides cleaved by different enzymes. The mature sequence presents 123 residues, the K23N mutation and forms seven disulfide bonds. (B) Top-down fragmented MS/MS spectrum of BATXPLA7 and the first eight N-terminal residues determined by de novo analysis.
(A) Top-down fragmented MS/MS spectrum of BATXPLA8 and the first seven N-terminal residues determined by de novo analysis. (B) Bottom-up MS/MS spectrum and de novo analysis of the digested peptide HLVQFEKLLQLLAGR, revealing the 15 N-terminal amino acids of BATXPL8.
In total, 105 native peptides were identified in the venom peptidome of B. atrox, in the mass range of 0.4 to 13.9 kDa (Table 2). Thirteen of the heavy sequences (> 7 kDa) were only confirmed after analysis of bottom-up or top-down MS/MS spectra of digested peptides or native peptides, respectively. Although many other proteins were identified in the digested samples of crude venoms, we only considered the peptides that assisted in the assembly of the native peptide structures. It is worth to mention that the B. atrox database did not contain glutaminyl-peptide cyclotransferases (GPC), that catalyzes N-terminal pyroglutamate formation. However, by searching the broader Bothrops database, we identified the homologous B. jararaca’s GPC (Q9YIB5) with 12 peptides and significant score (-10log(p) = 224).
Bothrops atrox quantitative peptidomics: females vs. males
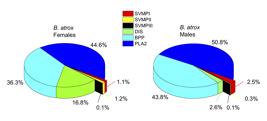
Quantitative analysis of B. atrox venom peptides showed that they belong to the following protein families, in decreasing order of abundance: PLA2, BPP, DIS and SVMP (Figure 7). Comparison of the profiles in female and male specimens showed a strong difference in the levels of disintegrins, with females presenting 16.8% of these peptides versus 2.6% in males (p < 0.05, Additional file 1 Additional file 1. Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens. ). A significant difference was also observed in the levels of SVMPII peptides, 1.2% of peptides in females versus 0.3% in males (p < 0.05, Additional file 1 Additional file 1. Peptide ions obtained by LC-MS/MS runs of Bothrops atrox venom peptidomic samples. A total of 16 runs were performed on the venoms of males and female specimens. ). Although all other peptide families presented statistically equivalent levels (Figure 7), on average, the lack of disintegrins in male venoms are occupied by the BPPs, showing 43.8% of the peptides, against 36.3% in females. Individually, from the 105 identified peptides, 26 peptides are differentially expressed at significant levels and 25 of these 26 are increased in females (Figure 8).
Percentage distribution of peptides by precursor protein families in the venoms of female and male specimens of B. atrox. Quantification based on native peptide ion intensities.
Volcano plot of the peptide ions quantified in the venom of B. atrox. Fold changes calculated as the average intensity ratios of female/male and expressed in the log2 basis. Abundances in log10 scale were proportional to the circle sizes. Filled red circles represent identified peptides and open black circles represent non-identified.
The overall quantitative profile of the disintegrin family is a reflection of the individual peptides. All seven significantly different DIS peptides are highly increased in the venoms of females (Table 2 and Figure 8). For instance, the peptides BATXDIS1:1-71 and BATXDIS1:3-71, on average the 3rd and 5th most intense ions of the peptidome, are 7 and 9 times more intense in the venoms of females, respectively. BATXDIS1:Z1-71, BATXDIS1:4-71 and BATXDIS1:2-71 are also among the most significant differential peptides. Nine of the 18 differential SVMP peptides contain the sequence SFGEWR, all increased in females. The only identified peptide increased in males is the BATXSVMPI1 fragment VEIWSNKDLINVQPAAP. There are other peptide ions increased in males, however these were not identified (Figure 8).
Discussion
Venoms of Bothrops snakes are rich in biologically active peptides that play important roles in the envenomation process. BPPs, for instance, target the cardiovascular system of the prey by inhibiting the angiotensin-converting enzyme (ACE) [5252. Camargo AC, Ianzer D, Guerreiro JR, Serrano SM. Bradykinin-potentiating peptides: beyond captopril. Toxicon. 2012 Mar 15;59(4):516-23. doi: 10.1016/j.toxicon.2011.07.013.
https://doi.org/10.1016/j.toxicon.2011.0...
]. ACE participates of blood pressure regulation by cleaving angiotensin I to angiotensin II [5353. Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002 Apr 26;277(17):14838-43. doi: 10.1074/jbc.M200581200.
https://doi.org/10.1074/jbc.M200581200...
], an hypertensive peptide, and by inactivating the hypotensive peptide bradykinin (Bk) [5454. Moreau ME, Dubreuil P, Molinaro G, Chagnon M, Müller-Esterl W, Lepage Y, et al. Expression of metallopeptidases and kinin receptors in swine oropharyngeal tissues: effects of angiotensin I-converting enzyme inhibition and inflammation. J Pharmacol Exp Ther. 2005 Dec;315(3):1065-74. doi: 10.1124/jpet.105.088005.
https://doi.org/10.1124/jpet.105.088005...
]. The synergistic action of endogenous Bk generation by venom proteases and inhibition of ACE by the BPPs may cause a vascular shock in mammal preys [5555. Hayashi MA, Camargo AC. The Bradykinin-potentiating peptides from venom gland and brain of Bothrops jararaca contain highly site specific inhibitors of the somatic angiotensin-converting enzyme. Toxicon. 2005 Jun 15;45(8):1163-70. doi: 10.1016/j.toxicon.2005.02.017.
https://doi.org/10.1016/j.toxicon.2005.0...
]. Although a higher percentage of BPPs was observed in males of B. atrox (44% vs. 36%), the difference was not statistically significant (Figure 7). BPPs seem to be equally important to both genders of B. atrox, presenting high percentages of the peptidomes. One characteristic of the BPPs is the pyroglutamate at the N-terminal [1717. Tashima AK, Zelanis A. Snake venom peptidomics. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Dordrecht: Springer Netherlands; 2016. p. 317-31. doi: 10.1007/978-94-007-6416-3_49.
https://doi.org/10.1007/978-94-007-6416-...
,3131. Serrano SMT, Zelanis A, Kitano ES, Tashima AK. Analysis of the snake venom peptidome. Methods Mol Biol. 2018;1719:349-58. doi: 10.1007/978-1-4939-7537-2_23.
https://doi.org/10.1007/978-1-4939-7537-...
], whose formation is catalyzed by GPC. The identification of the GPC (Q9YIB5) in the venom of B. atrox explains the high number of BPPs identified.
Snake venom disintegrins containing the RGD motif are potent inhibitors of aggregation responses due to the binding to platelet αIIbβ3 integrins [2121. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
https://doi.org/10.1016/0167-4889(90)900...
,5656. Kamiguti AS, Zuzel M, Theakston RD. Snake venom metalloproteinases and disintegrins: interactions with cells. Braz J Med Biol Res. 1998 Jul;31(7):853-62. doi: 10.1590/s0100-879x1998000700001.
https://doi.org/10.1590/s0100-879x199800...
]. In the Bothrops genus, these toxins have been reported in the venoms of several species as B. atrox [2121. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
https://doi.org/10.1016/0167-4889(90)900...
], B. cotiara [5050. Scarborough RM, Rose JW, Naughton MA, Phillips DR, Nannizzi L, Arfsten A, et al. Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J Biol Chem. 1993 Jan 15;268(2):1058-65. ,5757. Tashima AK, Sanz L, Camargo AC, Serrano SM, Calvete JJ. Snake venomics of the Brazilian pitvipers Bothrops cotiara and Bothrops fonsecai. Identification of taxonomy markers. J Proteomics. 2008 Oct 7;71(4):473-85. doi: 10.1016/j.jprot.2008.07.007.
https://doi.org/10.1016/j.jprot.2008.07....
], B. jararaca, B. jararacussu [5050. Scarborough RM, Rose JW, Naughton MA, Phillips DR, Nannizzi L, Arfsten A, et al. Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J Biol Chem. 1993 Jan 15;268(2):1058-65. ], B. asper [5858. Angulo Y, Castro A, Lomonte B, Rucavado A, Fernández J, Calvete JJ, et al. Isolation and characterization of four medium-size disintegrins from the venoms of Central American viperid snakes of the genera Atropoides, Bothrops, Cerrophidion and Crotalus. Biochimie. 2014 Dec;107(Pt B):376-84. doi: 10.1016/j.biochi.2014.10.010.
https://doi.org/10.1016/j.biochi.2014.10...
], B. insularis [4343. Modesto JC, Junqueira-de-Azevedo IL, Neves-Ferreira AG, Fritzen M, Oliva ML, Ho PL, et al. Insularinase A, a prothrombin activator from Bothrops insularis venom, is a metalloprotease derived from a gene encoding protease and disintegrin domains. Biol Chem. 2005 Jun;386(6):589-600. doi: 10.1515/BC.2005.069.
https://doi.org/10.1515/BC.2005.069...
] and B. colombiensis [5959. Sánchez EE, Rodríguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, et al. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009 Mar;83(3):271-9. doi: 10.1007/s00204-008-0358-y.
https://doi.org/10.1007/s00204-008-0358-...
], for instance. These type of disintegrins are proteolytically processed from PII SVMPs and released as stable proteins [6060. Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008 Jun;275(12):3016-30. doi: 10.1111/j.1742-4658.2008.06466.x.
https://doi.org/10.1111/j.1742-4658.2008...
]. We observed significantly higher levels of disintegrins in the B. atrox venoms of females, 6.5 times higher than males (Figure 7). Such higher level of disintegrins in females should reflect on higher inhibition of platelet aggregation on preys, and consequently to higher anticoagulant activity. However, minimum coagulant dose assays with crude venoms of B. atrox showed just the opposite effect, as the venoms of females presented high coagulant activity in citrated human plasma [1414. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.]. To interpret the result, it is important to consider that proteins as SVSP, SVMP, CTL, PLA2 and other toxins play roles in the coagulation process. Some are procoagulant and others are anticoagulant [6161. Alvarez-Flores MP, Faria F, Andrade SA, Chudzinski-Tavassi AM. Snake venom components affecting the coagulation system. In: Gopalakrishnakone P, Inagaki H, Vogel CW, Mukherjee AK, Rahmy, TR, editors. Snake venoms. Dordrecht: Springer Netherlands; 2016. p. 1-20. doi: 10.1007/978-94-007-6648-8_31-1.
https://doi.org/10.1007/978-94-007-6648-...
]. Thus, synergistic effect [1414. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.,6262. Xiong S, Huang C. Synergistic strategies of predominant toxins in snake venoms. Toxicol Lett. 2018 May 1;287:142-154. doi: 10.1016/j.toxlet.2018.02.004.
https://doi.org/10.1016/j.toxlet.2018.02...
] and a balance of actions produce the final venom activity. Possibly, specific platelet aggregation assays could be used to evaluate the activities of disintegrins of the different genders.
Several other SVMP peptide fragments are observed in B. atrox venoms. They represent smaller percentages of the peptidomes, but may play relevant biological roles. We identified the 15-aa peptide Ba1a containing the three histidines of the SVMP BATXSVMPII1 catalytic site. Computational fold simulation indicates slight positional shifts of the first two Ba1a histidines from His-142 and His-146 of the template SVMP leucurolysin-A (Figure 2B). However, the third His of Ba1a turns considerably in comparison to the corresponding His-152 of leucurolysin-A. This latter shift may affect the Zn affinity of the peptide and modify its biological action in comparison to the original protein. The biological activity of Ba1a can be explored in future experimental studies. Anyway, it is interesting to observe the SVMP catalytic site in this native peptide. The SVMP peptides may have been originated from proteolytic processing of proteases inside the venom glands, as the SVMPs and SVSPs [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,3131. Serrano SMT, Zelanis A, Kitano ES, Tashima AK. Analysis of the snake venom peptidome. Methods Mol Biol. 2018;1719:349-58. doi: 10.1007/978-1-4939-7537-2_23.
https://doi.org/10.1007/978-1-4939-7537-...
]. The hypothesis of peptides being produced endogenously is corroborated by the identification of the GPC (Q9YIB5) in the crude venom and the relatively high frequency of pyroglutamic acid in the N-terminal of the SVMP peptides (Table 2). Unexpectedly, L-amino acid oxidase peptides were not found in the venoms of B. atrox as opposed to other studies of Bothrops snake venom peptidomes [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
,3030. Nicolau CA, Carvalho PC, Junqueira-de-Azevedo IL, Teixeira-Ferreira A, Junqueira M, Perales J, et al. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca. J Proteomics. 2017 Jan 16;151:214-31. doi: 10.1016/j.jprot.2016.06.029.
https://doi.org/10.1016/j.jprot.2016.06....
,6363. Souza GH, Catharino RR, Ifa DR, Eberlin MN, Hyslop S. Peptide fingerprinting of snake venoms by direct infusion nano-electrospray ionization mass spectrometry: potential use in venom identification and taxonomy. J Mass Spectrom. 2008 May;43(5):594-9. doi: 10.1002/jms.1351.
https://doi.org/10.1002/jms.1351...
]. The use of protease inhibitors immediately after venom extraction may have prevented the generation of artefactual peptides, as previously reported [2929. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
https://doi.org/10.1074/mcp.M112.019331...
].
Although this was a peptidomic study, we also observed 13.8 kDa PLA2 toxins in our analyses. There is not an official definition of a peptide size, although most studies use the 10 kDa as an approximate cut-off value [6464. Schrader M. Origins, Technological Development, and Applications of Peptidomics. Methods Mol Biol. 2018;1719:3-39. doi: 10.1007/978-1-4939-7537-2_1.
https://doi.org/10.1007/978-1-4939-7537-...
]. Nevertheless, the B. atrox PLA2 toxins were sufficiently hydrophilic to be extracted in our peptide enrichment methods. PLA2 represented the most abundant peptide family of both genders with 45% and 51% of the total venom peptides in females and males, respectively. The higher percentage of PLA2 in males was also observed in the proteomic study and in the in vitro activity by colorimetric assay [1414. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.]. It is important to mention that the peptidomic quantification was based in native precursor ion intensities. While the quantitative methods were different, the corroboration of proteomic, peptidomic and biological activity data for PLA2 is noteworthy.
Conclusion
The venom peptidomes of male and female specimens of Bothrops atrox were uncovered by mass spectrometry-based approaches in the present work. New peptides were identified as well as known peptides processed at non-canonical sites were observed. The genders present abundant and statistically equivalent levels of BPPs and PLA2, but female venoms are significantly richer in disintegrins. This difference may result in biological implications on platelet function in preys; however, current experimental data do not point to differences in coagulation. Specific assays should be performed in future works to elucidate possible differences on platelet aggregation by male and female venoms. It was also shown that SVMP peptides are probably processed endogenously due to the presence of pyroglutamic fragments and of GPC in the venom. In summary, the differences in the venom peptidomes may reflect on distinct ecological needs of males and females and may have their potential pharmacological properties explored in future works.
Acknowledgments
The authors would like to thank Prof. Dr. Reinaldo Salomão from UNIFESP for the use of Progenesis QI for proteomics (FAPESP grant n. 2017/21052-0).
References
- 1. Calvete JJ. Venomics: integrative venom proteomics and beyond. Biochem J. 2017 Feb 20;474(5):611-34. doi: 10.1042/BCJ20160577.
» https://doi.org/10.1042/BCJ20160577 - 2. Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013 Apr;28(4):219-29. doi:10.1016/j.tree.2012.10.020.
» https://doi.org/10.1016/j.tree.2012.10.020 - 3. Núñez V, Cid P, Sanz L, de la Torre P, Angulo Y, Lomonte B, et al. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics. 2009 Aug 5;73(1):57-78. doi: 10.1016/j.jprot.2009.07.013.
» https://doi.org/10.1016/j.jprot.2009.07.013 - 4. Calvete JJ, Sanz L, Pérez A, Borges A, Vargas AM, Lomonte B, et al. Snake population venomics and antivenomics of Bothrops atrox: paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J Proteomics. 2011 Apr 1;74(4):510-27. doi: 10.1016/j.jprot.2011.01.003.
» https://doi.org/10.1016/j.jprot.2011.01.003 - 5. Santos Barreto GN, Oliveira SS, Anjos IV, Chalkidis HM, Mourão RH, Moura-da-Silva AM, et al. Experimental Bothrops atrox envenomation: efficacy of antivenom therapy and the combination of Bothrops antivenom with dexamethasone. PLoS Negl Trop Dis. 2017 Mar 17;11(3):e0005458. doi: 10.1371/journal.pntd.0005458.
» https://doi.org/10.1371/journal.pntd.0005458 - 6. Oliveira SS, Sampaio VS, Sachett JA, Alves EC, Silva VC, Lima JA, et al. Snakebites in the Brazilian Amazon: current knowledge and perspectives. In: Gopalakrishnakone P, editor. Clinical toxinology in Australia, Europe, and Americas. Dordrecht: Springer; 2018. p. 73-99. doi: 10.1007/978-94-017-7438-3_61.
» https://doi.org/10.1007/978-94-017-7438-3_61 - 7. Sousa LF, Portes JA Jr, Nicolau CA, Bernardoni JL, Nishiyama MY Jr, Amazonas DR, et al. Functional proteomic analyses of Bothrops atrox venom reveals phenotypes associated with habitat variation in the Amazon. J Proteomics. 2017 Apr 21;159:32-46. doi: 10.1016/j.jprot.2017.03.003.
» https://doi.org/10.1016/j.jprot.2017.03.003 - 8. Zelanis A, Menezes MC, Kitano ES, Liberato T, Tashima AK, Pinto AF, et al. Proteomic identification of gender molecular markers in Bothrops jararaca venom. J Proteomics. 2016 Apr 29;139:26-37. doi: 10.1016/j.jprot.2016.02.030.
» https://doi.org/10.1016/j.jprot.2016.02.030 - 9. Furtado MF, Travaglia-Cardoso SR, Rocha MM. Sexual dimorphism in venom of Bothrops jararaca (Serpentes: Viperidae). Toxicon. 2006 Sep 15;48(4):401-10. doi: 10.1016/j.toxicon.2006.06.005.
» https://doi.org/10.1016/j.toxicon.2006.06.005 - 10. Amorim FG, Costa TR, Baiwir D, Pauw E, Quinton L, Sampaio SV. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: influence of snake gender on venom composition. Toxins (Basel). 2018 Apr 26;10(5):177. doi: 10.3390/toxins10050177.
» https://doi.org/10.3390/toxins10050177 - 11. Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996 Feb 8;379(6565):537-40. doi: 10.1038/379537a0.
» https://doi.org/10.1038/379537a0 - 12. Barlow A, Pook CE, Harrison RA, Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci. 2009 Jul 7;276(1666):2443-9. doi: 10.1098/rspb.2009.0048.
» https://doi.org/10.1098/rspb.2009.0048 - 13. Zelanis A, Tashima AK, Rocha MM, Furtado MF, Camargo AC, Ho PL, et al. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J Proteome Res. 2010 May 7;9(5):2278-91. doi: 10.1021/pr901027r.
» https://doi.org/10.1021/pr901027r - 14. Hatakeyama DM, Tasima LJ, Bravo-Tobar CA, Serino-Silva C, Tashima AK, Rodrigues CF, et al. Venom complexity from Bothrops atrox (common lancehead) snake in siblings. J Venom Anim Toxins Incl Trop Dis. 2020;26:e20200018. Forthcoming 2020.
- 15. Escoubas P, Quinton L, Nicholson GM. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J Mass Spectrom. 2008 Mar;43(3):279-95. doi: 10.1002/jms.1389.
» https://doi.org/10.1002/jms.1389 - 16. McCleary RJ, Kini RM. Non-enzymatic proteins from snake venoms: a gold mine of pharmacological tools and drug leads. Toxicon. 2013 Feb;62:56-74. doi: 10.1016/j.toxicon.2012.09.008.
» https://doi.org/10.1016/j.toxicon.2012.09.008 - 17. Tashima AK, Zelanis A. Snake venom peptidomics. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Dordrecht: Springer Netherlands; 2016. p. 317-31. doi: 10.1007/978-94-007-6416-3_49.
» https://doi.org/10.1007/978-94-007-6416-3_49 - 18. Ferreira SH, Greene LH, Alabaster VA, Bakhle YS, Vane JR. Activity of various fractions of bradykinin potentiating factor against angiotensin I converting enzyme. Nature. 1970 Jan 1;225:379-80. doi: 10.1038/225379a0.
» https://doi.org/10.1038/225379a0 - 19. Ondetti MA, Williams NJ, Sabo E, Pluscec J, Weaver ER, Kocy O. Angiotensin-converting enzyme inhibitors from the venom of Bothrops jararaca Isolation, elucidation of structure, and synthesis. Biochemistry. 1971 Oct 26;10(22):4033-9. doi: 10.1021/bi00798a004.
» https://doi.org/10.1021/bi00798a004 - 20. Kloog Y, Ambar I, Sokolovsky M, Kochva E, Wollberg Z, Bdolah A. Sarafotoxin, a novel vasoconstrictor peptide: phosphoinositide hydrolysis in rat heart and brain. Science. 1988 Oct 14;242(4876):268-70. doi: 10.1126/science.2845579.
» https://doi.org/10.1126/science.2845579 - 21. Rucinski B, Niewiarowski S, Holt JC, Soszka T, Knudsen KA. Batroxostatin, an Arg-Gly-Asp-containing peptide from Bothrops atrox, is a potent inhibitor of platelet aggregation and cell interaction with fibronectin. Biochim Biophys Acta Mol Cell Res. 1990 Sep 24;1054(3):257-62. doi: 10.1016/0167-4889(90)90096-V.
» https://doi.org/10.1016/0167-4889(90)90096-V - 22. Konno K, Picolo G, Gutierrez VP, Brigatte P, Zambelli VO, Camargo AC, et al. Crotalphine, a novel potent analgesic peptide from the venom of the South American rattlesnake Crotalus durissus terrificus Peptides. 2008 Aug;29(8):1293-304. doi: 10.1016/j.peptides.2008.04.003.
» https://doi.org/10.1016/j.peptides.2008.04.003 - 23. Lomonte B, Calvete JJ. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J Venom Anim Toxins Incl Trop Dis. 2017;23(1):26. doi: 10.1186/s40409-017-0117-8.
» https://doi.org/10.1186/s40409-017-0117-8 - 24. Galizio NC, Serino-Silva C, Stuginski DR, Abreu PA, Sant’Anna SS, Grego KF, et al. Compositional and functional investigation of individual and pooled venoms from long-term captive and recently wild-caught Bothrops jararaca snakes. J Proteomics. 2018 Aug 30;186:56-70. doi: 10.1016/j.jprot.2018.07.007.
» https://doi.org/10.1016/j.jprot.2018.07.007 - 25. Morais-Zani K, Serino-Silva C, Galizio NC, Tasima LJ, Pagotto JF, Rocha MM, et al. Does the administration of pilocarpine prior to venom milking influence the composition of Micrurus corallinus venom? J Proteomics. 2018 Mar 1;174:17-27. doi: 10.1016/j.jprot.2017.12.010.
» https://doi.org/10.1016/j.jprot.2017.12.010 - 26. Zelanis A, Keiji Tashima A. Unraveling snake venom complexity with ‘omics’ approaches: challenges and perspectives. Toxicon. 2014 Sep;87:131-4. doi: 10.1016/j.toxicon.2014.05.011.
» https://doi.org/10.1016/j.toxicon.2014.05.011 - 27. Menin L, Perchuć A, Favreau P, Perret F, Michalet S, Schöni R, et al. High throughput screening of bradykinin-potentiating peptides in Bothrops moojeni snake venom using precursor ion mass spectrometry. Toxicon. 2008 Jun 1;51(7):1288-302. doi: 10.1016/j.toxicon.2008.02.019.
» https://doi.org/10.1016/j.toxicon.2008.02.019 - 28. Coutinho-Neto A, Caldeira CA, Souza GH, Zaqueo KD, Kayano AM, Silva RS, et al. ESI-MS/MS identification of a bradykinin-potentiating peptide from Amazon Bothrops atrox snake venom using a hybrid Qq-oaTOF mass spectrometer. Toxins (Basel). 2013 Feb 18;5(2):327-35. doi: 10.3390/toxins5020327.
» https://doi.org/10.3390/toxins5020327 - 29. Tashima AK, Zelanis A, Kitano ES, Ianzer D, Melo RL, Rioli V, et al. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 2012 Nov;11(11):1245-62. doi: 10.1074/mcp.M112.019331.
» https://doi.org/10.1074/mcp.M112.019331 - 30. Nicolau CA, Carvalho PC, Junqueira-de-Azevedo IL, Teixeira-Ferreira A, Junqueira M, Perales J, et al. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca J Proteomics. 2017 Jan 16;151:214-31. doi: 10.1016/j.jprot.2016.06.029.
» https://doi.org/10.1016/j.jprot.2016.06.029 - 31. Serrano SMT, Zelanis A, Kitano ES, Tashima AK. Analysis of the snake venom peptidome. Methods Mol Biol. 2018;1719:349-58. doi: 10.1007/978-1-4939-7537-2_23.
» https://doi.org/10.1007/978-1-4939-7537-2_23 - 32. Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896-906. doi: 10.1038/nprot.2007.261.
» https://doi.org/10.1038/nprot.2007.261 - 33. Abreu TF, Sumitomo BN, Nishiyama MY Jr, Oliveira UC, Souza GH, Kitano ES, et al. Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J Proteomics. 2017 Jan 16;151:232-42. doi: 10.1016/j.jprot.2016.07.012.
» https://doi.org/10.1016/j.jprot.2016.07.012 - 34. Lomazi RL, Nishiduka ES, Silva PI Jr, Tashima AK. Identification of peptides in spider venom using mass spectrometry. Methods Mol Biol. 2018;1719:359-67. doi: 10.1007/978-1-4939-7537-2_24.
» https://doi.org/10.1007/978-1-4939-7537-2_24 - 35. Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014 Feb;11(2):167-70. doi: 10.1038/nmeth.2767.
» https://doi.org/10.1038/nmeth.2767 - 36. Morano C, Zhang X, Fricker LD. Multiple isotopic labels for quantitative mass spectrometry. Anal Chem. 2008 Dec 1;80(23):9298-309. doi: 10.1021/ac801654h.
» https://doi.org/10.1021/ac801654h - 37. Tashima AK, Fricker LD. Quantitative peptidomics with five-plex reductive methylation labels. J Am Soc Mass Spectrom. 2018;29(5):866-78. doi: 10.1007/s13361-017-1852-3.
» https://doi.org/10.1007/s13361-017-1852-3 - 38. Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000 Sep 8;302(1):205-17. doi: 10.1006/jmbi.2000.4042.
» https://doi.org/10.1006/jmbi.2000.4042 - 39. Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016 Jul 8;44(W1):W449-54. doi: 10.1093/nar/gkw329.
» https://doi.org/10.1093/nar/gkw329 - 40. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. JVENN: an interactive Venn diagram viewer. BMC Bioinformatics. 2014 Aug 29;15(1):293. doi: 10.1186/1471-2105-15-293.
» https://doi.org/10.1186/1471-2105-15-293 - 41. Borges D, Perez-Riverol Y, Nogueira FC, Domont GB, Noda J, da Veiga Leprevost F, et al. Effectively addressing complex proteomic search spaces with peptide spectrum matching. Bioinformatics. 2013 May 15;29(10):1343-4. doi: 10.1093/bioinformatics/btt106.
» https://doi.org/10.1093/bioinformatics/btt106 - 42. Yen CY, Russell S, Mendoza AM, Meyer-Arendt K, Sun S, Cios KJ, et al. Improving sensitivity in shotgun proteomics using a peptide-centric database with reduced complexity: protease cleavage and SCX elution rules from data mining of MS/MS spectra. Anal Chem. 2006 Feb 15;78(4):1071-84. doi: 10.1021/ac051127f.
» https://doi.org/10.1021/ac051127f - 43. Modesto JC, Junqueira-de-Azevedo IL, Neves-Ferreira AG, Fritzen M, Oliva ML, Ho PL, et al. Insularinase A, a prothrombin activator from Bothrops insularis venom, is a metalloprotease derived from a gene encoding protease and disintegrin domains. Biol Chem. 2005 Jun;386(6):589-600. doi: 10.1515/BC.2005.069.
» https://doi.org/10.1515/BC.2005.069 - 44. Ferreira RN, Rates B, Richardson M, Guimarães BG, Sanchez EO, Pimenta AM, et al. Complete amino-acid sequence, crystallization and preliminary X-ray diffraction studies of leucurolysin-a, a nonhaemorrhagic metalloproteinase from Bothrops leucurus snake venom. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Aug 1;65(Pt 8):798-801. doi: 10.1107/S1744309109025767.
» https://doi.org/10.1107/S1744309109025767 - 45. Rodrigues VM, Soares AM, Guerra-Sá R, Rodrigues V, Fontes MRM, Giglio JR. Structural and functional characterization of neuwiedase, a nonhemorrhagic fibrin(ogen)olytic metalloprotease from Bothrops neuwiedi snake venom. Arch Biochem Biophys. 2000 Sep 15;381(2):213-24. doi: 10.1006/abbi.2000.1958.
» https://doi.org/10.1006/abbi.2000.1958 - 46. Bode W, Gomis-Rüth FX, Stöckler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993 Sep 27;331(1-2):134-40. doi: 10.1016/0014-5793(93)80312-i.
» https://doi.org/10.1016/0014-5793(93)80312-i - 47. Freitas-de-Sousa LA, Amazonas DR, Sousa LF, Sant’Anna SS, Nishiyama MY Jr, Serrano SM, et al. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie. 2015 Nov;118:60-70. doi: 10.1016/j.biochi.2015.08.006.
» https://doi.org/10.1016/j.biochi.2015.08.006 - 48. Ferreira SH, Bartelt DC, Greene LJ. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry. 1970 Jun 23;9(13):2583-93. doi: 10.1021/bi00815a005.
» https://doi.org/10.1021/bi00815a005 - 49. Ianzer D, Konno K, Marques-Porto R, Vieira Portaro FC, Stöcklin R, Martins de Camargo AC, et al. Identification of five new bradykinin potentiating peptides (BPPs) from Bothrops jararaca crude venom by using electrospray ionization tandem mass spectrometry after a two-step liquid chromatography. Peptides. 2004 Jul;25(7):1085-92. doi: 10.1016/j.peptides.2004.04.006.
» https://doi.org/10.1016/j.peptides.2004.04.006 - 50. Scarborough RM, Rose JW, Naughton MA, Phillips DR, Nannizzi L, Arfsten A, et al. Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J Biol Chem. 1993 Jan 15;268(2):1058-65.
- 51. Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, et al. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005 Jun 15;45(8):1063-74. doi: 10.1016/j.toxicon.2005.02.024.
» https://doi.org/10.1016/j.toxicon.2005.02.024 - 52. Camargo AC, Ianzer D, Guerreiro JR, Serrano SM. Bradykinin-potentiating peptides: beyond captopril. Toxicon. 2012 Mar 15;59(4):516-23. doi: 10.1016/j.toxicon.2011.07.013.
» https://doi.org/10.1016/j.toxicon.2011.07.013 - 53. Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002 Apr 26;277(17):14838-43. doi: 10.1074/jbc.M200581200.
» https://doi.org/10.1074/jbc.M200581200 - 54. Moreau ME, Dubreuil P, Molinaro G, Chagnon M, Müller-Esterl W, Lepage Y, et al. Expression of metallopeptidases and kinin receptors in swine oropharyngeal tissues: effects of angiotensin I-converting enzyme inhibition and inflammation. J Pharmacol Exp Ther. 2005 Dec;315(3):1065-74. doi: 10.1124/jpet.105.088005.
» https://doi.org/10.1124/jpet.105.088005 - 55. Hayashi MA, Camargo AC. The Bradykinin-potentiating peptides from venom gland and brain of Bothrops jararaca contain highly site specific inhibitors of the somatic angiotensin-converting enzyme. Toxicon. 2005 Jun 15;45(8):1163-70. doi: 10.1016/j.toxicon.2005.02.017.
» https://doi.org/10.1016/j.toxicon.2005.02.017 - 56. Kamiguti AS, Zuzel M, Theakston RD. Snake venom metalloproteinases and disintegrins: interactions with cells. Braz J Med Biol Res. 1998 Jul;31(7):853-62. doi: 10.1590/s0100-879x1998000700001.
» https://doi.org/10.1590/s0100-879x1998000700001 - 57. Tashima AK, Sanz L, Camargo AC, Serrano SM, Calvete JJ. Snake venomics of the Brazilian pitvipers Bothrops cotiara and Bothrops fonsecai. Identification of taxonomy markers. J Proteomics. 2008 Oct 7;71(4):473-85. doi: 10.1016/j.jprot.2008.07.007.
» https://doi.org/10.1016/j.jprot.2008.07.007 - 58. Angulo Y, Castro A, Lomonte B, Rucavado A, Fernández J, Calvete JJ, et al. Isolation and characterization of four medium-size disintegrins from the venoms of Central American viperid snakes of the genera Atropoides, Bothrops, Cerrophidion and Crotalus. Biochimie. 2014 Dec;107(Pt B):376-84. doi: 10.1016/j.biochi.2014.10.010.
» https://doi.org/10.1016/j.biochi.2014.10.010 - 59. Sánchez EE, Rodríguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, et al. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009 Mar;83(3):271-9. doi: 10.1007/s00204-008-0358-y.
» https://doi.org/10.1007/s00204-008-0358-y - 60. Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008 Jun;275(12):3016-30. doi: 10.1111/j.1742-4658.2008.06466.x.
» https://doi.org/10.1111/j.1742-4658.2008.06466.x - 61. Alvarez-Flores MP, Faria F, Andrade SA, Chudzinski-Tavassi AM. Snake venom components affecting the coagulation system. In: Gopalakrishnakone P, Inagaki H, Vogel CW, Mukherjee AK, Rahmy, TR, editors. Snake venoms. Dordrecht: Springer Netherlands; 2016. p. 1-20. doi: 10.1007/978-94-007-6648-8_31-1.
» https://doi.org/10.1007/978-94-007-6648-8_31-1 - 62. Xiong S, Huang C. Synergistic strategies of predominant toxins in snake venoms. Toxicol Lett. 2018 May 1;287:142-154. doi: 10.1016/j.toxlet.2018.02.004.
» https://doi.org/10.1016/j.toxlet.2018.02.004 - 63. Souza GH, Catharino RR, Ifa DR, Eberlin MN, Hyslop S. Peptide fingerprinting of snake venoms by direct infusion nano-electrospray ionization mass spectrometry: potential use in venom identification and taxonomy. J Mass Spectrom. 2008 May;43(5):594-9. doi: 10.1002/jms.1351.
» https://doi.org/10.1002/jms.1351 - 64. Schrader M. Origins, Technological Development, and Applications of Peptidomics. Methods Mol Biol. 2018;1719:3-39. doi: 10.1007/978-1-4939-7537-2_1.
» https://doi.org/10.1007/978-1-4939-7537-2_1 - 65. Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019 Jan 8;47(D1):D442-50. doi: 10.1093/nar/gky1106.
» https://doi.org/10.1093/nar/gky1106
-
Abbreviations
ACE: angiotensin-converting enzyme; ACN: acetonitrile; BEH: bridged ethyl hybrid; Bk: hypotensive peptide bradykinin; BPPs: bradykinin potentiating peptides; CID: collision induced dissociation; COBEA: Brazilian College of Animal Experimentation; CTL: C-type lectin; DIS: disintegrin; DTT: dithiothreitol; EDTA: ethylenediamine tetra acetic acid; ESI: electrospray ionization; FDR: false discovery rate; GPC: glutaminyl-peptide cyclotransferase; HDMS: high definition mass spectrometry; LC-MS/MS: liquid chromatography-mass spectrometry/mass spectrometry; PLA2: phospholipase A2; PMSF: phenylmethylsulfonyl fluoride; PSM: peptide-spectrum matches; PTM: post-translational modification; SDB-XC: styrenedivinylbenzene; SVMP: snake venom metalloproteinase; SVSP: snake venom serine proteinase; TFA: trifluoroacetic acid; UDMSE: ultra definition MSE; UPLC: ultra performance liquid chromatography.
-
Availability of data and materials
Mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE [6565. Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019 Jan 8;47(D1):D442-50. doi: 10.1093/nar/gky1106.
https://doi.org/10.1093/nar/gky1106... ] partner repository with the dataset identifier PXD018632. -
Funding
The present work was supported by the São Paulo Research Foundation (FAPESP grant n. 2017/20106-9 to AKT, n. 2018/25786-0 to AMTA and n. 2019/03502-3to AS), the Funding Authority for Studies and Projects (FINEP) and the Coordination for the Improvement of Higher Education Personnel (CAPES), Finance Code 001. -
Ethics approval
Experiments were approved by the Ethics Committee of Butantan Institute (number 6303280220) and the Ethics Committee of Federal University of São Paulo (number 3437250719). Moreover, all tests were performed in accordance with the Brazilian laws for the use of experimental animals and with the ethical principles adopted by the Brazilian College of Animal Experimentation (COBEA). -
Consent for publication
Not applicable.
Supplementary material
The following online material is available for this article:
Publication Dates
-
Publication in this collection
07 Oct 2020 -
Date of issue
2020
History
-
Received
18 Apr 2020 -
Accepted
09 Sept 2020