Abstracts
Chromosomes and nuclear DNA content variation in the eel Synbranchus marmoratus were studied in seven samples taken from rio Paraguay and rio Paraná basins to examine whether karyotype and nuclear DNA content variations could offer useful information for the characterization of independent evolutionary lineages in the genus and construction of evolutionary and biogeographical hypotheses. The occurrence of different karyotypes was already described for this species; however, a new cytotype from rio Miranda was discovered. Nuclear DNA content showed a wide variation among samples and individuals, ranging from 5.2 to 9.1 pg of DNA/nuclei. An analysis of variance confirmed the occurrence of significant differences among samples. In a deeper series of analyses, a multimodality was found in the distribution of nuclear DNA content, in which several, more or less discrete units arose. Finally, combining karyotype formulae with nuclear DNA content, a complex relationship among the rivers sampled was found. Based on the available data, we suggest that several independent evolutionary lineages of Synbranchus marmoratus occur in the rivers sampled. Biogeographical hypotheses are proposed and discussed.
Fish DNA; chromosomes; genome diversity; karyotype; Synbranchus marmoratus
Os cromossomos e a variação de conteúdo de DNA nuclear foram estudados em sete amostras de Synbranchus marmoratus das bacias dos rios Paraguai e Paraná com o objetivo de avaliar se diferenças no cariótipo e no conteúdo de DNA nuclear poderiam fornecer informações para a caracterização de linhagens evolutivas independentes no gênero e para a elaboração de hipóteses evolutivas e biogeográficas. A ocorrência de diferentes cariótipos já foi descrita para essa espécie, entretanto, um novo citótipo foi encontrado no rio Miranda. O conteúdo de DNA nuclear mostrou uma ampla variação entre as amostras e indivíduos, com valores entre 5,2 a 9,1 pg de DNA/núcleo. Uma análise de variância confirmou a ocorrência de diferenças significativas entre as amostras. Em uma série de análises, um padrão multimodal foi encontrado na distribuição do conteúdo de DNA nuclear, pelo qual várias unidades, mais ou menos discretas, foram identificadas. Combinando a fórmula cariotípica com o conteúdo de DNA nuclear, uma relação complexa entre os rios foi observada. Com base nos dados disponíveis, sugerimos que existem várias linhagens evolutivas independentes de Synbranchus marmoratus nos rios amostrados. Hipóteses biogeográficas são propostas e discutidas.
Surprising genomic diversity in the Neotropical fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species?
Rodrigo Augusto TorresI; James Joseph RoperII; Fausto ForestiIII; Claudio OliveiraIII
ILaboratório de Genômica Evolutiva e Ambiental, Departamento de Zoologia, Universidade Federal do Paraná, Centro Politécnico, Setor de Ciências Biológicas, Caixa Postal 19020, 81531-990 Curitiba, PR, Brazil. e-mail: ratorres@ufpr.br
IIDepartamento de Biologia, Universidade Tuiutí do Paraná, Curitiba, PR, Brazil. e-mail: jjroper@uol.com.br
IIILaboratório de Biologia e Genética de Peixes, Departamento de Morfologia, Universidade Estadual Paulista, Botucatu, SP, Brazil
ABSTRACT
Chromosomes and nuclear DNA content variation in the eel Synbranchus marmoratus were studied in seven samples taken from rio Paraguay and rio Paraná basins to examine whether karyotype and nuclear DNA content variations could offer useful information for the characterization of independent evolutionary lineages in the genus and construction of evolutionary and biogeographical hypotheses. The occurrence of different karyotypes was already described for this species; however, a new cytotype from rio Miranda was discovered. Nuclear DNA content showed a wide variation among samples and individuals, ranging from 5.2 to 9.1 pg of DNA/nuclei. An analysis of variance confirmed the occurrence of significant differences among samples. In a deeper series of analyses, a multimodality was found in the distribution of nuclear DNA content, in which several, more or less discrete units arose. Finally, combining karyotype formulae with nuclear DNA content, a complex relationship among the rivers sampled was found. Based on the available data, we suggest that several independent evolutionary lineages of Synbranchus marmoratus occur in the rivers sampled. Biogeographical hypotheses are proposed and discussed.
Key-words: Fish DNA, chromosomes, genome diversity, karyotype, Synbranchus marmoratus
RESUMO
Os cromossomos e a variação de conteúdo de DNA nuclear foram estudados em sete amostras de Synbranchus marmoratus das bacias dos rios Paraguai e Paraná com o objetivo de avaliar se diferenças no cariótipo e no conteúdo de DNA nuclear poderiam fornecer informações para a caracterização de linhagens evolutivas independentes no gênero e para a elaboração de hipóteses evolutivas e biogeográficas. A ocorrência de diferentes cariótipos já foi descrita para essa espécie, entretanto, um novo citótipo foi encontrado no rio Miranda. O conteúdo de DNA nuclear mostrou uma ampla variação entre as amostras e indivíduos, com valores entre 5,2 a 9,1 pg de DNA/núcleo. Uma análise de variância confirmou a ocorrência de diferenças significativas entre as amostras. Em uma série de análises, um padrão multimodal foi encontrado na distribuição do conteúdo de DNA nuclear, pelo qual várias unidades, mais ou menos discretas, foram identificadas. Combinando a fórmula cariotípica com o conteúdo de DNA nuclear, uma relação complexa entre os rios foi observada. Com base nos dados disponíveis, sugerimos que existem várias linhagens evolutivas independentes de Synbranchus marmoratus nos rios amostrados. Hipóteses biogeográficas são propostas e discutidas.
Introduction
South America is the worldwide richest region in freshwater fish species. Around 6,000 species have been estimated, but this number will certainly increase with new studies in poorly studied areas, especially those located in the headwaters of the great river complexes (Reis et al., 2003). Although, in some ways, Synbranchidae is a relatively well-known family, its biogeography and evolution are still poorly understood because of its simple morphology and life-history characteristics that are shared among many species (Favorito-Amorim, 1998). Currently only two genera, Ophisternon and Synbranchus, and four species, two in each genus, are currently recognized in the Neotropical region (Kullander, 2003). Synbranchus marmoratus Bloch, 1795, commonly known as the swamp eel or water snake, is widely distributed from Mexico to northern Argentina while S. madeirae is known only from the rio Madeira basin (Kullander, 2003). The long body, elongated skull, greatly reduced fins and scales (covered by a thick mucous) and a single ventral gill opening provide the reasons for the common names. Able to breathe air (Favorito-Amorim, 1998), resistant to variable salinity (Tyler & Feller, 1996) and capable of sex reversal (Liem, 1968; Lo Nostro & Guerrero, 1996; Favorito-Amorim, 1998), this species is often associated with extremely variable environmental conditions, enabling its wide ecological and geographical distribution (Favorito-Amorim, 1998).
Biochemical and genetic studies have identified a great diversity within the species S. marmoratus. Two electrophoretic patterns of hemoglobins were found, each of which was exclusive to a specific geographic region in the state of São Paulo, Brazil (Nakamoto et al., 1986a, b). Cytogenetic studies in this genus have revealed diploid numbers ranging from 2n=42 to 2n=46 (Table 1). Chromosome number and hemoglobin phenotype are associated. Thus, 2n=44 and 2n=46 are found in specimens with hemoglobin phenotype 1 while 2n=42 is found in specimens with hemoglobin phenotype 2 (Nakamoto et al., 1986a, b).
Variability in nuclear DNA content offers an additional tool for distinguish among local populations and species (Carvalho et al., 1998). The first studies of variability in nuclear DNA content began in the early 1950s, with the suggestion that an increase in the quantity of DNA might be important for the formation of new gene complexes (Hinegardner & Rosen, 1972). Increases and reductions in nuclear DNA content are common. Reductions contributed to speciation associated with the origin of the majority of the living teleosts (Hinegardner & Rosen, 1972). Thus, adaptation to variable environments may be associated with changes in nuclear DNA content (Goin et al., 1980).Understanding these variations can help elucidate the evolutionary and biogeographical histories of species (Chang et al., 1995). Based on the known genetic variability in Synbranchus marmoratus, we examined nuclear DNA content variations and karyotype diversity of seven samples of Synbranchus marmoratus looking for the characterization of independent evolutionary lineages in the genus and construction of evolutionary and biogeographical hypotheses.
Material and Methods
Specimens of Synbranchus marmoratus (4-6 specimens per sample site) were collected at seven localities of the rio Paraguay and rio Paraná basins: 1- Rio Miranda (state of Mato Grosso do Sul - MS LBP 2721; LBP 2723); 2- Rio Mogi-Guaçu (state of São Paulo SP1 LBP 2720); 3- Rio Tietê (state of São Paulo - SP2 LBP 2722); 4- Rio Paraná (state of São Paulo - SP3 LBP 2727); 5- Ribeirão Água do Caixão (state of Paraná - PR1 LBP 2726); 6- Rio Tibagi (state of Paraná - PR2 LBP 2724); and 7- Rio Paraná (state of Paraná PR3 LBP 2725) (Fig. 1). Voucher specimens were deposited in the collection of Laboratório de Biologia e Genética de Peixes (LBP), Departamento de Morfologia, UNESP, Botucatu.
Mitotic chromosomes were obtained following Bertollo et al. (1978) and Foresti et al. (1993) with increased mitotic indexes following Cucchi & Barufaldi (1990). Chromosome morphology was determined on the basis of arm ratio as proposed by Levan et al. (1964) and chromosomes were classified as metacentrics (M), submetacentrics (SM), subtelocentrics (ST) and acrocentrics (A). Nuclear DNA content was determined following Gold & Price (1985) modified by Carvalho et al. (1998). Representative nuclei were found in the blood smears and compared with three representative controls (chicken, common carp and rainbow trout) as described by Fenerich et al. (2004). Light absorbance for each nuclei was then standardized by comparison with controls and expressed as a percentage of the mean absorbance values of the controls. Nuclear DNA content was calculated by multiplying the standardized absorbance values by the known values of the standard species, and expressed in picograms.
To test whether samples formed populations with similar DNA content, nuclear DNA content (calculated as the average of 30 measurements per individual) was compared among samples using analysis of variance (ANOVA, followed by appropriate contrasts, depending upon assumptions of normality and homogeneity of variances). Cluster analysis was conducted using karyotype structure variables and nuclear DNA content as continuous variables with the average centroid method (Euclidean distances) (JMP, version 5, 2002). Chromosome variables were taken from the karyotypical formulae, providing five variables. For example, 2n=44 (4M+2SM+8ST+30A) provides five variables (2n = 44, M = 4, SM = 2, ST = 8 and A = 30).
Results
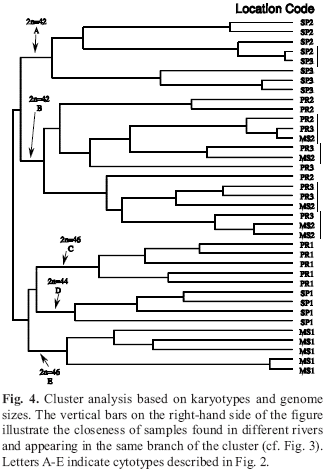
Differences in diploid numbers (with values ranging from 2n=42 to 2n-46) and chromosomal formulae (Table 1) among the samples resulted in the identification of five cytotypes of Synbranchus marmoratus (Fig. 2). A new cytotype was found in some specimens from the rio Miranda, with 2n=46 and the chromosome formula: 6M+2SM+6ST+32A (Fig. 2E).
In testing whether nuclear DNA content varied among samples and cytotypes (ANOVA) the residuals were not normally distributed. While Kruskal-Wallis ANOVA showed differences among groups (p < 0.05), closer analysis of the samples and residuals revealed multimodality in the distribution of nuclear DNA content, suggesting that nuclear DNA content was divided into more or less discrete groupings, both within and between samples (Fig. 3). By separating each of the five previously described samples based on their nuclear DNA content, 12 distinct groups exist in the rivers sampled (Fig. 3). Cluster analysis, with diploid numbers, karyotypical formulae and average individual nuclear DNA content as variables, grouped the individuals into five groups in which some individuals from different rivers were included within the same group (for example, SP2 and SP3 and PR2, PR3 and MS2). Nuclear DNA content of individuals from one locality may vary more than among individuals from different localities (SP3, PR2 and MS2, Fig. 3).
Discussion
The variation in diploid number found in this study (2n=42 to 2n=46) is similar to that observed in previous studies in Brazil and Argentina (Table 1). A new cytotype (identified here as MS1) from rio Miranda was found. Morphological studies showed that Ophisternon aenigmaticum is the basal sister species of the genus Synbranchus (Favorito-Amorim, 1998) and molecular analyses that not included the genus Ophisternon showed that the genus Mastacembelus is the basal sister group of Synbranchus marmoratus (Miya et al., 2003). Cytogenetic studies with Mastacembelus armatus (Oliveira et al., 1997) and M. aculeatus (Liu et al., 2002) showed that both species have 2n=48 chromosomes. Yet Ophisternon bengalense has 2n=46, 6 M + 40 ST (Arkhipchuk, 1999). For these reasons we suggest that the most primitive diploid number in Synbranchus is 2n=46 and subsequent rearrangements (possibly chromosome fusions) reduced the diploid number to 2n=44 and 2n=42.
Additionally, a surprising variation in nuclear DNA content was found in the present study. Cohesiveness within both data sets suggests that some cytotypes of Synbranchus marmoratus have wide distributions and exist in sympatry with other cytotypes. The occurrence of different cytotypes living in sympatry, as seen in the rio Miranda (Figs. 2A-E), and the absence of natural hybrids, reinforce the hypothesis that these cytotypes represent reproductively isolated units.
Besides the fact that the two cytotypes found in the rio Miranda are sympatric and syntopic, they are very different. MS1 has a new and unique karyotype (unlike other 2n=46 cytotypes previously recorded), while MS2 has a karyotype identical as those found in PR2 and PR3 samples (2n=42 cytotype; Figs. 2-3). Therefore, MS1 seems to be isolated, and might have been so for a relatively long time, possibly even reaching the status of a new species. On the other hand, MS2, PR2 and PR3 may represent three reproductively connected samples given their shared diploid number, karyotypical formula and DNA content. Within these last three samples, however, we may also find subdivisions at a finer scale, based on similar DNA content distributions (showed by the dotted lines in Fig. 3). This supports the hypothesis of advancing and receding ocean levels, allowing incursions of the 2n=42 samples, while leaving the other samples (2n=44 and 46) in the headwaters. Similar patterns were found with sympatric species of the genus Hypostomus (Artoni & Bertollo, 1996), sympatric cytotypes of Astyanax scabripinnis (Maistro et al., 2000) and more recently with sympatric and syntopic species of genus Characidium (Centofante et al., 2003a). Thus, the occurrence of different cytotypes in S. marmoratus may represent cryptic species originated during isolation episodes occurred during sea level fluctuations. The new cytotype (MS1) is probably related to the karyotype macrostructure observed from PR1. A single pericentric inversion in one of the four subtelocentric pairs of PR1 sample may have given rise to the third metacentric pair in the MS1 cytotype.
Since shared chromosome numbers and karyotypical formulae are the results of heredity-based processes, the eight local cytotypes were divided in five basic cytotypes (Fig. 2). Next, nuclear DNA content suggested further subdivisions within those eight local cytotypes (Figs. 3-4). However, these divisions did not follow predicted patterns, in that each region would show similarities in DNA content, but rather suggested sympatric processes that allowed differentiation in situ.
This wide variation in karyotype and nuclear DNA content (Figs. 2-4) provides evidence for a hypothesis about the biogeographical patterns of distribution of Synbranchus marmoratus. Thus, karyotypes are grouped by similarities into the following groupings: the samples SP2 and SP3; the samples PR2, PR3, and MS2; with the samples SP1, PR1, and MS1 appear as independent groups belonging to a particular branch (Fig. 4). Indeed one of the most interesting relationships is the close relationship between samples PR3 and MS2. This relationship suggests that there was a recent range expansion, since these samples have the same karyotype (Fig. 2A) and nuclear DNA content (Fig. 3). Cluster analysis (Fig. 4) supports this last hypothesis, given the similarities of the PR3 and MS2 samples. A similar pattern might also have occurred in the samples SP2 and SP3(Figs. 3-4). The relationship between the samples PR3 - MS2 and SP2 - SP3 may be explained by a similar metapopulation structure where these samples may interact by genetic flow (Baguette, 2004).
Following this reasoning, S. marmoratus may be subdivided into as many as 12 groups due to similarities in nuclear DNA content and karyotypes. These groups form easily interpreted clusters (Fig. 4), the exact details of which will only be determined by more detailed studies of nuclear DNA content in these populations. These clusters suggest cohesiveness between geographically distant areas (SP2, SP3, PR3 and MS2) and reproductive isolation in sympatry (MS samples). We suggest that these data can be explained by the last great marine incursion into South America (~5 mya - Frailey, 2002), since the distribution of the 2n=46 and 44 cytotypes (SP1, PR1 and MS1) are closest to the headwaters of the rivers, possibly isolated during the marine incursion events in the past (Table 1, Fig. 1; but see Sanchez & Fenocchio, 1996). Also, populations far from the headwaters (2n=42 cytotypes) underwent more drastic selection regimes, causing more variable karyotypes and nuclear DNA content. Headwater populations remained stable in allopatric and static condition (i.e., SP1, MS1 and PR1). This trend suggests a possible radiation of the 2n=42 samples. Additional support for this hypothesis is found in Nakamoto et al. (1986a, b), in which chromosome number and hemoglobin type are associated (2n=44 and 2n=46 are associated with hemoglobin electrophoretic phenotype 1 while 2n=42 is associated with hemoglobin electrophoretic phenotype 2). Thus, samples with 2n=44 and 46 are the least variable in their nuclear DNA content (as seen by the variability of the points along the nuclear DNA content axis in Fig. 3), while those with 2n=42 are the most variable (Figs. 3-4).
We return to the question of how many species of S. marmoratus exist. We show that seven sampled regions, presumably populations, in different rivers were divisible into several other groups, depending upon the criteria used. With classification based on karyotype and nuclear DNA content, some of these groups are probably cryptic species, especially in the 2n=42 group (see bars on right side of Fig. 4). Also, since sympatric samples may differ more among themselves than to allopatric samples, these data suggest possible sympatric speciation, or biogeographic events that lead to the sympatric distributions of very similar species. The exact nature of the species structure of these rivers as well as a reconstruction of the biogeographic processes giving rise to this structure, will require data gathered specifically to test this issue. Further sampling in MS1 and MS2 would be especially productive since these contain at least two distinct karyotypes as well as perhaps four nuclear DNA content sizes (Fig. 3). If these hypotheses are correct, then S. marmoratus is a species complex whose taxonomic status requires further study. Also, this surprising genomic diversity implies that the region contains more species than hitherto recognized, with many ecological, evolutionary and conservation implications (Ryder, 1986; Moritz, 1994). Since these data were not collected with this complexity in mind, we cannot attempt to answer the question of how many species, but we suggest that it is at least five, based on karyotypes (Fig. 4). Also, based on the data provided in the table 1, it would be possible to suggest a wide species diversity hosted by the name Synbranchus marmoratus. Among the cytotypes with 42 chromosomes, four different genetic identities (karyotypic formula) may identified: 4M/SM+38ST/A, 6M/SM+36ST/A, 4M+6SM+8ST+24A and 4M+2SM+8ST+28A. In addition, among the cytotypes with 44 chromosomes, two different genetic identities may be observed (4M/SM+40ST/A and 4M+2SM+8ST+30A). Yet, among the cytotypes with 46 chromosomes, three different genetic constitutions are observed: 4M/SM+42ST+A, 4M+2SM+8ST+32A and 6M+2SM+6ST+32A. Such hidden diversity seems to be not a privilege of Synbranchus marmoratus given its occurrence in another fish groups such as Erythrinus erythrinus (Bertollo et al., 2004), Astyanax altiparanae (Fernandes & Martins-Santos, 2004), Characidium spp. (Centofante et al., 2003a), Hyphessobrycon anisitsi (Centofante et al., 2003b). If the species is further divided based on DNA content, then the evolutionary complexity of those river systems will offer additional insights in evolution and in the biogeography of southern South America.
Acknowledgements
The authors are very grateful to Jorge Dergam, Orlando Moreira Filho, Guaracy Tadeu Rocha, Sandra Favorito, Walter A. P. Boeger and Marcio Pie for their contributions to the manuscript. This paper is part of the Doctoral thesis of R.A. Torres, supported by grants from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico Ministry of Science and Technology Brazilian Federal Government), FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo State Government of São Paulo) and PICD-CAPES (Ministry of Education Brazilian Federal Government).
Literature Cited
Received May 2004
Accepted May 2005
- Arkhipchuk, V.V. 1999. Database of Fish chromosomes. Http://www.fishbase.org
- Artoni, R. F. & L. A. C. Bertollo. 1996. Cytogenetic studies in Hypostominae (Pisces, Siluriformes, Loricariidae). Considerations on the karyotype evolution in the genus Hypostomus Caryologia, 49: 81-90.
- Baguette, M. 2004. The classical metapopulation theory and the real, natural world: a critical appraisal. Basic and Applied Ecology, 5: 312-224.
- Bertollo, L. A. C., C. S. Takahashi & O. Moreira-Filho. 1978. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics, 1: 103-120.
- Bertollo, L. A. C., C. Oliveira, W. F. Molina, V. P. Margarido, M. S. Fontes, M. C. Pastori, J. N. Falcão & A. S. Fenocchio. 2004. Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei, Characiformes). Heredity, 93(2): 228-233.
- Carvalho, M.L., C. Oliveira & F. Foresti. 1998. Nuclear DNA content of thirty species of neotropical fishes. Genetics and Molecular Biology, 21: 47-54.
- Centofante, L., L. A. C. Bertollo, P.A. Buckup & O. Moreira-Filho. 2003a. Chromosomal divergence and maintenance of sympatric Characidium fish species (Crenuchidae, Characidiinae). Hereditas, 138: 213-218.
- Centofante, L., L. A. C. Bertollo, C. S. Miyazawa & O. Moreira-Filho. 2003b. Chromosomal differentiation among allopatric populations of Hyphessobrycon anisitsi (Pisces, Tetragonopterinae). Cytologia, 68: 283-288.
- Chang, H. Y., T. K. Sang, K. Y. Jan & C. T. Chen. 1995. Cellular DNA contents and cell volumes of Batoids. Copeia, 3: 571-576.
- Cucchi, C. & A. Barufaldi. 1990. A new method for karyological studies in teleost fishes. Journal of Fish Biology, 37: 71-75.
- Favorito-Amorim, S. A. 1998. Relações filogenéticas da ordem Synbranchiformes e revisão sistemática da família Synbranchidae. Doctoral Thesis. Instituto de Biociências, Universidade de São Paulo, São Paulo, 217 p.
- Fernandes, C. A. & I. C. Martins-Santos. 2004. Cytogenetic studies in two populations of Astyanax altiparanae (Pisces, Characiformes) Hereditas, 141: 328-332.
- Fenerich, P. C., F. Foresti & C. Oliveira. 2004. Nuclear DNA content in 20 species of Siluriformes (Teleostei: Ostariophysi) from Neotropical region. Genetics and Molecular Biology, 27: 350-354.
- Foresti, F., C. Oliveira & O. S. Tien. 1992. Cytogenetic studies of the genus Synbranchus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 17: 129-138.
- Foresti, F., C. Oliveira & L. F.Almeida-Toledo. 1993. A method for chromosome preparations from large specimens of fishes using in vitro short treatment with colchicine. Experientia, 49: 810-813.
- Frailey, C. D. 2002. Neogene paleogeography of the Amazon basin. TER-QUA Symposium Series, 3: 71-97.
- Gold, J. R. & H. J. Price. 1985. Nuclear DNA content variation among North American minnows (Cyprinidae). I. Distribution of the variation in five species. Heredity, 54: 297-305.
- Goin, O. B., J. C. Goin & K. Backmann. 1980. DNA and amphibians life history. Nature, 284: 601-603.
- Hinegardner, R. & D. E. Rosen. 1972. Cellular DNA content and the evolution of teleostean fishes. American Naturalist, 106: 621-644.
- Kullander, S. O. 2003. Family Synbranchidae (Swamp-eels). In: Check list of the freshwater fishes of South America, R. E. Reis, S. O. Kullander and C. J. Ferraris (eds.). Edipucrs, Porto Alegre, pp. 594-595.
- Levan, A., K. Fredga & A. A. Sandberg. 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52: 201-220.
- Liem, K. F. 1968. Geographical and taxonomic variation in the pattern of natural sex reversal in the teleost fish order Synbranchiformes. Journal of Zoology, 156: 225-238.
- Liu, J. D., M. S. Yi, G. Zhao, F. Zhou, D. Q. Wang & Q. X. Yu. 2002. Sex chromosomes in the spiny eel (Mastacembelus aculeatus) revealed by mitotic and meiotic analysis. Cytogenetic and Genome Research, 98: 291-297.
- Lo Nostro, F. L. & G.A. Guerrero. 1996. Presence of primary an secondary males in a population of protogynous Synbranchus marmoratus Journal of Fish Biology, 49: 788-800.
- Maistro, E. L., C. Oliveira & F. Foresti. 2000. Sympatric occurrence of two cytotypes of Astyanax scabripinnis (Characiformes, Characidae). Genetics and Molecular Biology, 23: 365-369.
- Melillo, I. F. M., F. Foresti & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.
- Miya, M., H .Takeshima, H. Endo, N. B. Ishiguro, J. G. Inoue, T. Mukai, T. P. Satoh, M. Yamaguchi, A. Kawaguchi, K. Mabuchi, S. M. Shirai & M. Nishida. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 26: 121138.
- Moritz, C. 1994. Defining 'Evolutionarily Significant Units' for conservation. Trends in Ecology and Evolution,9: 373-375.
- Nakamoto, W., P. E. A. Machado & F. Foresti. 1986a. Hemoglobins patterns in different populations of Synbranchus marmoratus (Pisces, Synbranchidae). Comparative Biochemical and Physiology, 84: 377-382.
- Nakamoto, W. & P. E. A. Machado. 1986b. Characterization of the hemoglobins and globins of Synbranchus marmoratus (Pisces, Synbranchidae). Comparative Biochemical and Physiology, 84: 383-386.
- Oliveira, C., R. A. Torres, S. Favorito & F. Foresti. 1997. Cytogenetic studies of Mastecembelus armatus (Pisces, Mastacembelidae). Cytobios, 92: 83-89.
- Reis, R. E., S. O. Kullander & C. J. Ferraris Jr. 2003. Check list of the freshwater fishes of South and Central America. Porto Alegre, EDIPUCRS. 742 p.
- Ryder, O. A. 1986. Species conservation and systematics: the dilemma of subspecies. Trends in Ecology and Evolution, 1: 9-10.
- Sanchez, S. & A. S. Fenocchio. 1996. Karyotypic analysis in three populations of the South-American eel like fish Synbranchus marmoratus Caryologia, 49: 65-71.
- Tyler, J.C. & I. Feller. 1996. Caribbean marine occurrence in the mangroves of a typically fresh water synbranchiform fish. Gulf of Mexico Science, 1: 26-30.
Publication Dates
-
Publication in this collection
17 Dec 2007 -
Date of issue
June 2005
History
-
Accepted
May 2005 -
Received
May 2004