Abstracts
The Paraná-Paraguay basin encompasses central western Brazil, northeastern Paraguay, eastern Bolivia and northern Argentina. The Pantanal is a flooded plain with marked dry and rainy seasons that, due to its soil characteristics and low declivity, has a great water holding capacity supporting abundant fish fauna. Piaractus mesopotamicus, or pacu, endemic of the Paraná-Paraguay basin, is a migratory species economically important in fisheries and ecologically as a potential seed disperser. In this paper we employ eight microsatellite loci to assess the population structure of 120 pacu sampled inside and outside the Pantanal of Mato Grosso. Our main objective was to test the null hypothesis of panmixia and to verify if there was a different structuring pattern between the Pantanal were there were no physical barriers to fish movement and the heavily impounded Paraná and Paranapanema rivers. All loci had moderate to high levels of polymorphism, the number of alleles varied from three to 18. The average observed heterozygosity varied from 0.068 to 0.911. After the Bonferroni correction three loci remained significant for deviations from Hardy-Weinberg, and for those the frequency of null alleles was estimated. F ST and R ST pairwise comparisons detected low divergence among sampling sites, and differentiation was significant only between Paranapanema and Cuiabá and Paranapanema and Taquari. No correlation between genetic distance and the natural logarithm of the geographic distance was detected. Results indicate that for conservation purposes and for restoration programs small genetic differences detected in the Cuiabá and Paranapanema rivers should be taken in consideration.
A bacia Paraná-Paraguai compreende o oeste do Brasil, nordeste do Paraguai, leste da Bolívia e o norte da Argentina. O Pantanal do Mato Grosso é uma planície inundada com estações de chuva e seca bem definidas, as características do solo e baixa declividade favorecem a retenção de água proporcionando abrigo para uma abundante ictiofauna. O Piaractus mesopotamicus, ou pacu, endêmico da bacia do Paraná-Paraguai, é uma espécie migratória com importância econômica na pesca e ecológica como potencial dispersor de sementes. Neste estudo utilizamos oito loci de microssatélites para verificar a estrutura populacional de 120 pacus coletados dentro e fora do Pantanal do Mato Grosso. Nosso principal objetivo foi testar a hipótese de panmixia e verificar se haviam diferentes padrões de estruturação entre o Pantanal onde não existem barreiras físicas ao movimento migratório desses peixes em relação aos rios Paraná e Paranapanema com suas inúmeras barragens. Todos os loci apresentaram níveis de polimorfismo de moderado a alto e o número de alelos variou de três a 18. A heterozigosidade média observada variou de 0,068 a 0,911. Depois da correção usando o método de Bonferroni três loci permaneceram estatisticamente significantes para desvios de Hardy-Weinberg, para estes a frequência de alelos nulos foi calculada. Comparações par a par de F ST e R ST detectaram baixa divergência genética entre os locais de coleta e as diferenças foram significantes apenas entre amostras do Paranapanema e Cuiabá e Paranapanema e Taquari. Não foi detectada correlação entre a diversidade genética e o logaritmo natural da distância geográfica. Os resultados indicam que as pequenas diferenças genéticas encontradas nos rios Cuiabá e Paranapanema devem ser levadas em consideração quando se tratar de programas de conservação desta espécie.
Neotropical fish; Pantanal of Mato Grosso; Population genetics; Migratory fish; Serrasalmidae
Population genetic structuring in pacu (Piaractus mesopotamicus) across the Paraná-Paraguay basin: evidence from microsatellites
Daniela CalcagnottoI, II; Rob DeSalleII
IInstituto de Biociências, Departamento de Genética e Biologia Evolutiva, Universidade de São Paulo. Rua do Matão 277, 05508-900 São Paulo, SP, Brazil
IIDivision of Invertebrate Zoology, American Museum of Natural History, Central Park West at 79 St. New York, NY 10024-5192 USA
ABSTRACT
The Paraná-Paraguay basin encompasses central western Brazil, northeastern Paraguay, eastern Bolivia and northern Argentina. The Pantanal is a flooded plain with marked dry and rainy seasons that, due to its soil characteristics and low declivity, has a great water holding capacity supporting abundant fish fauna. Piaractus mesopotamicus, or pacu, endemic of the Paraná-Paraguay basin, is a migratory species economically important in fisheries and ecologically as a potential seed disperser. In this paper we employ eight microsatellite loci to assess the population structure of 120 pacu sampled inside and outside the Pantanal of Mato Grosso. Our main objective was to test the null hypothesis of panmixia and to verify if there was a different structuring pattern between the Pantanal were there were no physical barriers to fish movement and the heavily impounded Paraná and Paranapanema rivers. All loci had moderate to high levels of polymorphism, the number of alleles varied from three to 18. The average observed heterozygosity varied from 0.068 to 0.911. After the Bonferroni correction three loci remained significant for deviations from Hardy-Weinberg, and for those the frequency of null alleles was estimated. FST and RST pairwise comparisons detected low divergence among sampling sites, and differentiation was significant only between Paranapanema and Cuiabá and Paranapanema and Taquari. No correlation between genetic distance and the natural logarithm of the geographic distance was detected. Results indicate that for conservation purposes and for restoration programs small genetic differences detected in the Cuiabá and Paranapanema rivers should be taken in consideration.
Key words: Neotropical fish, Pantanal of Mato Grosso, Population genetics, Migratory fish, Serrasalmidae.
RESUMO
A bacia Paraná-Paraguai compreende o oeste do Brasil, nordeste do Paraguai, leste da Bolívia e o norte da Argentina. O Pantanal do Mato Grosso é uma planície inundada com estações de chuva e seca bem definidas, as características do solo e baixa declividade favorecem a retenção de água proporcionando abrigo para uma abundante ictiofauna. O Piaractus mesopotamicus, ou pacu, endêmico da bacia do Paraná-Paraguai, é uma espécie migratória com importância econômica na pesca e ecológica como potencial dispersor de sementes. Neste estudo utilizamos oito loci de microssatélites para verificar a estrutura populacional de 120 pacus coletados dentro e fora do Pantanal do Mato Grosso. Nosso principal objetivo foi testar a hipótese de panmixia e verificar se haviam diferentes padrões de estruturação entre o Pantanal onde não existem barreiras físicas ao movimento migratório desses peixes em relação aos rios Paraná e Paranapanema com suas inúmeras barragens. Todos os loci apresentaram níveis de polimorfismo de moderado a alto e o número de alelos variou de três a 18. A heterozigosidade média observada variou de 0,068 a 0,911. Depois da correção usando o método de Bonferroni três loci permaneceram estatisticamente significantes para desvios de Hardy-Weinberg, para estes a frequência de alelos nulos foi calculada. Comparações par a par de FST e RST detectaram baixa divergência genética entre os locais de coleta e as diferenças foram significantes apenas entre amostras do Paranapanema e Cuiabá e Paranapanema e Taquari. Não foi detectada correlação entre a diversidade genética e o logaritmo natural da distância geográfica. Os resultados indicam que as pequenas diferenças genéticas encontradas nos rios Cuiabá e Paranapanema devem ser levadas em consideração quando se tratar de programas de conservação desta espécie.
Introduction
The Paraná-Paraguay basin encompasses central western Brazil, northeastern Paraguay, eastern Bolivia and northern Argentina (Swarts, 2000). The rio Paraná, the 10th longest river in the world, runs towards the rio De La Plata in Argentina, and is divided into upper, high, middle and lower sections (Agostinho et al., 1995). The rio Paraguay is its main west tributary.
The Paraguay can be divided into upper and lower sections. The upper Paraguay comprises two different geological regions, the highlands and the Pantanal. The latter remained isolated from the Paraná until the Holocene (10.000 ya to today) therefore the fish fauna from upper Paraguay is slightly different from the lower Paraguay and Paraná rivers (Lundberg et al., 1998; Britski, 1999).
The Pantanal is a flooded plain with marked dry (March to September) and rainy (October to February) seasons, due to its soil characteristics and low declivity, it has a great water holding capacity and as a result supports abundant fish fauna. The lower Paraguay flows in a much shallower and wider bed. To the west of lower Paraguay and middle Paraná lies the Gran Chaco, a huge lowland plain that expands into Argentina, Paraguay and Bolivia (Resende, 2003). There are no dams in this portion of the river.
In the upper rio Paraná, the Salto das Sete Quedas or Guaira Falls was the historical barrier to upriver movement until the flooding of the Itaipú reservoir in 1983, since then the functional barrier is the Itaipú dam (Resende, 2003; Agostinho et al., 1995). On the upper rio Paraná and its major tributaries dams are much more common, the first one was built in 1901 in the rio Tietê, however, the majority of the dams were constructed after 1960's (Agostinho et al., 1995).
The serrasalmins, which include 80 valid species in 15 genera among them the fruit eating pacu (Piaractus mesopotamicus) and tambaqui (Colossoma macropomum) and the voraciuos piranhas, were traditionally considered a subfamily of Characidae. Recent phylogenies based on molecular characters suggest the serrasalmins should be elevated to family level (Calcagnotto et al., 2005; Orti et al., 2008). Their distribution is strictly Neotropical, the largest diversity is found on the Amazon basin, in the Paraná-Paraguay basin there are just six genera. The serrasalmids occupy all fresh water biotopes except for small streams and benthic portions of large rivers (Jégu, 2003). Colossoma, Piaractus and Mylossoma are the most important serralsalmid genera in fishing and in fish farming in South America. The genus Piaractus has two species, P. brachypomus that is restricted to the Amazon basin and P. mesopotamicus, known as pacu-caranha or just pacu, which is widely distributed in the Paraná-Paraguay basin. The pacu grows to around 60 cm in length and is characterized by strong molariform dentition capable of breaking hard fruit and seeds. As most of economically important fish of the Paraná-Paraguay pacu is a migratory species, and along with other characins, is one of the first to start the up river spawning migration, known as piracema, in September by the end of the dry season. After spawning adult fish migrate back down river and the larvae enter the flooded areas carried by the current (Resende, 2003). Although migratory routes are still poorly known, some trends have been suggested based on tagging studies. Makrakis et al. (2007) conducted a mark-recapture study from 1997-2005 to investigate the migration patterns of pacu in the upper rio Paraná. The authors tagged and released 2,976 pacu and recaptured 367. Their results indicate that although pacu travels at a maximum speed of 26.4 km/day, which is similar to other species in the basin, their movements are not as wide ranged. However, Makrakis et al. (2007) believe that releasing in the reservoir, sexually immature fish raised in captivity might not exactly reproduces the movement of wild fish.
Beginning in the 90's sport fishing became the second most important economic activity in Pantanal and an increasing number of towns nowadays depend solely on sport fishing (Resende, 2003). The pacu is considered overexploited in the rio Paraná in Argentina and in the Pantanal where 125 tones were harvested in the year 2002 alone (Catella et al., 2005). To minimize the effects of over fishing in 1994 the minimum capture size was increased from 40 to 45 cm in the State of Mato Grosso do Sul and in the lower Paraná fishing pacu is entirely prohibited (Resende, 2003). Besides its value for fisheries, in the Pantanal the pacu also has an important ecological role as seed disperser. One of the major items in its diet is the fruit from the palm tree Bactris glaucences (Galetti et al., 2008). Their results show that bigger fish are better seed dispersers, thus by allowing only the capture of adult fish past a certain size, on one hand may protect young fish that did not yet reproduced, but on the other hand puts an increasing pressure on the dispersal of palm and other species that depend on the fish for seed dispersal.
Despite of its great economic importance, there is only very limited information about the migration patterns of this species (Agostinho et al., 2003; Resende, 2003, and references therein) and there is only one published study on the population structure of pacu (Povh et al., 2008). To the best of our knowledge our study is one of the very few to use microsatellite markers to assess fish population structure in the Neotropical region and the first to employ this technique on an economically important fish in the Paraná/Paraguay basin. Previously, Beheregaray & Sunnucks (2001) studied the genetic structure of marine and estuarine populations of the silverside fish Odontesthes argentinensis using nine microsatellite loci in southern Brazil, Uruguay and Argentina. Barroso et al. (2005) employed seven microsatellites to investigate the genetic structure of Brycon opalinus in the Paraíba do Sul basin, an isolated riverine system in southeastern Brazil. Herbk et al. (2007) analyzed the distribution of the giant fish Arapaima gigas in the Amazonian várzea employing 14 microsatellite loci and mtDNA. More recently, Yazbeck & Kalapothakis (2007) described five microsatellite loci for Prochilodus lineatus another economically important migratory Neotropical species.
In this study we employed allelic variation at eight microsatellite loci to: (i) test the null hypothesis of panmixia among collection sites against the alternative hypothesis of population structuring, (ii) compare samples from Pantanal where there are no physical barriers to migration and the samples from Paraná/Paranapanema with its several hydroelectric dams.
Material and Methods
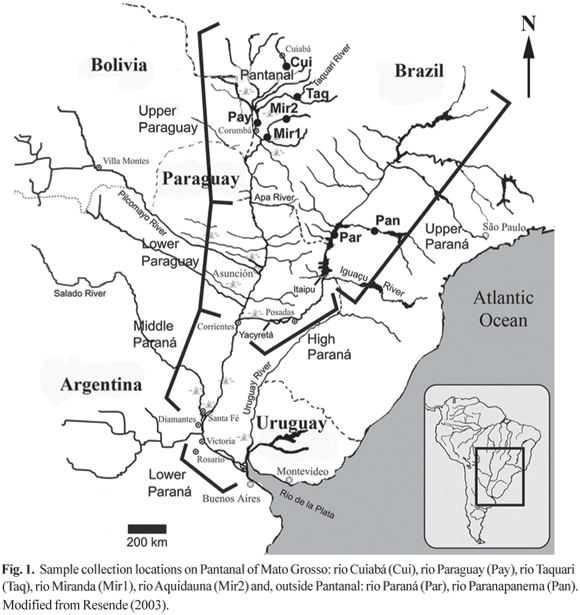
Collection sites and sample preparation. For the microsatellite analysis collections were carried out on four sites within the Pantanal of Mato Grosso, rio Cuiabá (Cui), rio Paraguay (Pay), rio Aquidauana (Mir1), rio Miranda (Mir2), and rio Taquari (Taq) and on two locations outside the Pantanal on rio Paraná (Par) and rio Paranapanema (Pan). The samples from Aquidauana and Miranda were pooled together for the statistical analysis. Geographical distribution of sampling sites is shown on Fig. 1 and Table 1 and totaled 120 specimens from six locations. All sampling was done in the years of 1997, 1998 (Pantanal) and 2000 (Paraná, Paranapanema) by the authors accompanied by an IBAMA agent or by IBAMA agents. Blood samples obtained from adult or juvenile fish were taken from the caudal artery/vein using heparinized syringes. Blood samples and fin clippings, taken from the caudal fin, were immediately transferred to Eppendorf 1.5 ml tubes containing 95% EtOh. Both, blood samples and fin clippings were archived at room temperature. After sampling the fish were released.
Genotyping. Genomic DNA was extracted from ethanol fixed tissues (fin clippings or blood samples) with DNeasy Tissue kit (Qiagen Inc.). Polymerase chain reaction (PCR) to amplify eight microsatellite loci developed for Piaractus mesopotamicus were carried out using a PE 9600 or 9700 thermal cycler (PE Biosystems) in a total volume of 12.5 µl containing: 0.5 µl DNA extract, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 µM dNTPs, 7.5 µg BSA, 0.8 µM each primer and 0.5 U of Amplitaq DNA Polymerase (PE Biosystems) (Calcagnotto et al., 2001). Microsatellite amplified fragments were analyzed on an ABI 377 DNA Sequencer (PE Biosystems) or on an Applied Biosystems 3700 DNAAnalyzer along with a fluorescently labeled size standard (TAMRA550). To score, bin and output allelic and genotypic data, GENESCAN version 2.1 and GENOTYPER version 2.1 fragment analysis softwares (PE Biosystems) were used.
Microsatellite statistical analysis. Descriptive statistics, such as allele number and heterozygosis for each locus were estimated using GENEPOP (Raymond & Rousset, 1995, 2004). Samples were tested for conformance to Hardy-Weinberg equilibrium expectations at each locus as FIS and the nullhypothesis of FIS = 0 tested at each locus with the Markov chain Monte Carlo and 10,000 permutations following Guo & Thompson (1984) were used as implemented in GENEPOP. Expected and observed heterozygosity and its p-value were estimated with exact test using a Markov chain with a 10,000 permutations (Guo & Thompson, 1992; Levene, 1949) implemented in ARLEQUIN (Excoffier et al., 1992). For the loci that did not conform with Hardy-Weinberg expectations the frequency of null alleles (r) was estimated following Brookfield (1996) equation 3, r = D/(2 - D), where D = (He - Ho)/He and He is the expected heterozygosity and Ho is the observed heterozygosity. Linkage disequilibrium was estimated across all pairs of loci. The level of significance for Hardy-Weinberg and linkage disequilibrium tests was adjusted using nonsequential Bonferroni corrections (Rice, 1989). Number or migrants (Nm) was estimated using the private allele method (Slatkin, 1985) as implemented in GENEPOP.
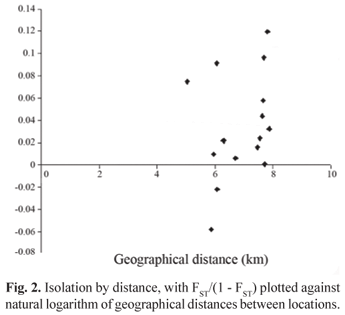
To estimate the connectivity between populations Wright's FST and Slatkin's RST were used. FST was estimated following Weir & Cockerham (1984) as implemented in ARLEQUIN. RST is an FST analogue assuming a stepwise mutation model (SMM), and is thought to reflect more accurately the mutation pattern of microsatellites (Balloux & Lugon-Moulin, 2002). The parameter ρ of RST was estimated with ARLEQUIN. Significance levels were calculated using 10,000 permutations. Hierarchical partitioning of genetic variation among groups of populations, population samples within groups and variating within sampling sites was evaluated using AMOVA in ARLEQUIN. To analyze the relationship of genetic distance and the natural logarithm of geographical distance (shortest waterway distance between pairs of sampling sites in kilometers) the program ISOLDE implemented in GENEPOP was used. Regression significance was tested with 10,000 permutations using the Mantel test. Under an isolation by distance hypothesis the relationship of genetic similarity and distance is expected to be linear (Rousset, 1997).
Results
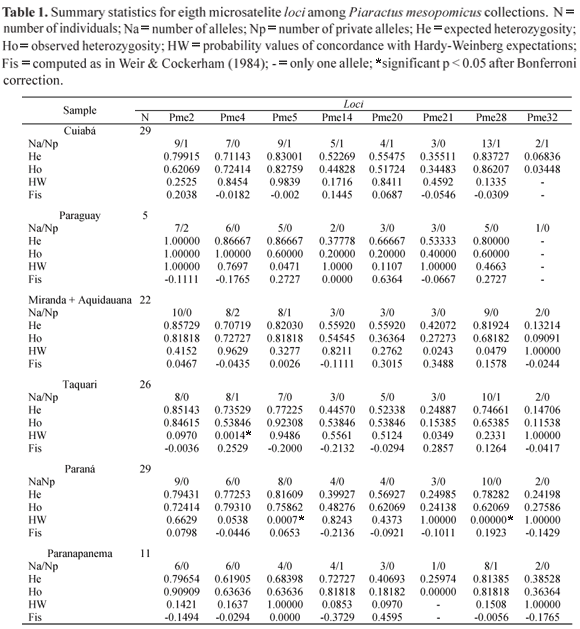
Microsatellite variability and private alleles. Genotypic data were analyzed for 120 pacu from four locations within the Pantanal of Mato Grosso and two outside the Pantanal (Fig. 1). Of the eight loci, one is a compound and interrupted repeat array (Pme5), two are relatively short trinucleotide repeat arrays (Pme14 and Pme32), others are dinucleotide (GT) repeats. All loci had moderate to high levels of polymorphism (Table 1), the number of alleles per microsatellite locus varied from three at loci Pme32 and Pme21, to 18 at locus Pme28, the mean number of alleles per locus was 8.5. Levels of microsatellite variability were similar to those reported for fresh water fish, for example the average number of alleles per locus is 9.1 in other freshwater species (DeWoody & Avise, 2000). Among the analyzed loci, the two trinucleotide repeats, Pme14 and Pme32, showed smaller allelic diversity 6 and 3 alleles, respectively. Trinucleotide microsatellite are often found within exons and could be presumably subjected to some selective pressure to limit their size (Sutherlands & Richards, 1995). The most common repeat found was GT corroborating the idea that the animal kingdom is very rich in this type of microsatellite (Jarne & Lagoda, 1996).
One to three high frequency alleles characterized most loci, the remaining alleles were detected at low frequencies (Table 2). Among the eight loci the total amount of genetic variation ranged from FIT = -0.096 (Pme14) to 0.12 (Pme21). The highest level of genetic divergence among sampling sites was detected in Pme32 (FST 0.041) (Table 2).
All loci, but one (Pme21), presented private alleles, their frequencies however were low between 0.017 and 0.1, their mean frequency was 0.036. Private alleles were detected in all sampling sites, except for Paraná, and ranged from two in the Paraguay, Taquari e Paranapanema rivers, three in the Miranda and six in the rio Cuiabá (Table 1).
Hardy-Weinberg equilibrium and null alleles. The average observed heterozygosity varied from 0.068 in rio Cuiabá at locus Pme32 to 0.911 in rio Paraguay at locus Pme2 and average expected heterozygosity ranged from 0.835 at locus Pme2 to 0.296 at locus Pme32 (Table 1). Of the 45 estimates of FIS, 25 were negative (heterozygote excess), 18 were positive (heterozygote deficiency), two were zero. In three loci there was only one allele (Pme21-Paranapanema and Pme32Paraguay and Cuiabá). Over all loci and sites the average FIS is positive (0.035) but nonsignificant, indicating that although there were more sites with negative values the positive ones were higher. Seven of the 48 single locus tested for Hardy-Weinberg genotype proportions were statistically significant at the α = 0.05 level, from those, two were only marginally significant p = 0.047 (Table 1). However, after the Bonferroni correction for multiple tests only three remained significant (Table 1). The genotypic distribution at locus Pme4 deviated significantly from Hardy-Weinberg expectations (heterozygote deficiency) in the Taquari sample. In the sample from the Paraná sample two loci presented genotypic distributions that deviated from Hardy-Weinberg expectations, Pme5 and Pme28, both showing heterozygote deficiency (Table 1). Testing jointly over all eight loci only the Paraná sample deviated significantly from Hardy-Weinberg, at this site locus Pme28 showed highly significant heterozygote deficit.
To further investigate the Hardy-Weinberg deviations we examined the occurrence of non amplification in our samples. Most common reasons for nonamplification are tissue and/or DNA degradation and random PCR failure. However, in some cases the presence of null alleles, believed to be the result of lack of amplification due to mutations in the primer site that prevent the primer/complement annealing, produces the same pattern (Callen et al., 1993). In our study only in three instances were bands and/or peaks not visualized, two in locus Pme14 and one in Pme21. Therefore, the frequency of null alleles was estimated using Brookfield's (1996) equation 3. The estimated frequency of null alleles (r) for loci that did not conform to Hardy-Weinberg expectations was 0.1545 (Pme4/Taquari), 0.036 (Pme5/Paraná) and 0.1155 (Pme28/ Paraná). When averaged over all populations locus Pme21 has the highest frequency of null alleles (r = 0.1741) followed by Pme20 (r = 0.1506) while other loci presented considerably lower frequencies (r = -0.0002 in Pme14 to 0.0623 in Pme28). Brookfield (1996) assumes that null alleles, not population subdivision, are the main cause of heterozygote deficiency. No evidence of linkage disequilibrium was observed in any of the 8 loci investigated allowing allelic variation to be treated as independent. The number of migrants after correction for size was 5.926.
Population genetic structure. To test the genetic structure the sampling sites were artificially divided into two groups. Group 1: Cuiabá, Paraguay, Taquari and Miranda, and group 2: Paraná and Paranapanema. Using the number of different alleles (FST) as distance measure most of genetic variation resides within sampling sites (98.1%) followed by among sampling sites within groups (1.13%) and among groups (0.77%). When we used the sum of squared allele size difference (RST) the genetic variation within sampling sites was slightly lower (96.78%) and the variation among sampling sites within groups doubled (2.52%) the variation among groups remained practically unchanged (0.70%).
The global estimate of FST over all populations and loci was not significantly different from zero (FST = 0.0145) suggesting that there is no population differentiation in our sample. Pairwise FST values ranged between -0.00008 and 0.07315, and all pairwise comparisons involving the Paranapanema sample were significant (Table 3, bellow diagonal). After Bonferroni sequential correction, however, only two pairwise comparisons involving samples from Cuiabá e Paranapanema rivers (p = 0.0000) and Taquari and Paranapanema rivers (p = 0.00010) remained significant. Pairwise RST values ranged from -0.05451 to 0.10801, most comparisons were not significant (Table 3, above diagonal). The ones involving the sampling sites Taquari and Miranda, Taquari and Paraná and, Taquari and Paranapanema were significant. After Bonferroni sequential correction, however, none of these estimates remained significant. It has been argued that the Bonferroni method is not suited for cases where more than five comparisons are made, when it would become highly conservative (Altman, 1991; Hutchinson et al., 2001). The regression of pairwise (FST/FST - 1) against the natural logarithm of geographical distances between sampling sites showed a slightly positive association, however when tested, using the one tailed p-value the probability of correlation > observed correlation was not significant, p = 0.1386 under null hypothesis (Fig. 2).
Discussion
Heterozygote deficiency and null alleles. Heterozygote deficiency when compared to Hardy-Weinberg expectations is common in fish and these deficiencies could arise either by population subdivision (Wahlund effect) or by inbreeding (O'Connell & Wright, 1997). An alternative explanation for heterozygote deficiency is the presence of null alleles (Chakraborty et al., 1994; Pemberton et al., 1995). Heterozygote deficits that are locus specific are generally an indicative of the presence of null alleles. On the other hand, when there is inbreeding or other population processes their signature usually can be seen across loci (Endersby et al., 2006). In our study, of the three loci, Pme5 and Pme28 in the Paraná sample and Pme4 in Taquari, that did not conform to Hardy-Weinberg expectations after Bonferroni corrections, Pme28 and Pme4 presented high frequency of null alleles 0.1155 and 0.1545, respectively, and Pme5 showed moderate value 0.0365. When we compare the frequency of null alleles for these three loci in other samples only Pme28 show moderate to high r values in other sampling sites: 0.0663 (Taquari), 0.0915 (Miranda) and 0.1429 (Paraguay). In Pme5 a moderate r value 0.036 was detected in the Paranapanema sample. Pme4, on the other hand, showed negative values of r in all sampling sites, with the exception of Taquari. These results suggest that possibly the significant deviations from Hardy-Weinberg proportions involving Pme28 could be due to the presence of null alleles and when we removed locus Pme28 from the analysis none of the sampling sites deviated significantly from Hardy-Weinberg expectations.
Nevertheless, when we consider the upriver, reproduction related migration pattern of pacu it seems difficult to rule out that populations processes, not just null alleles, play a part in the deviations from Hardy-Weinberg. A subdivided population, where gene flow is restricted can show heterozygote deficiency proportional to the magnitude of genetic subdivision (Ruzzante et al., 2000). The rio Paraná is heavily impounded including some 130 dams that are over 10 m high (Makrakis et al., 2007) therefore its not surprising to find two loci that were heterozygotes deficient. The presence of many migratory fish species has been severely diminished due to overfishing and dam construction (Agostinho et al., 1999). Pacu, once a common species downstream the Itaipú Dam, have practically disappeared from the region and in the last decade hatchery stocks of pacu have been used in a restoration strategy (Agostinho et al., 1994). Therefore, inbreeding could be one of the causes of the heterozygote deficit detected among pacus from the Paraná sample. Interestingly, Pme14 where we detected two possible null homozygotes, one in the Miranda and the other in the Aquidauana sample, showed the lowest frequency of null alleles when averaged across all populations (r = -0.0002).
Structure in Paraná/Paranapanema. The microsatellite data suggest that there is low, though significant, genetic differentiation between the pacu population from the Paranapanema and populations from the Pantanal (Table 3, FST estimates). One reason for this could be the many dams built in both in Paranapanema and in Paraná rivers, which starting in 1901 isolated that population, which is now showing an incipient degree of differentiation. Yamamoto et al. (2004) using five microsatellite loci to analyze 11 populations of the white-spotted char (Salvelinus leucomaenis) detected population differentiation and loss of genetic variability when comparing populations above and bellow dams. Between the Paranapanema collection site in Quebra Canoa (Fig. 1) and the Paraná sampling site at the confluence with the rio Ivaí there are three dams. Between the Paraná and the Pantanal there are also a few dams being Itaipú the largest one.
Makrakis et al. (2007) in the first tagging-recapture study detected that most pacu released in the Itaipú Reservoir moved upstream and downstream. While upstream movement for other characins ranged from 450-700 km for Prochilodus (Godoy, 1972) to 1,400 km for Salminus (Sverlij & Espinach-Ros, 1986) only 10 (4.11%) of the pacus released by Makrakis et al. (2007) showed long distance migration (422 km). No marked fish was captured upstream the Porto Primavera Dam, most of them, 95.88%, moved upstream inside the reservoir or into its major bays and tributaries. Downstream migration have also been recorded for migratory species, reaching a maximum of about 1000 km for Salminus and Prochilodus lineatus both in the rio Paraná stretch that runs through Argentina (Bonetto et al., 1981). The downstream migration of pacu detected by Makrakis et al. (2007) indicated the movement of only a few individuals swimming much shorter distances and, again, most of them remained inside the reservoir and its tributaries. The authors, however, recognize that the fact of releasing hatchery reared immature fish inside the reservoir could have influenced the results. Povh et al. (2008) using RAPD markers detected moderate population differentiation between 29 pacu from the broodstock of the Estação de Aquicultura e Hidrobiologia of the Duke Energy power plant (formed by 250 fish from rio Paraná) and 30 pacu captured on the fish ladders of the Canoas I and II reservoirs located in the mid rio Paranapanema. On the other hand, in the upper rio Paraná, in a region that is free of dams, Revaldares et al. (1997) analysed 19 isozymes in Prochilodus lineatus from the rios Paraná, Ivinheima and Baia and although the authors detected high polymorphims and heterozygosity the Nei's values for genetic similarity were also high indicating that they form a panmitic population. Therefore, even though most of the dams have means to allow for fish movement such as fish ladders and elevators, the extensive impounding of these two rivers is probably contributing to a population differentiation when compared to pacu from the Pantanal of Mato Grosso.
Structure in the Pantanal. We did not obtain a significant positive correlation between the genetic differentiation and the natural logarithm of geographical distances and when the sampling sites were compared there were also no significant genetic differentiation among them. Inside the Pantanal the geographical distances ranged from 155 km (Paraguay/ Miranda) to 813 km (Cuiabá/Taquari) with a mean distance between sites of 544 km. Pacu was tentatively classified as a short distance migrant, traveling less than 100 km (Agostinho et al., 2003). Migration is a necessary step for gonadal maturation and spawning habitats are mainly the headwaters of large tributaries such as Cuiabá but reproducing adults were already captured in the headwaters of the rio Taquari. The low levels of genetic differentiation found among sampling sites in the Pantanal, where the pacu is not as overexploited as it is in the rio Paraná and tributaries, could be an indicative that pacu is capable of longer migrations specially if we take into account that it swims at about the same speed (26.4 km/day) as other fish such as Salminus (Bonetto et al., 1971). It is important, however, to keep in mind that these were rough estimates that considered only linear distances versus the time between release and capture (Makrakis et al., 2007). Another interesting aspect to consider is the putative role of the floodplain establishing shorter waterways, when compared to river pathway, connecting sampling locales during the wet season.
When we examined the genetic diversity, with microsatellites, it was larger in the Cuiabá sample. We detected higher allele richness in the Cuiabá sample 52 alleles versus 46 in Miranda, Taquari and Paraná, when comparing only samples of similar size (Table 1). The number of private alleles found in Cuiabá was six while in the other samples the maximum number was three in Miranda (Table 1). Calcagnotto (1998) using restriction fragment length polymorphism of mitochondrial DNA detected only three different haplotypes in hatchery and wild stocks of pacu. One of them was common to all sampled localities and the two different and rarer haplotypes were detected only in the stock formed between 1980-1982 by fish from Coxim, Taquari and Cuiabá rivers.
Conservation issue. Most population studies on Neotropical fish species seem to point out to some level of population structuring (Hatanaka & Galetti Jr., 2003; Barroso et al., 2005; Povh et al., 2008). Sanches & Galetti Jr. (2007) used RAPDs to assess the genetic variability of 80 individuals of the migratory characin Brycon hilarii at four sites of the rio Miranda subbasin (Paraná-Paraguay basin) and found significant population differences. Those differences, however, were more evident during the dry season when fish movement is limited. However it is important to emphasize that there is still very little knowledge about the population structure of Neotropical fish and, that most studies conducted so far used RAPDs, markers of poor reliability for population studies (Pérez et al., 1998). In the light of theses results is interesting that pacu only shows an insipient population structuring in the Paranapanema sub-basin.
While in the Paraná/Paranapanema dams are among the biggest treats, in the upper Paraguay agriculture in high lands is main impact on migratory fish. However, the construction of the only hydroelectric dam in the Pantanal, finished in the year 2000 in the rio Manso, had according to Resende (2003) a major effect on fish populations and she argues that its effects on the long run are still largely unpredictable.
The present work was the pilot study for a larger project involving several laboratories in collaboration with the Brazilian environmental agency IBAMA with the objective to describe the genetic variation in Piaractus mesopotamicus from the Pantanal to create ex-situ genetic banks that could be used as a resource for restorations projects. Based on the microsatellite data two separated ex-situ banks could be suggested, one with fish from the rio Cuiabá and other from rio Paranapanema. However, a larger sampling effort from the rio Paraguay and other large tributaries it also recommended to further protect this highly exploited and endangered food resource.
Acknowledgements
This work is dedicated to the memory of Silvio de Almeida Toledo Filho. We thank Geraldo Bernardino for his valuable help and support in field trips to the Pantanal of Mato Grosso, José Senhorine (CEPTA/IBAMA) provided samples of Piaractus mesopotamicus from Paraná and Paranapanema rivers, and the Estação de Piscicultura Peixe Vivo kindly let us sample their broodstock from the rio Aquidauana. We are grateful to Diogo Meyer for his comments on the statistical analysis, to Emiko Resende (EMBRAPA-Pantanal) and Joachim Carolsfeld (World Fisheries Trust) for allowing the use of the map in Fig. 1. Two anonymous reviewers also contributed valuable suggestions. This work was supported by a postdoctoral fellowship from FAPESP (99/06207-7) to D.C.
Literature Cited
Accepted August 24, 2009
Published December 18, 2009
- Agostinho A. A., J. R. Borghetti, A. E. A. M. Vazzoler & L. C. Gomes. 1994. Itaipú Reservoir: impacts on the ichthyofauna and biological bases for its management. Pp. 135-148. In: United Nations Centre for Regional development. Environmental and social dimensions of reservoir development and management in the La Plata River basin. Nagoya, Japan, UNCRD Research Report Series.
- Agostinho, A. A., L. C. Gomes, H. I. Suzuki & H. F. Júlio. 2003. Migratory fishes of upper Paraná River Basin. Pp. 19-89. In: Carolsfeld, J., B. Harvey, C. Ross & A. Baer (Eds.). Migratory fishes of South America: biology, fisheries, and conservation states. Victoria, World Fisheries Trust/IDRC/World Bank, 372p.
- Agostinho, A. A., L. E. Miranda, L. M. Bini, L. C. Gomes, S. M. Thomaz & H. I. Suzuki. 1999. Patterns of colonization in neotropical reservoirs, and progress on aging. Pp. 227-265. In: Tundisi, J. G. & M. Straskraba (Eds.). Theoretical reservoir ecology and its applications. São Carlos, International Institute of Ecology, Brazilian Academy of Sciences and Backhuys Publishers, 585p.
- Agostinho, A. A., A. E. A. M. Vazzoler & S. M. Thomaz. 1995. The high river Paraná Basin: limnological and ichtyological aspects. Pp. 59-103. In: Tundisi, J. G., C. E. M. Bicudo & T. M. Tundisi (Eds.). Limnology in Brazil. Rio de Janeiro, ABC/SBL, 384p.
- Altman, D. G. 1991. Practical statistics for medical research. London, Chapman and Hall, 610p.
- Balloux, F. & N. Lugon-Moulin. 2002. The estimation of population differentiation with microsatellite markers. Molecular Ecology, 11: 155-165.
- Barroso, R. M., A. W. S. Hilsdorf, H. L. M. Moreira, P. H. Cabello & Y. M. Traub-Cseko. 2005. Genetic diversity of wild and cultured populations of Brycon opalinus (Cuvier, 1819) (Characiformes, Characidae, Bryconinae) using microsatellites. Aquaculture, 247: 51-65.
- Beheregaray, L. & P. Sunnucks. 2001. Fine scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis Molecular Ecology, 10: 2849-2866.
- Bonetto, A. A., C. Pignalberi, E. Cordiviola de Yuan & O. Oliveros. 1971. Informaciones complementarias sobre migraciones de peces en la Cuenca del Plata. Physis, 30(81): 505-520.
- Bonetto, A. A., D. Roldán & M. C. Veron. 1981. Algunos aspectos estructurales y ecológicos de la ictiofauna del sistema del Iberá (Corrientes, Argentina). Ecosur, 8: 79-89.
- Britski, H. A., K. Z. S. Silimon & B. S. Lopes. 1999. Peixes do Pantanal. Manual de Identificação. Brasília, Embrapa, 184p.
- Brookfield, J. F. Y. 1996. A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology, 5: 453-455.
- Calcagnotto, D. 1998. Caracterização de bancos genéticos selvagens de pacu (Piaractus mesopotamicus) e de tambaqui (Colossoma macropomum) através da análise do DNA mitocondrial. Unpublished Ph.D. Dissertation, Universidade de São Paulo, São Paulo, 131p.
- Calcagnotto, D., M. Russello & R. DeSalle. 2001. Isolation and characterization of microsatellite loci in Piaractus mesopotamicus and their applicability in other Serrasalminae fish. Molecular Ecology Notes, 1(4): 245-247.
- Calcagnotto, D., S. A. Schaefer & R. DeSalle. 2005. Relationships among characiform fishes inferred from analysis of nuclear and mitochondrial gene sequences. Molecular Phylogenetics and Evolution, 36(1): 135-153.
- Callen, D. F., A. D. Thompson, Y. Shen, H. Phillips, R. I. Richards, J. C. Mulley & G. R. Sutherland. 1993. Incidence and origin of 'null' alleles in the (AC)n microsatellite marker. American Journal of Human Genetics, 52: 922-927.
- Catella, A. C., F. Albuquerque & F. L. R. Campos. 2005. Sistema de controle da pesca de Mato Grosso do Sul SCPESCA/MS - 6 1999. Corumbá, Embrapa Pantanal, 60p.
- Chakraborty, R., Y. Zhong, L. Jin & B. Budowle. 1994. Nondetectability of restriction fragments and independence of DNA fragments sizes within and between loci in RFLP typing of DNA. American Journal of Human Genetics, 55: 391-401.
- DeWoody, J. A. & J. C. Avise. 2000. Microsatellite in marine, freshwater and anadromous fishes compared to other animals. Journal of Fish Biology, 56: 461-473.
- Endersby, N. M., S. W. Mckechnie, P. M. Ridland & A. R. Weeks. 2006. Microsatellite reveal a lack of structure in Australian populations of the diamondback moth, Plutella xylostella (L.). Molecular Ecology, 15: 107-118.
- Excoffier, L., P. E. Smouse & J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA data analysis. Evolutionary Bioinformatics Online, 1: 47-50. Available from: http://anthro.unige.ch/arlequin/
- Galleti, M., C. I. Donatti, M. A. Pizo & H. C. Giacomini. 2008. Big fish are the best: seed dispersal of Bactris galucescens by the pacu fish (Piaractus mesopotamicus) in the Pantanal, Brazil. Biotropica, 40(3): 386-389.
- Godoy, M. P. 1972. Brazilian tagging experiments: fish migration and Upper Paraná River ecosystem. Revista Brasileira de Biologia, 32: 473-484.
- Guo, S. W. & E. A. Thompson. 1992. Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics, 48: 361-372.
- Hatanaka, T. & P. M. Galetti Jr. 2003. RAPD markers indicate the occurrence of structured populations in a migratory freshwater fish species. Genetics and Molecular Biology, 26(1): 19-25.
- Hrbek, T., M. Crossa & I. P. Farias. 2007. Conservation strategies for Arapaima gigas (Schinz, 1822) and the Amazonian várzea ecosystem. Brazilian Journal of Biology, 67(Suppl. 4): 909-917.
- Hutchinson, W. F., G. R. Carvalho & S. I. Rogers. 2001. Marked genetic structuring in localized spawning populations of cod Gadus morhua in the North Sea and adjoining waters as revealed by microsatellites. Marine Ecology Progress Series, 223: 251-260.
- Jarne, P. & P. J. L. Lagoda. 1996. Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution, 11: 424-429.
- Jégu, M. 2003. Subfamily Serrasaminae. Pp.182-196. In: Reis, R. E., S. O. Kullander & C. J. Ferraris Jr. (Eds.). Check list of the freshwater fishes of South and Central America. Porto Alegre, Edipucrs, Porto Alegre, 729p.
- Lundberg, J. G., L. G. Marshall, J. Guerrero, B. Horton, M. C. S. L. Malabarba & F. Wesselingh. 1998. The stage for neotropical fish diversification: a history of tropical South American rivers. Pp. 13-48. In: Malabarba, L. R., R. E. Reis, R. P. Vari, Z. M. S. Lucena & C. A. S. Lucena (Eds.). Phylogeny and Classification of Neotropical Fishes. Porto Alegre, Edipucrs, 603p.
- Makrakis, M. C., L. E. Miranda, S. Makrakis, A. M. M. Xavier, H. M. Fontes & W. G. Morlis. 2007. Migratory movements of pacu Piaractus mesopotamicus, in highly impounded Paraná River. Journal Applied Ichthyology, 23: 700-704.
- O'Connell, M. & J. M. Wright. 1997. Microsatellite DNA in fishes. Reviews in Fish Biology and Fisheries, 7: 331-363.
- Ortí, G., A. Sivasundar, K. Dietz & M. Jégu. 2008. Phylogeny of the Serrasalmidae (Characiformes) based on mitochondrial DNA sequences. Genetics and Molecular Biology, 31(1): 343-351.
- Pemberton, J. M., J. Slate, D. R. Bancroft & J. A. Barret. 1995. Non-amplifying alleles at microsatellite loci: caution for parentage and population studies. Molecular Ecology, 4: 249-252.
- Pérez, T., J. Albornoz & A. Dominguez. 1998. An evaluation of RAPD fragment reproducibility and nature. Molecular Ecology, 7: 1347-1357.
- Povh, J. A., R. P. Ribeiro, R. N. Sirol, D. P. Streit Jr., N. M. Lopera-Barrero, L. Vargas, P. C. Gomes & T. S. Lopes. 2008. Diversidade genética de pacu do rio Paranapanema e do estoque de um programa de repovoamento. Pesquisa Agropecuária Brasileira, 43(2): 201-206.
- Raymond, M. & F. Rousset. 1995. GENEPOP (version 1.2). Population genetics software for exact tests and ecumenicism. Journal of Heredity, 86: 248-249.
- Raymond, M. & F. Rousset. 2004. GENEPOP (version 3.4). Population genetics software for exact tests and ecumenicism. Montpellier, France, Laboratoire de Genetique et Environment.
- Resende, E. K. 2003. Migratory fishes of the Paraguay-Paraná Basin excluding the upper Paraná River. Pp. 99-155. In: Carolsfeld, J., B. Harvey, C. Ross & A. Baer (Eds.). Migratory fishes of South America: biology, fisheries, and conservation status. Victoria, World Fisheries Trust/IDRC/World Bank, 380p.
- Revaldaves, E., E. Renesto & M. F. P. S. Machado. 1997. Genetic variability of Prochilodus lineatus (Characiformes, Prochilodontidae) in the Upper Paraná River. Brazilian Journal of Genetics, 20(3): 381-388.
- Rice, W. R. 1989. Analyzing tables of statistical tests. Evolution, 43(1): 223-225.
- Rousset, F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145: 1219-1228.
- Sanches, A. & P. M. Galetti Jr. 2007. Genetic evidence of population structuring in the neotropical freshwater fish Brycon hilarii (Valenciennes, 1850). Brazilian Journal of Biology, 67(4): 889-895.
- Slatkin, M. 1985. Rare alleles as indicators of gene flow. Evolution, 39: 53-65.
- Sutherlands, G. R. & R. I. Richards. 1995. Simple tandem DNA repeats and human genetic disease. Proceedings of the National Academy of Science USA, 92: 3636-3641.
- Sverlij, S. B. & A. Espinach-Ros. 1986. El dorado Salminus maxillosus (Pisces, Characiformes), em el rio de la Plata y rio Uruguay inferior. Revista de Investigacíon y Desarrollo Pesquero, 6: 57-75.
- Swarts, F. A. 2000. The Pantanal in the 21st century. In: Swarts, F. A. (Ed.). The Pantanal: Understanding and Preserving the World's Largest Wetland. St. Paul, MinnesotaParagon House, 336p.
- Weir, B. & C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358-1370.
- Yazbeck, G. M. & E. Kalapothakis. 2007. Isolation and characterization of microsatellite DNA in the piracema fish Prochilodus lineatus (Characiformes). Genetics and Molecular Research, 6(4): 1026-1034.
- Yamamoto, S., K. Morita, I. Koizume & K. Maekawa, 2004. Genetic differentiation of white-spotted char (Salvelinus leucomaenis) populations after habitat fragmentation: spatio-temporal changes in gene frequencies. Conservation Genetics, 5(4): 529-538.
Publication Dates
-
Publication in this collection
18 Jan 2010 -
Date of issue
2009
History
-
Accepted
18 Dec 2009 -
Received
24 Aug 2009