Abstracts
The fish species Synbranchus marmoratus has been reported to exist as a species complex due to high intraspecific karyotypic variability in spite of the difficulty or impossibility to distinguish them using morphological traits alone. The goal of this work was to use cytogenetic and molecular methods to determine the species delimitations and understand the karyoevolution ofS. marmoratus using samples collected from distinct Brazilian localities. Among the analyzed specimens, a large degree of cytogenetic variation related to diploid numbers and karyotype structure was observed, with karyotypes showing 2n=42, 44 and 46 chromosomes. In addition, using sequences of three mitochondrial genes, the phylogenetic relationships between every sample with a known karyotype were determined, which revealed significant nucleotide divergence among the karyomorphs. Also, the analyses indicate that chromosomal rearrangements occurred independently within the distinct lineages of S. marmoratus complex, which resulted in the appearance of distinct karyotypic variants in a non-linear fashion related to diploid numbers and in the appearance of similar non-homologous chromosomes. Finally, the integration of both molecular cytogenetic and phylogenetic approaches allowed the determination of specific chromosomes possibly involved in rearrangements and a better understanding about the evolutionary processes involved in the differentiation ofSynbranchus genus.
Chromosomal diversity; Fish cytogenetics; karyotype evolution; Repetitive DNA; Species complex
A espécie de peixe Synbranchus marmoratus tem sido reportada como um complexo de espécies devido à elevada variabilidade cariotípica intraespecífica a despeito da dificuldade ou impossibilidade de distingui-las usando apenas caracteres morfológicos. O objetivo deste trabalho foi utilizar métodos citogenéticos e moleculares para determinar a delimitação das espécies e compreender a carioevolução de S. marmoratus utilizando amostras coletadas em distintas localidades brasileiras. Dentre os espécimes analisados, um alto grau de variação citogenética relativo aos números diploides e estrutura cariotípica foi observado, com cariótipos mostrando 2n=42, 44 e 46 cromossomos. Adicionalmente, utilizando sequências de três genes mitocondriais, as relações filogenéticas entre cada amostra com cariótipo conhecido foram determinadas, revelando uma divergência nucleotídica significativa entre os cariomorfos. Além disso, as análises indicam que rearranjos cromossômicos ocorreram independentemente nas distintas linhagens do complexo S. marmoratus, o que resultou no aparecimento de distintas variantes cariotípicas de forma não linear em relação aos números diploides e no surgimento de cromossomos similares e não homólogos. Finalmente, a integração de uma abordagem citogenética molecular e filogenética permitiu a determinação de cromossomos específicos que, possivelmente, estão envolvidos em rearranjos e um melhor entendimento sobre os processos evolutivos envolvidos na diferenciação do gênero Synbranchus.
Introduction
Cryptic species are defined as two or more morphologically indistinguishable species that are incapable of interbreeding (Bickfordet al., 2007Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. L. Ng, R. Meier, K. Winker, K. K. Ingram & I. Das. 2007. Cryptic species as a window on diversity and conservation. Trends Ecology and Evolution, 22: 148-155.). Such species likely arise through interespecific reproductive isolation, which can be caused by pre-zygotic means, such as gametic incompatibility and/or ecological isolation (Miyatake et al., 1999Miyatake, T. & T. Shimizu. 1999. Genetic correlations between life-history and behavioral traits can cause reproductive isolation. Evolution, 53: 201-208.; Landry et al., 2003Landry, C., L. B. Geyer, Y. Arakak, T. Uehara & S. R. Palumbi. 2003. Recent speciation in the Indo-West Pacific: Rapid evolution of gamete recognition and sperm morphology in cryptic species of sea urchin. Journal of the Academy of Natural Sciences of Philadelphia, 270: 1839-1847.), or by post-zygotic means, such as hybrid inviability and/or sexual selection against hybrids (Orr, 1995Orr, H. A. 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics, 139: 1805-1813.; Nooret al., 2001Noor, M. A. F., K. L. Grams, L. A. Bertucci & J. Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences, 98: 12084-12088.; Presgraves et al., 2002Presgraves, D. C., L. Balagopalan, S. M. Abmayr & H. A. Orr. 2002. Adaptative evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature, 423: 715-719.). Generally, these types of speciation events occur in organisms with little or no motility, making certain plants (Presgraves et al., 2002Presgraves, D. C., L. Balagopalan, S. M. Abmayr & H. A. Orr. 2002. Adaptative evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature, 423: 715-719.), insects (Campbell et al., 1994Campbell, B. C., J. D. Steffen-Campbell & J. H. Werren. 1994. Phylogeny of the Nasonia species complex (Hymenoptera: Pteromalidae) inferred from an internal transcribed spacer (ITS2) and 28S rDNA sequences. Insect Molecular Biology, 2: 225-237.), fungi (Theodoroet al., 2008Theodoro, R. C., E. Bagagli & C. Oliveira. 2008. Phylogenetic analysis of PRP8 intein in Paracoccidioides brasiliensis species complex. Fungal Genetics and Biology, 45: 1284-1291.), small mammals (Green et al., 1980Green, C. A., H. Keogh, D. H. Gordon, M. Pinto & E. K. Hartwig. 1980. The distribution, identification, and naming of the Mastomys natalensis species complex in southern Africa (Rodentia: Muridae). Journal of Zoology, 192: 17-23.), amphibians (Kozak et al., 2006Kozak, K. H., R. A. Blaine & A. Larson. 2006. Gene lineages and eastern North American palaeodrainage basins: phylogeography and speciation in salamanders of the Eurycea bislineata species complex. Molecular Ecology, 15: 191-207.) and fishes (Moreira-Filho & Bertollo, 1991Moreira-Filho, O. & L. A. C. Bertollo. 1991. Astyanax scabripinnis (Pisces, Characidae): a species complex. Revista Brasileira de Genética, 14: 331-357.) suitable model organisms for studying this phenomenon. Although morphological differences between cryptic species are minimal, other traits can be used to detect these species complexes, including behaviour (Crossley, 1986Crossley, S. A. 1986. Courtship-sounds and behavior in the four species of the Drosophila bipectinata complex. Animal Behaviour, 34: 1146-1159.), karyotype structure (Moreira-Filho & Bertollo, 1991Moreira-Filho, O. & L. A. C. Bertollo. 1991. Astyanax scabripinnis (Pisces, Characidae): a species complex. Revista Brasileira de Genética, 14: 331-357.; Dobigny et al., 2002Dobigny, G., V. Aniskin & V. Volobouev. 2002. Explosive chromosome evolution and speciation in the gerbil genus Taterillus (Rodentia, Gerbillinae): a case of two new cryptic species. Cytogenetic and Genome Research, 96: 117-124.; Amaro et al., 2012Amaro, R. C. L., M. T. Rodrigues, Y. Yonenaga-Yassuda & A. C. Carnaval. 2012. Demographic processes in the montane Atlantic rainforest: Molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Molecular Phylogenetics and Evolution, 62: 880-888.) and protein (Nakamoto et al., 1986Nakamoto, W., P. E. A. Machado & F. Foresti. 1986. Hemoglobins patterns in different populations of Synbranchus marmoratus (Pisces, Synbranchidae). Comparative Biochemistry and Physiology B, 84: 377-382.; Fong & Garthwaite, 1994Fong, P. P. & R. L. Garthwaite. 1994. Allozyme electrophoretic analysis of the Hediste limnicola - H. diversicolor - H. japonica species complex (Polychaeta: Nereididae). Marine Biology, 118: 463-470.) and DNA (Kazan et al., 1993Kazan, K., J. M. Manners & D. F. Camero. 1993. Genetic relationships and variation in the Stylosanthes guianensis species complex assessed by random amplified polymorphic DNA. Genome, 36: 43-49.; Hebert et al., 2004Hebert, P. D. N., E. H. Penton, J. M. Burns, D. H. Janzen & W. Hallwachs. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astrapte fulgerator. Journal of the Academy of Natural Sciences of Philadelphia, 101: 14812-14817.) sequences.
Neotropical fishes are excellent models for studying cryptic species, as they are distributed widely across continents and have a propensity to form endemic and isolated populations, often culminating in allopatric differentiation (Lundberg et al., 1998Lundberg, J. G., L. G. Marshall, J. Guerrero, B. Horton, M. C. S. L. Malabarba & F. Wesselingh. 1998. The stage of Neotropical fish diversification: a history of tropical South American rivers. Pp 13-48. In: Malabarba, L. R., R. E. Reis, R. P. Vari, Z. M. S. Lucena & C. A. S. Lucena. Phylogeny and Classification of Neotropical Fishes. EDIPUCRS, Porto Alegre.; Ribeiro, 2006Ribeiro, A. C. 2006. Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: an example of faunal evolution associated with a divergent continental margin. Neotropical Ichthyology, 4: 225-246.). In addition, natural or unnatural random events, such as headwater capture or changes in watercourses, can lead to secondary contacts between previously separated species (Blanco et al., 2009Blanco, D. R., R. L. Lui, L. A. C. Bertollo, D. Diniz & O. Moreira-Filho. 2009. Characterization of invasive fish species in a river transposition region: evolutionary chromosome studies in the genus Hoplias (Characiformes, Erythrinidae). Reviews in Fish Biology and Fisheries, 20: 1-8.; Peres et al., 2009Peres, W. A. M., P. A. Buckup, D. L. Z. Kantek, L. A. C. Bertollo & O. Moreira-Filho. 2009. Chromosomal evidence of downstream dispersal of Astyanax fasciatus (Characiformes, Characidae) associated with river shed interconnection. Genetica, 137: 305-311.), providing ideal scenarios to study novel species complexes. Indeed, such complexes have already been studied in a variety of fish orders, including Characiformes, Synbranchiformes, and Gymnotiformes (Moreira-Filho & Bertollo, 1991Moreira-Filho, O. & L. A. C. Bertollo. 1991. Astyanax scabripinnis (Pisces, Characidae): a species complex. Revista Brasileira de Genética, 14: 331-357.; Torres et al., 2005Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.; Milhomemet al., 2008Milhomem, S. S. R., J. C. Pieczarka, W. G. R. Crampton, D. S. Silva, A. C. P. Souza, J. R. Carvalho Jr. & C. Y. Nagamachi. 2008. Chromosomal evidence for a cryptic species in the Gymnotus carapo species-complex (Gymnotiformes, Gymnotidae). BMC Genetics, 9: 75.).
Fishes belonging to the genus Synbranchus (Synbranchiformes, Synbranchidae) are currently divided into three recognized species: (1)S. madeirae Rosen & Rumney, 1972Rosen, D. E. & A. Rumney. 1972. Evidence of second species of Synbranchus (Pisces, Teleostei) in South America. American Museum Novitates, 2497: 1-45., which is restricted to the Madeira River basin; (2) S. lampreia Favorito, Zanata & Assumpção, 2005Favorito, S. E., A. M. Zanata & M. I. Assumpção. 2005. A new Synbranchus (Teleostei: Synbranchiformes: Synbranchidae) from ilha de Marajó, Pará, Brazil, with notes on its reproductive biology and larval development. Neotropical Ichthyology, 3: 319-328., which is restricted to Marajó Island; and (3) S. marmoratusBloch, 1795, which is widely distributed throughout South and Central America (Rosen & Rumney, 1972Rosen, D. E. & A. Rumney. 1972. Evidence of second species of Synbranchus (Pisces, Teleostei) in South America. American Museum Novitates, 2497: 1-45.). AlthoughS. marmoratus specimens appear to belong to a single taxonomic group, some populations may display an extensive karyotype diversity, with diploid numbers ranging from 42 to 46 chromosomes; furthermore, variations in chromosome morphology as well as in the distribution of constitutive heterochromatin and rDNA genes have also been observed (Foresti et al., 1992Foresti, F., C. Oliveira & O. S. Tien. 1992. Cytogenetic studies of the genus Synbranchus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 17: 129-138.; Melilo et al., 1996Melilo, I. F. M., F. Foresti F & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.; Sanchez & Fenocchio, 1996Sanchez, S. & A. S. Fenocchio. 1996. Karyotypic analysis in three populations of the South-American eel like fish Synbranchus marmoratus. Caryologia, 49: 65-71.; Torres et al., 2005Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.). Although distinguishing the intraspecific S. marmoratus groups is relatively easy with cytogenetic tools, distinguishing between these groups based solely on morphology is often impossible (Rosen & Rumney, 1972Rosen, D. E. & A. Rumney. 1972. Evidence of second species of Synbranchus (Pisces, Teleostei) in South America. American Museum Novitates, 2497: 1-45.), making the identification of new species difficult. Furthermore, one must say that karyotypic structural analyses are limited in their utility; for example, whereas these analyses can identify chromosomal diversity and suggest possible rearrangements leading to this diversity, the chronological order in which such events occurred and the evolutionary relationships among the karyomorphs cannot be directly determined.
Considering the wide distribution of S. marmoratusthroughout the waters of South and Central America, the purpose of this study was to characterize the karyotypes of distinct groups within this species and determine whether divergent species exist within the current S. marmoratusgrouping, to analyze the relationships between the sampled taxa and to investigate the history of chromosomal rearrangements in this species as a whole.
Material and Methods
A total of 75 S. marmoratus specimens were collected from distinct Brazilian localities (Fig. 1, Table 1) and analyzed in the present study. The specimens were identified as previously reported (Rosen & Greenwood, 1976Rosen, D. E. & P. H. Greenwood. 1976. A fourth Neotropical species of Synbranchid eel and the phylogeny and systematics of Synbranchiform. American Museum of Natural History, 157: 1-70.). Following analysis, the fish were fixed in 10% formalin, stored in 70% ethanol and deposited in the fish collection of the Laboratório de Biologia e Genética de Peixes - UNESP, Botucatu, São Paulo, Brazil. Voucher information is shown in Table 1. Two specimens of Ophisternon aenigmaticum Rosen & Greenwood, 1976, basal sister species ofSynbranchus (Miya et al., 2003Miya, M., H. Takeshima, H. Endo, N. B. Ishiguro, J. G. Inoue, T. Mukai, T. P. Satoh, M. Yamaguchi, A. Kawaguchi, K. Mabuchi, S. M. Shirai & M. Nishida. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 26: 121-138.), from Isla Margarita, Venezuela were included as out-groups for the molecular analyses.
A map showing the Synbranchus marmoratus specimen collection sites. Numbers indicate the sample locality, whereas symbols represent the karyomorphs found at each locality.
Specimens of Synbranchus marmoratus analysed. N, Number of samples; LBP, deposit number at the fish collection of the Laboratório de Biologia e Genética de Peixes, Instituto de Biociências de Botucatu, UNESP.
Cell suspensions and chromosome banding. Fish were anaesthetized with a benzoncaine solution before being sacrificed for cytogenetic analyses. Mitotic chromosomes were obtained from cell suspensions of anterior kidney tissue using standard methods (Foresti et al., 1981Foresti, F., L. F. Almeida-Toledo & S. A. Toledo-Filho. 1981. Polymorphic nature of nucleolus organizer regions on fishes. Cytogenetic and Cell Genetics, 31: 137-144.). In addition to Giemsa staining, chromosomes were analyzed using a C-banding procedure to visualize constitutive heterochromatin (Sumner, 1972Sumner, A. T. 1972. A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research, 75: 304-306.) and Ag-NOR staining to detect active nucleolar regions (Howell & Black, 1980Howell, W. M. & D. A. Black. 1980. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia, 36: 1014-1015.).
Fluorescence in situ hybridization (FISH) was performed using the method described by Pinkel et al., (1986)Pinkel, D., T. Straume & J. W. Gray. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences, 83: 2934-2938.. Ribosomal 5S and 18S probes, isolated from the genome ofS. marmoratus (karyomorph B) were labeled during secondary PCR by incorporating the nucleotide biotin-16-dUTP (Roche Applied Science) (5S rDNA) and digoxigenin-11-dUTP (Roche Applied Science) (18S rDNA) and the detection of hybridization signals was obtained with avidin-FITC and anti-digoxigenin-rhodamine, respectively. Chromosomes were counterstained with DAPI (4', 6-diamidino-2-phenylindole, Vector Laboratories).
Chromosomal morphology was determined as described previously (Levan et al., 1964), and samples were classified as metacentric (m), submetacentric (sm), subtelocentric (st) or acrocentric (a).
Molecular analyses. For the molecular analyses, 42 samples were selected to represent all known karyomorphs from every sampled locality, and DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Heidelberg, Germany), as suggested by the manufacturer. Partial sequences from the mitochondrial genes COI, CytB, and 16S were obtained for each sample using the primers described by Ward et al. (2005), Kocheret al. (1989)Kocher, T. D., W. K. Thomas, A. Meyer, S. V. Edwards, S. Pääbo, F. X. Villablanca & A. C. Wilson. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Journal of the Academy of Natural Sciences of Philadelphia, 86: 6196-6200. and Palumbi (1996)Palumbi, S. R. 1996. Nucleic acids II: the polymerase chain reaction. In: Molecular systematics. Edited by Hillis, D., C. Moritz & B. Mable. Massachusetts: Sinauer Associates Inc. 205-247. The PCR products were purified using ExoSap-IT (USB Corporation, Cleveland, OH, USA), according to the manufacturer's instruction, and the fragments were sequenced using an ABI Prism 3110 DNA sequencer (Applied Biosystems, Foster City, CA, USA) and the Big DyeTM Terminator v 3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA). The sequences were aligned using the software program BioEdit, version 7.0.9 (Hall, 1999Hall, T. 1999. BioEdit. a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98.), and deposited in the GenBank database under the accession numbers KC880197-KC880242 (COI), KC880243-KC880288 (CytB) and KC880289-KC880334 (16S).
For the phylogenetic analyses, the gene sequences were aligned using the MUSCLE algorithm (Edgar, 2004Edgar, R. C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32: 1792-1797.) and concatenated into a single matrix, which was separated into seven partitions: one for the noncoding 16S gene and six corresponding to each of the three codon reading frames for the two protein-coding genes CytB and COI. A matrix saturation test was conducted using the DAMBE software program, version 5.1.1, as described previously (Xia et al., 2003Xia, X., Z. Xie, M. Salemi, L. Chen & Y. Wang. 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution, 26: 1-7.). A search for the best model of nucleotide evolution for each partition was performed using Modeltest, version 3.6 (Posada & Crandall, 1998Posada, D. & K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14: 817-818.). The phylogenetic analysis based on Maximum Likelihood (ML) was performed using the RaxML-HPC2 tool on XSEDE (Stamatakis et al., 2008Stamatakis, A., P. Hoover & J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML Web Servers. Systematic Biology, 57: 758-771.). A bootstrap test using 1,000 pseudo-replicates (Felsenstein, 1985Felsenstein, J. 1985. Confidence limits on phylogenies an approach using the bootstrap. Evolution, 39: 783-791.) was used as a statistical test of phylogeny.
Ethics statement. Samples were collected in accordance with Brazilian environmental protection legislation (collection permission MMA/IBAMA/SISBIO - number 3245), and the procedures for collection, maintenance and analysis of the fish were performed in compliance with the Brazilian College of Animal Experimentation (COBEA) and was approved (protocol 503) by the BIOSCIENCE INSTITUTE/UNESP ETHICS COMMITTEE ON USE OF ANIMALS (CEUA).
Results
Cytogenetic analyses. Conventional Giemsa staining revealed the existence of five distinct groups, which could be described using the following karyotypic formulae (Figs. 2a-f): 2n=42 (4m + 12st + 26a), referred to as karyomorph A; 2n=42 (6m + 10sm + 30a), referred to as karyomorph B; 2n=44 (4m + 10st + 30a), referred to as karyomorph C; 2n=46 (6m + 10st + 30a), referred to as karyomorph D; and 2n=46 (4m + 10st + 32a), referred to as karyomorph E. Each karyomorph showed a distinct distribution among the collection sites, with sympatric karyomorphs being found at Igaraçu do Tietê (karyomorphs A and E), Pirassununga (karyomorphs A, D and E) and Icém (karyomorphs A and D). Information concerning each karyomorph and the localities in which specimens were collected is summarized in Table 1.
Karyotypes of Synbranchus marmoratus after FISH with 5S (green) and 18S (red) rDNA, and metaphase plates after silver staining and C-banding of Karyomorph A (a, g, m), B (b, h, n), C-Cerrito (c, i, o), C-Rio Branco (d, j, p), D (e, k, q) and E (f, l, r). The interindividual polymorphisms of 18S rDNA distribution are highlighted as "variation" in the karyotypes. The arrows indicate the Ag-NOR bearing sites chromosomes and the positive C-banding in these chromosomes. Bar = 10 mm.
Ag-NORs were revealed at the terminal position in the 15th pair of chromosomes in karyomorph A, in the 15th pair in karyomorph B, in the 15th pair and in one of the homologues of pair 9 in karyomorph C, in the 2nd pair in karyomorph D and in the 3rd pair in karyomorph E (Figs. 2g-l). Furthermore, interstitial heterochromatic blocks associated with the active NOR sites were detected (Figs. 2m-r).
The C-banding technique revealed centromeric constitutive heterochromatin, as well as some interstitial blocks on particular chromosome pairs, such as in pair 2 of karyomorphs A and B and pair 3 of karyomorphs B, C and E (Figs. 2m-r; Supplementary file 1). These characteristics allowed us to infer certain chromosomal homologies between the different karyomorphs, which are represented in the ideogram in Fig. 3 constructed using organized karyotypes (Supplementary file 1).
Representative ideograms of the analyzed karyomorphs of Synbranchus marmoratus showing the heterochromatic blocks, as determined by C-banding, and hybridization patterns of ribosomal sites.
FISH experiments with 5S rDNA probes revealed that all of the analyzed samples had only two clusters of the 5S ribosomal gene in the interstitial position of a homologous acrocentric chromosomal pair (Figs. 2a-f), except samples of karyomorph C collected at Rio Branco, which presented an additional 5S rDNA-bearing pair (Fig. 2d). In contrast, the 18S rDNA probes revealed distinct hybridization patterns between the karyomorphs. In addition, the hybridizations demonstrated intrapopulational variability at the 18S rDNA sequences, revealing that these populations are polymorphic in the number and chromosomal distribution of these sites. Remarkably, each karyomorph had two constant chromosomal clusters present in all individuals, which corresponded to the Ag-NOR-bearing pairs: pair 15 (karyomorph A), pair 15 (karyomorph B), pair 15 (karyomorph C-Cerrito), pair 3 (karyomorph C-Rio Branco), pair 2 (karyomorph D) and pair 3 (karyomorph E) (Figs. 2a-f).
Molecular analyses. Molecular data consisted of 2,150 nucleotides, of which 414 were polymorphic sites and 389 parsimony informative. The matrix saturation test indicated that there are not saturations in the genes. Appropriate evolutionary models for the genes were investigated in Modeltest 3.6 (Posada & Crandall, 1998) and the best fit-model to each partition was GTR+I (16S), TrNef+I (Codon 1 COI), F81 (Codon 2 COI), GTR (Codon 3 COI), TrN+I (Codon 1 CytB), TrN+I (Codon 2 CytB), HKY+I (Codon 3 CytB).
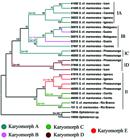
Phylogenetic analysis yielded the dendrogram shown in Fig. 4, which provided significant statistical support for each node. Furthermore, the analysis revealed complex biogeographical relationships, with certain localities bearing only single karyomorphs and other localities showing different karyomorphs living in sympatry. In addition, it was also shown that karyomorphs with the same diploid number do not necessarily constitute monophyletic groups. Also, the analyses provided evidence for the existence of two main clades within S. marmoratus (I and II), one of which can be further subdivided into four subclades (IA, IB, IC and ID). Thus, subclades IA, IB and IC contain specimens sharing the same karyomorph (A); subclade IB is composed of specimens belonging to different karyomorphs (A and B); subclade ID is composed solely of karyomorph D specimens; and clade II is composed by specimens belonging to karyomorphs C and E (Fig. 4).
A dendrogram representing the relationship between the sampled Synbranchus marmoratus specimens based on the mitochondrial 16S, COI and Cyt B genes. The colors represent each of the characterized karyomorphs, and the groups (IA, IB, IC, ID and II) used as references are shown on the right side. Bootstrap support (>50%) are given above the branches. Diploid numbers of the samples are given along the branches. 2n=46* Diploid number of Ophisternon aenigmaticum (Nirchio et al., 2011).
Discussion
Morphological differences between species in the Synbranchiformes order are commonly used in systematic and biogeographical studies (Rosen & Rumney, 1972Rosen, D. E. & A. Rumney. 1972. Evidence of second species of Synbranchus (Pisces, Teleostei) in South America. American Museum Novitates, 2497: 1-45.; Rosen & Greenwood, 1976Rosen, D. E. & P. H. Greenwood. 1976. A fourth Neotropical species of Synbranchid eel and the phylogeny and systematics of Synbranchiform. American Museum of Natural History, 157: 1-70.; Favorito et al., 2005Favorito, S. E., A. M. Zanata & M. I. Assumpção. 2005. A new Synbranchus (Teleostei: Synbranchiformes: Synbranchidae) from ilha de Marajó, Pará, Brazil, with notes on its reproductive biology and larval development. Neotropical Ichthyology, 3: 319-328.). However, many morphological traits can be subtle or ambiguous, which often makes it difficult to recognize and describe new species. Therefore, it is necessary to increase the number of tools and characteristics available for differentiating species from this group (Nakamoto et al., 1986Nakamoto, W., P. E. A. Machado & F. Foresti. 1986. Hemoglobins patterns in different populations of Synbranchus marmoratus (Pisces, Synbranchidae). Comparative Biochemistry and Physiology B, 84: 377-382.; Foresti et al., 1992Foresti, F., C. Oliveira & O. S. Tien. 1992. Cytogenetic studies of the genus Synbranchus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 17: 129-138.; Melilo et al., 1996Melilo, I. F. M., F. Foresti F & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.; Sanchez & Fenocchio, 1996Sanchez, S. & A. S. Fenocchio. 1996. Karyotypic analysis in three populations of the South-American eel like fish Synbranchus marmoratus. Caryologia, 49: 65-71.; Perdices et al., 2005Perdices, A., I. Doadrio & E. Bermingham. 2005. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Molecular and Phylogenetics Evolution, 37: 460-473.; Torres et al., 2005Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.; Nirchio et al., 2011Nirchio, M., T. C. Mariguela, I. A. Ferreira, F. Foresti & C. Oliveira. 2011. Karyotype and nucleolus organizer regions of Ophisternon aenigmaticum (Teleostei: Synbranchiformes: Synbranchidae) from Venezuela. Interciencia, 3: 229-233.; Carvalho et al., 2012Carvalho, N. D. M., M. C. Gross, C. H. Schneider, M. L. Terencio, J. Zuanon & E. Feldberg. 2012. Cytogenetics of Synbranchiformes: a comparative analysis of two Synbranchus Bloch, 1795 species from the Amazon. Genetica, 140: 149-158.). In the present study, several karyomorphs of S. marmoratus collected in distinct Brazilian river basins were described. From these, sympatric karyomorphs were sampled in three distinct localities and the lack of reports of hybrids reinforces the hypothesis that exists as a species complex, which point the need of a deep taxonomic review in this group.
The phylogenetic analyses evidenced the existence of five distinct groups (clades IA, IB, IC, ID and II) within S. marmoratus. However, these results suggest that such grouping do not correspond to the five described karyomorphs reciprocally, indicating that not every karyomorph correspond to a unique species (e.g., karyomorph C), nor a single species is represented by one karyomorph (e.g., karyomorphs A and B). Therefore we propose the existence of, at least, five species withinS. marmoratus complex. Thus, karyomorphs D and E represent one species each; karyomorph C is split in two species; and karyomorph A and B represent one single species in initial stages of differentiation.
Although more detailed information concerning the geographic distribution ofS. marmoratus specimens is needed, acquiring such data is often problematic; for example, these species are widely used as fisheries bait, which can result in the widespread accidental introduction of these organisms into new environments. As a consequence of fish transposition, the specimens from karyomorph A sampled at Cáceres (Paraguay River basin) should be noted, as this karyomorph is positioned between two distinct branches of the dendrogram that includes specimens from the Parana River basin (Icém and Igaraçu do Tietê) (Fig. 4). Considering that many recreational fishermen frequent the Cáceres region, it is possible that these animals originated as bait fish from the Paraná River. Therefore, sample transposition events can be very harmful and make biogeographical and historical studies in this group difficult.
The debate concerning the relationship between chromosomal rearrangements and speciation has been long and controversial (Trickett & Butlin, 1994Trickett, A. J. & R. K. Butlin. 1994. Recombination suppressors and the evolution of new species. Heredity, 73: 339-345.; Rieseberg, 2001Rieseberg, L. H. 2001. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution, 16: 351-358.; Noor et al., 2005; Hoffman & Rieseberg, 2008Hoffman, A. A. & L. H. Rieseberg. 2008. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annual Review of Ecology, Evolution, and Systematics, 39: 21-42.; Faria & Navarro, 2010Faria, R. & A. Navarro. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends in Ecology & Evolution, 25: 660-669.). Although the data presented here are not strong enough to determine whether chromosomal alterations were the cause or consequence of speciation in Synbranchus, it was possible to demonstrate that within subclade IB, two distinct karyomorphs (A and B) from nearby localities constitute the same haplotype and constitute a monophyletic group (Fig. 4). Namely, karyomorph B contained a small pair of metacentric chromosomes, whereas karyomorph A lacked these chromosomes, a difference which most likely arose after the occurrence of a pericentric inversion in a submetacentric chromosome in karyomorph A (Fig. 3). Although small metacentric chromosomes can be observed in all other karyomorphs (with the exception of karyomorph A), the phylogenetic tree highlighted that this specific chromosome pair present in karyomorph B likely arose independently from the small metacentric chromosomes observed in the other samples, since this is a more parsimonious hypothesis than one claiming a common origin for these chromosomes at the base of the Synbranchus lineage with a more recent loss of these chromosomes in all karyomorph A lineages.
The occurrence of species with distinct diploid numbers in Synbranchidae reveals that fusion/fission rearrangements are fundamental mechanisms in the karyoevolution of this group (Torres et al., 2005Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.; Carvalho et al., 2012Carvalho, N. D. M., M. C. Gross, C. H. Schneider, M. L. Terencio, J. Zuanon & E. Feldberg. 2012. Cytogenetics of Synbranchiformes: a comparative analysis of two Synbranchus Bloch, 1795 species from the Amazon. Genetica, 140: 149-158.). Hypotheses concerning the primitive diploid number inSynbranchus have already been proposed, but lead to opposite predictions (Melilo et al., 1996Melilo, I. F. M., F. Foresti F & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.; Torres et al., 2005Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.). Herein, based on the karyotypic information plotted on a phylogenetic tree of S. marmoratus an alternative hypothesis concerning chromosomal differentiation patterns inSynbranchus can be suggested.
Although the deduction of an ancestral karyomorph was not possible, our phylogenetic analyses support the hypothesis that homoplastic cytogenetic events occurred inS. marmoratus and are related to the origins of the karyomorphs showing 2n=44 (karyomorph C-Rio Branco and karyomorph C-Cerrito) and 2n=46 (karyomorphs D and E) chromosomes. Therefore, a chromosome fusion event could have led to the karyomorph showing 2n=44 chromosomes, with a subsequent reversion event (via chromosome fission) returning the diploid number to 2n=46 (karyomorph E); another hypothesis states that both karyomorph C samples (2n=44) could have originated in parallel by independent fusion events. Despite the absence of a clear evolutionary pathway, one should note that one of these homoplasies did occur. In this way, we hypothesize that independent and potentially bidirectional rearrangements, such as fusion and fission events, were responsible for the appearance of distinct diploid numbers and karyotype arrangements observed in this species complex.
Variations related to NORs have already been detected in Synbranchusand indicate that microstructural rearrangements occur frequently and contribute to the karyotypic differentiation of these fish (Foresti et al., 1992Foresti, F., C. Oliveira & O. S. Tien. 1992. Cytogenetic studies of the genus Synbranchus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 17: 129-138.; Melillo et al., 1996Melilo, I. F. M., F. Foresti F & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.; Sanchez & Fenocchio, 1996Sanchez, S. & A. S. Fenocchio. 1996. Karyotypic analysis in three populations of the South-American eel like fish Synbranchus marmoratus. Caryologia, 49: 65-71.; Carvalho et al., 2012Carvalho, N. D. M., M. C. Gross, C. H. Schneider, M. L. Terencio, J. Zuanon & E. Feldberg. 2012. Cytogenetics of Synbranchiformes: a comparative analysis of two Synbranchus Bloch, 1795 species from the Amazon. Genetica, 140: 149-158.). Herein, an extensive intrapopulational polymorphism of 18S rDNA sites in several karyomorphs was detected and seems to be related to the association of these sequences with transposable elements or with their telomeric position, which would facilitate the transference of this material during interphase (Forestiet al., 1981Foresti, F., L. F. Almeida-Toledo & S. A. Toledo-Filho. 1981. Polymorphic nature of nucleolus organizer regions on fishes. Cytogenetic and Cell Genetics, 31: 137-144.; Mantovani et al., 2005Mantovani, M., L. D. S. Abel & O. Moreira-Filho. 2005. Conserved 5S and variable 45S rDNA chromosomal localization revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica, 123: 211-216.). Furthermore, these polymorphic differences have also been detected in other Synbranchus species (Carvalho et al., 2012Carvalho, N. D. M., M. C. Gross, C. H. Schneider, M. L. Terencio, J. Zuanon & E. Feldberg. 2012. Cytogenetics of Synbranchiformes: a comparative analysis of two Synbranchus Bloch, 1795 species from the Amazon. Genetica, 140: 149-158.), suggesting that this is a common characteristic in this group of organisms. Thus, 18S rDNA dispersion was likely already present before the diversification of these species/karyomorphs. Moreover, intrapopulational polymorphism would be potentiated by gametic combination and therefore contribute to the diversification of these sites.
Remarkably, from all analyzed individuals, only one pair of 18S rDNA-bearing chromosomes was constant, in contrast to the occurrence of several random variants. In most cases, Ag-NORs were located in these constant pairs; this finding is likely a consequence of an epigenetic phenomenon that controls the effective dosage of rRNA genes, such as nucleolar dominance (Pikaard, 2000Pikaard, C. S. 2000. The epigenetics of nucleolar dominance. Trends in Genetics, 16: 495-500.; Hashimoto et al., 2009Hashimoto, D. T., A. Laudicina, J. Bortolozzi, F. Foresti & F. Porto-Foresti. 2009. Chromosomal features of nucleolar dominance in hybrids between the Neotropical fish Leporinus macrocephalus and Leporinus elongatus (Characiformes, Anostomidae). Genetica, 137: 135-140.). An interesting feature observed here is the co-localization of heterochromatic blocks with the active NORs, unlike the other 18S rDNA variants. Thus, the mechanism responsible for this situation may be related to the associated heterochromatin that is likely located in the intergenic spacers of 45S rDNA.
Finally, the identification of specific marker chromosomes in certain karyomorphs, which could be tracked by unique morphologies, banding characteristics or repetitive DNAs distribution patterns, could be informative with respect to the genomic rearrangements that occurred during the evolutionary history ofSynbranchus and help understand the phylogenetic relationships between the karyomorphs. Thus, karyomorphs A and B share the exclusive second metacentric pair containing one interstitial heterochromatic block within the long arm and the first submetacentric pair containing two interstitial heterochromatic blocks within the long arm. Besides that, both had the same constant 18S rDNA sites-bearing chromosomes (pair 15). Marker chromosomes also confirm the proximity between karyomorphs C and E, which share the first subtelocentric pair containing one interstitial heterochromatic block within the long arm. Moreover, karyomorph C (Rio Branco) and E have constant sites of 18S rDNA in this same pair. However, the presence of similar chromosomes in distinct lineages can be misleading; for example, a small metacentric pair was identified in karyomorphs B, C, D, and E, although the origin of this pair in karyomorph B was independent of the others. Besides, the detection of an additional 5S rDNA-bearing acrocentric pair observed in individuals belonging to karyomorph C-Rio Branco and some 18S rDNA chromosomal variants of karyomorphs C-Cerrito and E, that evidence sinteny between both ribosomal sites also casts doubt on the real homology of the 5S rDNA-bearing chromosomes in all karyomorphs. Therefore, the nature of the homology between these specific chromosomes remains unclear, and it highlights the importance of phylogenetic and/or chromosome painting analyses to reduce possible misinterpretations of karyoevolution in organisms.
Acknowledgments
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The authors would like to thank D. T. Hashimoto, G. T. Rocha and M. R. Vicari for their substantial contributions to the manuscript.
Literature Cited
- Amaro, R. C. L., M. T. Rodrigues, Y. Yonenaga-Yassuda & A. C. Carnaval. 2012. Demographic processes in the montane Atlantic rainforest: Molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Molecular Phylogenetics and Evolution, 62: 880-888.
- Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. L. Ng, R. Meier, K. Winker, K. K. Ingram & I. Das. 2007. Cryptic species as a window on diversity and conservation. Trends Ecology and Evolution, 22: 148-155.
- Blanco, D. R., R. L. Lui, L. A. C. Bertollo, D. Diniz & O. Moreira-Filho. 2009. Characterization of invasive fish species in a river transposition region: evolutionary chromosome studies in the genus Hoplias (Characiformes, Erythrinidae). Reviews in Fish Biology and Fisheries, 20: 1-8.
- Campbell, B. C., J. D. Steffen-Campbell & J. H. Werren. 1994. Phylogeny of the Nasonia species complex (Hymenoptera: Pteromalidae) inferred from an internal transcribed spacer (ITS2) and 28S rDNA sequences. Insect Molecular Biology, 2: 225-237.
- Carvalho, N. D. M., M. C. Gross, C. H. Schneider, M. L. Terencio, J. Zuanon & E. Feldberg. 2012. Cytogenetics of Synbranchiformes: a comparative analysis of two Synbranchus Bloch, 1795 species from the Amazon. Genetica, 140: 149-158.
- Crossley, S. A. 1986. Courtship-sounds and behavior in the four species of the Drosophila bipectinata complex. Animal Behaviour, 34: 1146-1159.
- Dobigny, G., V. Aniskin & V. Volobouev. 2002. Explosive chromosome evolution and speciation in the gerbil genus Taterillus (Rodentia, Gerbillinae): a case of two new cryptic species. Cytogenetic and Genome Research, 96: 117-124.
- Edgar, R. C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32: 1792-1797.
- Faria, R. & A. Navarro. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends in Ecology & Evolution, 25: 660-669.
- Favorito, S. E., A. M. Zanata & M. I. Assumpção. 2005. A new Synbranchus (Teleostei: Synbranchiformes: Synbranchidae) from ilha de Marajó, Pará, Brazil, with notes on its reproductive biology and larval development. Neotropical Ichthyology, 3: 319-328.
- Felsenstein, J. 1985. Confidence limits on phylogenies an approach using the bootstrap. Evolution, 39: 783-791.
- Fong, P. P. & R. L. Garthwaite. 1994. Allozyme electrophoretic analysis of the Hediste limnicola - H. diversicolor - H. japonica species complex (Polychaeta: Nereididae). Marine Biology, 118: 463-470.
- Foresti, F., C. Oliveira & O. S. Tien. 1992. Cytogenetic studies of the genus Synbranchus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 17: 129-138.
- Foresti, F., L. F. Almeida-Toledo & S. A. Toledo-Filho. 1981. Polymorphic nature of nucleolus organizer regions on fishes. Cytogenetic and Cell Genetics, 31: 137-144.
- Green, C. A., H. Keogh, D. H. Gordon, M. Pinto & E. K. Hartwig. 1980. The distribution, identification, and naming of the Mastomys natalensis species complex in southern Africa (Rodentia: Muridae). Journal of Zoology, 192: 17-23.
- Haga, T. & S. Noda. 1976. Cytogenetics of the Scilla scilloides complex. Genetica, 46: 161-176.
- Hall, T. 1999. BioEdit. a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98.
- Hashimoto, D. T., A. Laudicina, J. Bortolozzi, F. Foresti & F. Porto-Foresti. 2009. Chromosomal features of nucleolar dominance in hybrids between the Neotropical fish Leporinus macrocephalus and Leporinus elongatus (Characiformes, Anostomidae). Genetica, 137: 135-140.
- Hebert, P. D. N., E. H. Penton, J. M. Burns, D. H. Janzen & W. Hallwachs. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astrapte fulgerator. Journal of the Academy of Natural Sciences of Philadelphia, 101: 14812-14817.
- Hoffman, A. A. & L. H. Rieseberg. 2008. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annual Review of Ecology, Evolution, and Systematics, 39: 21-42.
- Howell, W. M. & D. A. Black. 1980. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia, 36: 1014-1015.
- Kazan, K., J. M. Manners & D. F. Camero. 1993. Genetic relationships and variation in the Stylosanthes guianensis species complex assessed by random amplified polymorphic DNA. Genome, 36: 43-49.
- Kocher, T. D., W. K. Thomas, A. Meyer, S. V. Edwards, S. Pääbo, F. X. Villablanca & A. C. Wilson. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Journal of the Academy of Natural Sciences of Philadelphia, 86: 6196-6200.
- Kozak, K. H., R. A. Blaine & A. Larson. 2006. Gene lineages and eastern North American palaeodrainage basins: phylogeography and speciation in salamanders of the Eurycea bislineata species complex. Molecular Ecology, 15: 191-207.
- Landry, C., L. B. Geyer, Y. Arakak, T. Uehara & S. R. Palumbi. 2003. Recent speciation in the Indo-West Pacific: Rapid evolution of gamete recognition and sperm morphology in cryptic species of sea urchin. Journal of the Academy of Natural Sciences of Philadelphia, 270: 1839-1847.
- Levan, A., K. Fredga & A. A. Sandberg. 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52: 201-220.
- Lundberg, J. G., L. G. Marshall, J. Guerrero, B. Horton, M. C. S. L. Malabarba & F. Wesselingh. 1998. The stage of Neotropical fish diversification: a history of tropical South American rivers. Pp 13-48. In: Malabarba, L. R., R. E. Reis, R. P. Vari, Z. M. S. Lucena & C. A. S. Lucena. Phylogeny and Classification of Neotropical Fishes. EDIPUCRS, Porto Alegre.
- Mantovani, M., L. D. S. Abel & O. Moreira-Filho. 2005. Conserved 5S and variable 45S rDNA chromosomal localization revealed by FISH in Astyanax scabripinnis (Pisces, Characidae). Genetica, 123: 211-216.
- Melilo, I. F. M., F. Foresti F & C. Oliveira. 1996. Additional cytogenetic studies on local populations of Synbranchus marmoratus (Pisces, Synbranchiformes, Synbranchidae). Naturalia, 21: 201-208.
- Milhomem, S. S. R., J. C. Pieczarka, W. G. R. Crampton, D. S. Silva, A. C. P. Souza, J. R. Carvalho Jr. & C. Y. Nagamachi. 2008. Chromosomal evidence for a cryptic species in the Gymnotus carapo species-complex (Gymnotiformes, Gymnotidae). BMC Genetics, 9: 75.
- Miya, M., H. Takeshima, H. Endo, N. B. Ishiguro, J. G. Inoue, T. Mukai, T. P. Satoh, M. Yamaguchi, A. Kawaguchi, K. Mabuchi, S. M. Shirai & M. Nishida. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 26: 121-138.
- Miyatake, T. & T. Shimizu. 1999. Genetic correlations between life-history and behavioral traits can cause reproductive isolation. Evolution, 53: 201-208.
- Moreira-Filho, O. & L. A. C. Bertollo. 1991. Astyanax scabripinnis (Pisces, Characidae): a species complex. Revista Brasileira de Genética, 14: 331-357.
- Nakamoto, W., P. E. A. Machado & F. Foresti. 1986. Hemoglobins patterns in different populations of Synbranchus marmoratus (Pisces, Synbranchidae). Comparative Biochemistry and Physiology B, 84: 377-382.
- Nirchio, M., T. C. Mariguela, I. A. Ferreira, F. Foresti & C. Oliveira. 2011. Karyotype and nucleolus organizer regions of Ophisternon aenigmaticum (Teleostei: Synbranchiformes: Synbranchidae) from Venezuela. Interciencia, 3: 229-233.
- Noor, M. A. F., K. L. Grams, L. A. Bertucci & J. Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences, 98: 12084-12088.
- Orr, H. A. 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics, 139: 1805-1813.
- Palumbi, S. R. 1996. Nucleic acids II: the polymerase chain reaction. In: Molecular systematics. Edited by Hillis, D., C. Moritz & B. Mable. Massachusetts: Sinauer Associates Inc. 205-247.
- Perdices, A., I. Doadrio & E. Bermingham. 2005. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Molecular and Phylogenetics Evolution, 37: 460-473.
- Peres, W. A. M., P. A. Buckup, D. L. Z. Kantek, L. A. C. Bertollo & O. Moreira-Filho. 2009. Chromosomal evidence of downstream dispersal of Astyanax fasciatus (Characiformes, Characidae) associated with river shed interconnection. Genetica, 137: 305-311.
- Pikaard, C. S. 2000. The epigenetics of nucleolar dominance. Trends in Genetics, 16: 495-500.
- Pinkel, D., T. Straume & J. W. Gray. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences, 83: 2934-2938.
- Posada, D. & K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14: 817-818.
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr & H. A. Orr. 2002. Adaptative evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature, 423: 715-719.
- Ribeiro, A. C. 2006. Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: an example of faunal evolution associated with a divergent continental margin. Neotropical Ichthyology, 4: 225-246.
- Rieseberg, L. H. 2001. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution, 16: 351-358.
- Rosen, D. E. & A. Rumney. 1972. Evidence of second species of Synbranchus (Pisces, Teleostei) in South America. American Museum Novitates, 2497: 1-45.
- Rosen, D. E. & P. H. Greenwood. 1976. A fourth Neotropical species of Synbranchid eel and the phylogeny and systematics of Synbranchiform. American Museum of Natural History, 157: 1-70.
- Sanchez, S. & A. S. Fenocchio. 1996. Karyotypic analysis in three populations of the South-American eel like fish Synbranchus marmoratus. Caryologia, 49: 65-71.
- Stamatakis, A., P. Hoover & J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML Web Servers. Systematic Biology, 57: 758-771.
- Steven, G. N. & R. Subramanyam. 2009. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Molecular and Ecology Research, 9: 172-180.
- Sumner, A. T. 1972. A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research, 75: 304-306.
- Theodoro, R. C., E. Bagagli & C. Oliveira. 2008. Phylogenetic analysis of PRP8 intein in Paracoccidioides brasiliensis species complex. Fungal Genetics and Biology, 45: 1284-1291.
- Torres, R. A., J. J. Roper, F. Foresti & C. Oliveira. 2005. Surprising genomic diversity in the Neotropical Fish Synbranchus marmoratus (Teleostei: Synbranchidae): how many species? Neotropical Ichthyology, 3: 277-284.
- Trickett, A. J. & R. K. Butlin. 1994. Recombination suppressors and the evolution of new species. Heredity, 73: 339-345.
- Xia, X., Z. Xie, M. Salemi, L. Chen & Y. Wang. 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution, 26: 1-7.
Publication Dates
-
Publication in this collection
09 Jan 2015 -
Date of issue
Oct-Dec 2014
History
-
Received
24 Mar 2014 -
Accepted
04 Sept 2014