ABSTRACT
The Caribbean Red Snapper (Pargo) Lutjanus purpureus is the most economically important snapper in Brazil, which is sold, among other forms, as frozen fillets. During the process of transformation into fillets there is the removal of the distinctive morphological traits, being able to favor the substitution by less valued species. In addition, there is no national legislation requiring the insertion of the specific name on the product label. However, according to a Normative Instruction (IN N ° 29/2015 MAPA) that correlates the common and specific names of the products destined to the national trade, in Brazil only L. purpureus and L. campechanus can be denominated “Pargo”. Thus, the DNA barcode tool was used to identify the fillets sold in north of Brazil, labeled “Pargo”, with the aid of sequences from the public and control databases. The results showed that among 142 fillets examined, 78% was identified as L. purpureus and 22% as Rhomboplites aurorubens, a snapper with low commercial value in the country, revealing commercial fraud. The molecular identification method successfully used in this study to authenticate fillets snappers may also be used by surveillance authorities in the quality control of processed fish products, towards ensuring consumer rights.
Keywords:
Commercial fraud; DNA barcode; Fillets; Processed fish; Snapper
RESUMO
O Pargo Lutjanus purpureus, lutjanídeo mais importante economicamente no Brasil, é vendido, entre outras formas, como filés congelados. Durante a transformação em filés, há a remoção das características morfológicas distintivas, podendo favorecer a substituição por espécies menos valorizadas. Além disso, não há legislação nacional que exija a inserção do nome específico no rótulo. Porém, de acordo com uma Instrução Normativa (IN N° 29 /2015 MAPA) que correlaciona os nomes comuns e específicos dos produtos destinados ao comércio nacional, no Brasil somente L. purpureus e L. campechanus podem ser denominados “Pargo”. Assim, a ferramenta DNA barcode foi usada para identificar os filés vendidos no norte do Brasil, rotulados como “Pargo”, com o auxílio de sequências dos bancos de dados públicos e banco controle. Os resultados mostraram que entre os 142 filés examinados, 78% foi identificado como L. purpureus e 22% como Rhomboplites aurorubens, um lutjanídeo com baixo valor comercial no país, revelando fraude comercial. O método de identificação molecular, utilizado com êxito neste estudo para autenticar filés de lutjanídeos, pode também ser utilizado pelas autoridades de vigilância no controle de qualidade de produtos processados derivados de peixes em geral, para garantir os direitos dos consumidores.
Palavras-Chave:
DNA barcode; Filés; Fraude comercial; Pargo; Peixe processado
Introduction
Global fish consumption has increased remarkably, driving the commercial sectors to offer more diversified products (FAO, 2016Food and Agriculture Organization FAO. (2016). The state of world fisheries and aquaculture. Rome: FAO ; 2016.). Technological progress and logistical improvements combined with changes in consumer preferences are driving the modernization of the fishing industry, converging in the supply of innovative products that provide convenient handling, uniformity, and quality as exemplified by canned, smoked, sliced, and frozen fillets (FAO, 2016Food and Agriculture Organization FAO. (2016). The state of world fisheries and aquaculture. Rome: FAO ; 2016.).
Useful body parts for species identification are taken out during the manipulation of fishery products, for example, in preparation of fillets, which has led to the species replacement practices, since the visual distinction of species becomes unfeasible after this procedure (Ward, 2000Ward RD. Genetics in fisheries management. Hydrobiologia. 2000; 420:191-201.). The removal of these important structures is aggravated in cases of a group of species with a wide range of similar morphological traits, such as the Family Lutjanidae, or snappers, as they are commonly known (Allen, 1985Allen G-R. Snappers of the World. An annotated and illustrated catalogue of Lutjanid species known to date. Rome: FAO; 1985.; Cervigón et al., 1993Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al. FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.; Nelson et al., 2016Nelson JS, Grande TC, Wilson MVH. Fishes of the World. 5th ed. New Jersey: John Wiley & Sons; 2016.). Such fish are key fishery resources throughout their occurrence areas, including the Brazilian coast, wherein numerous species are targets of commercial fisheries (MPA, 2010Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura Brasil 2008-2009; 2010.; 2012Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2010; 2012.; 2013Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2011; 2013.).
The “Pargo” (Caribbean Red Snapper), as the species Lutjanus purpureus (Poey, 1867) is commonly known in Brazil, has always stood out among snappers consumed domestically, and the highest commercial value, as shown in the official records of the country’s fish catches (MPA, 2010Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura Brasil 2008-2009; 2010.; 2012Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2010; 2012.; 2013Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2011; 2013.). Nevertheless, the evidence suggests that the populations of “pargo” are overfished (Dias-Neto, Dornelles, 1996Dias Neto J, Dornelles LDC. Diagnóstico da pesca marítima do Brasil. Brasília: IBAMA; 1996.; Paiva, 1997Paiva MP. Recursos pesqueiros estuarinos e marinhos do Brasil. Fortaleza: UFC Edições; 1997.; Souza, 2002Souza RFC. Dinâmica Populacional do pargo, Lutjanus purpureus Poey, 1875 (Pisces: Lutjanidae) na Plataforma Norte do Brasil. [Dissertação de mestrado]. Belém, PA: Universidade Federal do Pará; 2002.) and, thus, species previously less targeted have gained importance in fisheries and represent potential candidates to replace the actual Caribbean Red Snapper, especially when this fish is processed to prepare fillet products.
Such species may also have been processed into fillets, sold in supermarkets only under the name “Pargo”, without including the scientific name of the raw material in the product label. On the other hand, in Brazil, exclusively the species L. purpureus and L. campechanus can be called “Pargo”, according to Normative Instruction (IN) N° 29 /2015, of Agriculture, Livestock and Food Supply Ministry (Ministério da Agricultura, Pecuária e Abastecimento - MAPA), which establishes the correlation between common and scientific names of products intended for domestic trade.
Fish sold after processing have shown high rates of replacement of the most commercially valued species by less valuable species. These labeling issues may result in economic losses (Filonzi et al., 2010Filonzi L, Chiesa S, Vaghi M, Marzano FN. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res Int . 2010; 43(5):1383-88.; Cawthorn et al., 2012Cawthorn DM, Steinman HA, Witthuhn RC. DNA barcoding reveals a high incidence of fish species misrepresentation and substitution on the South African Market. Food Res Int . 2012; 46(1):30-40.; Di Pinto et al., 2013Di Pinto A, Di Pinto P, Terio V, Bozzo G, Bonerba E, Ceci E et al. DNA barcoding for detecting market substitution in salted cod fillets and battered cod chunks. Food Chem. 2013; 141(3):1757-62.; Cutarelli et al., 2014Cutarelli A, Amoroso MG, De Roma A, Girardi S, Galiero G, Guarino A et al. Italian market fish species identification and commercial frauds revealing by DNA sequencing. Food Control. 2014; 37:46-50.; de Brito et al., 2015Brito MA, Schneider H, Sampaio I, Santos S. DNA barcoding reveals high substitution rate and mislabeling in croaker fillets (Sciaenidae) marketed in Brazil: The case of “pescada branca” (Cynoscion leiarchus and Plagioscion squamosissimus). Food Res Int. 2015; 70:40-46.) and human health hazards (Guallar et al., 2002Guallar E, Sanz-Gallardo I, Veer PV, Bode P, Aro A, Gómez-Aracena J et al. Mercury, fish oils, and the risk of myocardial infarction. N Engl J Med. 2002; 347 (22):1747-54.; Cohen et al., 2009Cohen J, Deeds JE, Wong E, Hanner RH, Yanci HF, White KD et al. Public health response to puffer fish (Tetrodotoxin) poisoning from mislabeled product. J Food Protect. 2009; 72(4):810-17.; Van Leeuwen et al., 2009Van Leeuwen SPJ, Van Velzen MJM, Swart CP, Van Der Veen I, Traag WA, De Boer J. Halogenated contaminants in farmed salmon, trout, tilapia, pangasius, and shrimp. Environmental science & technology. 2009; 43(11):4009-15.; Maralit et al., 2013Maralit BA, Aguila RD, Ventolero MFH, Perez SKL, Willette DA, Santos MD. Detection of mislabeled commercial fishery by-products in the Philippines using DNA barcodes and its implications to food traceability and safety. Food Control . 2013; 33(1):119-25.). Moreover, the misidentification of species may jeopardize the monitoring of fishery stocks, thus increasing the vulnerability of threatened species (Ardura et al., 2010Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv. 2010; 143 (6):1438-43.; Palmeira et al., 2013Palmeira CAM, Rodrigues-Filho LFS, Sales JBL, Vallinoto M, Schneider H, Sampaio I. Commercialization of a critically endangered species large tooth sawfish, Pristis perotteti in fish markets of northern Brazil: authenticity by DNA analysis. Food Control . 2013; 34(1):249-252.).
The unambiguous identification of taxa is the main requirement to prevent/control this type of activity, and forensic molecular approaches using the cytochrome oxidase subunit I (COI) gene have successfully addressed this issue (Filonzi et al., 2010Filonzi L, Chiesa S, Vaghi M, Marzano FN. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res Int . 2010; 43(5):1383-88.; Cawthorn et al., 2012Cawthorn DM, Steinman HA, Witthuhn RC. DNA barcoding reveals a high incidence of fish species misrepresentation and substitution on the South African Market. Food Res Int . 2012; 46(1):30-40.; Carvalho et al., 2017Carvalho DC, Guedes D, Trindade MG, Coelho RMS, Araujo PHL. Nationwide Brazilian governmental forensic programme reveals seafood mislabelling trends and rates using DNA barcoding. Fisheries Research. 2017; 191:30-35.). Such an assessment is crucial to alert the relevant authorities regarding the need for effective measures of organization, standardization, and surveillance of the fishery sector for combating replacement practices and safeguarding consumer rights. Despite the issues found throughout the supply chain involving the Caribbean Red Snapper, which is a key fishery resource in Brazil, no research study has been conducted to date, exclusively aimed to assess the authenticity of labeling in fillets of this species.
Considering the existence of numerous fraud reports regarding snappers (Marko et al., 2004Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS et al. Mislabelling of a depleted reef fish. Nature. 2004; 430:309-310.; Logan et al., 2008Logan CA, Alter E, Haupt AJ, Tomalty K, Palumbi SR. An impediment to consumer choice: Overfished species are sold as Pacific red snapper. Biol Conserv . 2008; 141(6):1591-1599.; Jacquet, Pauly 2008Jacquet JL Pauly D. Trade secrets: Renaming and mislabeling of seafood. Mar Policy. 2008; 32:309-18.; Wong, Hanner, 2008Wong EHK, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Research International. 2008; 41(8):828-37.; Hellberg, Morrissey 2011Hellberg RS, Morrisey MT. Advances in DNA-based techniques for the detection of seafood species substitution on the commercial market. Journal Lab Autom. 2011; 16(4):308-21.; Warner et al., 2013Warner K, Timme W, Lowell B, Hirshfield M. Oceana Study Reveals Seafood Fraud Nationwide. Oceana; 2013.; Stiles et al., 2013Stiles ML, Kagan A, Lahr HJ, Pullekines E, Walsh A. Seafood Sticker: Shock Why you may be paying too much for your fish. Oceana; 2013.) and knowing the efficacy of COI gene as an applicable bioidentification tool to the authentication of Lutjanidae species (Veneza et al., 2014Veneza I, Felipe B, Oliveira J, Silva R, Sampaio I, Schneider H et al. A barcode for the authentication of the snappers (Lutjanidae) of the western Atlantic: rDNA 5S or mitochondrial COI? Food Control . 2014; 38:116-23.); here, DNA barcode tool was used to determine which species are being traded under the popular designation “Pargo” and thereby verify the occurrence of commercial fraud.
Material and Methods
Sampling. The samples were fillets labeled as “Pargo” processed by a single supplier and marketed as frozen fillets with skin, in supermarkets in the State of Pará, North Brazil. A single supplier was used because it is the only one that makes the processing of this species in the region. The sampling was conducted from March 2013 to October 2014, during this period were bought 22 packages with a variable number of fillets, usually from five to eight units per package, weighing 500 g or 1 Kg. In total, 142 samples from nine different lots were obtained.
One muscle tissue sample was collected from each fillet piece, and these samples were individually stored in microtubes containing 90% alcohol, received a registration code and stored at -20°C. Only products with original packaging from the fillet supplier company were analyzed.
Ethics Statement. All the tissues used here were obtained from fillets. During the sampling, the species were not endangered or protected along the Brazilian Coast. Therefore, there was no need to apply for a license for collection or approval by the Animal Ethics Committee.
Isolation, amplification and sequencing of genetic material. Total genomic DNA was isolated using the Wizard Genomic® Kit (PROMEGA), according to the manufacturer’s instructions. Following DNA isolation, the samples were mixed on a solution of buffer and GelRed dye “blue juice” (2 μL of the mixture and 2 μL of DNA), then, loaded into 1% agarose gels and electrophoresed for 30 minutes/60V. After that, the samples were exposed under ultraviolet light to assess the quality of the extracted DNA.
The COI gene fragment was amplified by the Polymerase Chain Reaction (PCR) method with COIFishF2, and COIFishR2 primers (Ward et al., 2005Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005; 360(1462):1847-57.). The reactions were conducted in a final volume of 15 μL containing 2.4 μL DNTP (1.25 mM), 1.5 μL buffer (10x), 0.6 μL MgCl2 (50 mM), 0.6 μL of each primer (50 ng/μL), approximately 100 ng total DNA, 0.1 μL Taq DNA polymerase (5 U/μL), and ultrapure water to complete the final reaction volume. The following amplification conditions were: an initial denaturation at 95ºC for 3 minutes; followed by 35 cycles of 40 seconds at 94°C, 40 seconds at 55ºC, and 1 minute at 72ºC; and a final extension of 3 minutes at 72º C. The positive PCRs were sequenced using the dideoxy-terminal method (Sanger et al., 1977Sanger F, Nichlen S, Coulson AR. DNA Sequencing with Chain-Termination Inhibitors. Proceeding of the National Academy of Science of the USA. 1977; 74(12):5463-68.), with the Big Dye Kit (ABI PrismTM Dye Terminator Cycle Sequencing Reading Reaction), and employing an ABI 3500 XL automated capillary sequencer (Life Technologies).
Data analysis. The sequences were edited with the BioEdit software (Hall, 1999Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symp ser. 1999; 41:95-98.), and aligned using the parameter default of ClustalW (Thompson et al., 1994Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994; 22(22):4673-80.) available in BioEdit. After creating a database of fillet sequences, some corrections were performed manually in sequence positions with errors or doubts regarding the nucleotide present.
A list of haplotypes was generated using the DnaSP5 software (Librado, Rozas, 2009Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25(11):1451-52.) to guide the samples identification process. The haplotype sequences listed were compared with barcode fragments available in GenBank (National Center for Biotechnology Information - http://www.ncbi.nlm.nih.gov), via the BLASTn Search Tool (Altschul et al., 1997Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25 (17):3389-402.) and BOLD (Barcoding of Life Database - http://www.barcodinglife.org), using the Species Level database (Ratnasingham, Hebert, 2007Ratnasingham S, Hebert PDN. BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecology Notes. 2007; 7:355-64.). Only comparisons returning from the GenBank/BOLD search with values of similarity ≥ 99% were considered for further assessment.
Furthermore, the COI sequences of fillets were also compared with a control database consisting of sequences from several snappers, including L. purpureus and eight other potential replacement species of Lutjanidae (L. synagris, L. analis, L. jocu, L. buccanella, L. vivanus, L. cyanopterus, Ocyurus chrysurus, and Rhomboplites aurorubens), previously identified morphologically using specialized literature (Cervigón et al., 1993Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al. FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.).
The database was analyzed using the MEGA 7 software (Kumar et al. 2016Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016; 33:1870-74.), wherein a neighbor-joining (NJ) tree was constructed with the Kimura 2-parameter (K2P) model of nucleotide substitution (Kimura, 1980Kimura MA. Simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111-20.). The significance of the tree clusters was estimated using a bootstrap analysis, generated from 1,000 pseudoreplicates. Conodon nobilis (Haemulidae-Perciformes) sequences from GenBank (Accession codes: KF633261 and KF633260) was included in the database to root the tree. A corrected distance matrix was generated to assess intra and interspecific genetic distances, using the K2P evolution model and the MEGA 7 software. The polymorphic sites were identified, and then were used to evaluate the mutations that differ the species in order to corroborate the clustering pattern found in the phylogenetic tree and which is observed in the genetic distance values.
Results
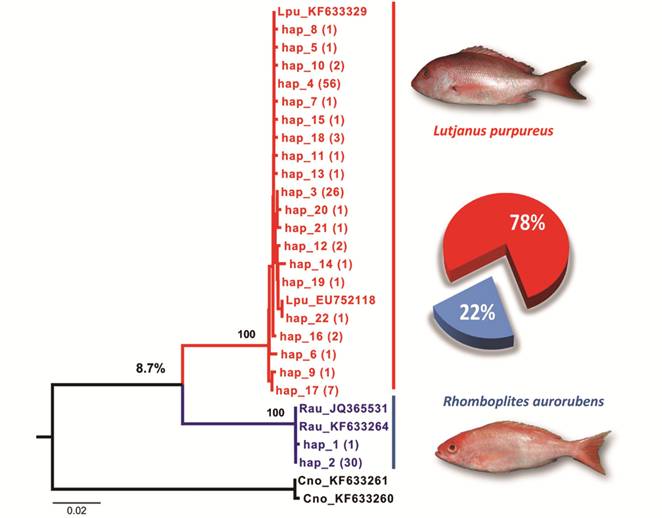
A 600-bp fragment corresponding to the barcode portion of COI gene was sequenced from the 142 fillet samples. The list of haplotypes created revealed three most common haplotypes, of which, were shared by approximately 80% of fillet samples. The other samples were all unique haplotypes (Fig. 1). The sequences data of all haplotypes have been submitted to the GenBank databases under accession numbers KU313734 - KU313755. Regarding the comparisons performed against the GenBank sequences and the control database, partitioning into two groups was observed, being the first one group formed by the most fillet sequences and the L. purpureus controls, and the second one formed by the controls of the Rhomboplites aurorubens species and remaining fillets. For L. purpureus fillets were observed 20 haplotypes, and only two for R. aurorubens.
Neighbor-joining (NJ) tree constructed from a 600-bp of the COI gene, showing two clades, which indicates the presence of two species among the fillets labeled as “Pargo”; the nucleotide divergence between the species (the value of 8.7%) is indicated above the outermost node. Within parentheses the frequency of each haplotype is described and the replacement rate present in the fillets is shown in plot to the right.
Similarly, to the list generated by DNAsp, the NJ tree clearly showed the formation of two groups, two clades statistically well supported, one clustering most samples with known sequences of the L. purpureus species (Haplotypes - EU752118; KF633329) and another cluster formed by 22% of fillets and control sequences of Rhomboplites (Haplotypes - JQ365531; KF633264) in addition to the outgroup (Fig. 1). Most packages sampled consisted of L. purpureus fillets, with two packages and replacement of one or two fillet pieces, also, three of these packages with their content completely replaced. Such replacements were observed under two different lots.
The genetic divergence calculated based on the comparison among and within the groups observed in the phylogenetic tree confirmed the presence of two species among all fillets analyzed. The mean interspecific distance was 8.7%, which is much higher than the value found for the mean intraspecific distance of merely 0.3%. The genetic distance among haplotypes ranged from 0.2% to 0.8% for L. purpureus and 0.2% for R. aurorubens. The presence of 60 variable sites, corresponded to 10% of polymorphism.
Discussion
Accidental Replacement or Commercial Fraud? The results together show the sold fillets as “Pargo” include the species L. purpureus, and another snapper species, namely R. aurorubens, suggesting commercial frauds. The IN/N° 29/2015 of MAPA established common names for the main species of fishes marketed in Brazil, in order to be inserted in packaging labels of the processed products. According to this governmental list, “Pargo” should be used only for two lutjanids: L. purpureus and L. campechanus.
The analysis of de Brito et al. (2015Brito MA, Schneider H, Sampaio I, Santos S. DNA barcoding reveals high substitution rate and mislabeling in croaker fillets (Sciaenidae) marketed in Brazil: The case of “pescada branca” (Cynoscion leiarchus and Plagioscion squamosissimus). Food Res Int. 2015; 70:40-46.) about authentication of croaker fillets (Sciaenidae) reported alarming results with high replacement rates ranging from 76.6 - 100%. Among the replaced species, there are some specimens from Serranidae and Lutjanidae families, including L. purpureus.
Numerous taxa of the family Lutjanidae are known to share morphological traits (Allen, 1985Allen G-R. Snappers of the World. An annotated and illustrated catalogue of Lutjanid species known to date. Rome: FAO; 1985.; Cervigón et al., 1993Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al. FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.; Moura, Lindeman, 2007Moura RL, Lindeman KC. A new species of snapper (Perciformes: Lutjanidae) from Brazil, with comments on the distribution of Lutjanus griseus and L. apodus. Zootaxa. 2007; 1422(3):31-43.), which hinders their identification exclusively based on anatomical traits. Several fish from that group are targeted by commercial fisheries in the Northern and Northeastern Brazil, including L. purpureus, L. synagris, L. analis, and Ocyurus chrysurus (MPA, 2010Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura Brasil 2008-2009; 2010.; 2012Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2010; 2012.; 2013Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2011; 2013.). These species have a sympatric widely distribution in the Western Atlantic (Cervigón et al., 1993Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al. FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.) and, therefore, are most likely to be caught together. However, Caribbean Red Snapper, L. purpureus is not only the most consumer-appreciated, and the most caught snapper, but also has the highest economic relevance in Brazil. Moreover, it may be sold fresh, whole gutted or frozen fillets. In case of frozen fillets, it’s impossible to recognize morphological features, leading to be an aggravating factor to the occurrence of replacement between species.
Additionally, R. aurorubens comprises enough different morphometric characteristics to build a monotypic genus, namely eye shape, caudal fin, number of dorsal spines, and stripes (Moura, Lindeman, 2007Moura RL, Lindeman KC. A new species of snapper (Perciformes: Lutjanidae) from Brazil, with comments on the distribution of Lutjanus griseus and L. apodus. Zootaxa. 2007; 1422(3):31-43.). All these differences become subtle when superficially compared to L. purpureus specimens, with both species being easily confused due to similarities in color pattern, overlapping morphometric traits, and habitat (Allen, 1985Allen G-R. Snappers of the World. An annotated and illustrated catalogue of Lutjanid species known to date. Rome: FAO; 1985.; Cervigón et al., 1993Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al. FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.; Moura, Lindeman, 2007Moura RL, Lindeman KC. A new species of snapper (Perciformes: Lutjanidae) from Brazil, with comments on the distribution of Lutjanus griseus and L. apodus. Zootaxa. 2007; 1422(3):31-43.). The species used for replacement have small sizes and low commercial values in Brazil, and they are usually caught accidentally with other snappers.
Other studies have already reported species replacements among snappers. Marko et al. (2004Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS et al. Mislabelling of a depleted reef fish. Nature. 2004; 430:309-310.) using a fragment of Cyt b gene detected that approximately 80% of the fish sold as L. campechanus in eight US states truly correspond to another species from the same family, all with lower commercial importance than red snapper. Wong, Hanner (2008Wong EHK, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Research International. 2008; 41(8):828-37.) found similar results, using DNA barcode, while identifying seafood from North American markets. These authors demonstrated that 25% of the samples analyzed were misidentified, which included replacements within Lutjanidae.
All sampling packages had only included vernacular names of the species on their labels, of which, are often confused by others. This lack of information on the labels reflect on the great variety of common names among regions. Beyond that, although illegal, the terms “Caribbean Red Snapper” or “Red Snapper” are commonly used as a category, referring to a set of similar snappers.
Vernacular names are shared by several species in the official Brazilian fishery records. The concern is even greater when noticing the existence of snappers listed without confirmed taxonomy in the fish landing data, which are therefore grouped as Lutjanus spp., a term referring to four different common names at least, namely, Cubera Snapper (“Caranha”) - Lutjanus griseus, Lutjanus jocu, Mutton Snapper (“Caranha-vermelha”) - Lutjanus analis, Lutjanus vivanus, Brazilian Snapper (“Carapitanga”) - Lutjanus alexandrei, Lutjanus cyanopterus, and Caribbean Red Snapper (“Pargo/Vermelho”) - Lutjanus purpureus (MPA, 2010Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura Brasil 2008-2009; 2010.; 2012Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2010; 2012.; 2013Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2011; 2013.).
Ardura et al. (2010Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv. 2010; 143 (6):1438-43.) have discussed the issues caused by the lack of regulation of common names of species sold in the Amazon. Using the 12S rDNA and COI as molecular markers, these authors identified fish purchased in local marketplaces and concluded that the group generically sold as “acará” encompasses seven different species, which compromises the estimates of exploitation and conservation of Amazonian fishery resources.
Thus, similar features between L. purpureus and R. aurorubens, their overlapped distribution, and the fact they are caught together may explain the replacement cases reported herein. However, misidentifications can easily occur considering aggravating factors (i.e., common names without standardization, wrong statistical records or practices of marketing), and yet, they can still suggest that these replacements may not be intentional in all cases.
On the other hand, some results still became quite questionable, for example, even though the most of the replaces were not significant (either one or two fillet pieces from the total present in a package), other packages had their contents substituted completely.
Furthermore, the use of the species R. aurorubens as a substitute of L. purpureus entails high economic benefits to the suppliers, due to this species holds lower economic value than the species stated in the label or any species closer related to “Caribbean Red Snapper” in Brazil.
For the last, the absence of scientific names in the labels seems to be an attempt to camouflage the irregularity of this practice, since the term “Pargo”, as discussed above, is applied to innumerous snappers, including R. aurorubens.
Replacement activities had not only negative implications for fishery management, but for the consumer as well, resulting in economic losses. So, an alternative should be to apply the labeling of products in accordance with legal requirements, avoiding these problems. Nonetheless, there is not in Brazilian any law that requires the inclusion of the scientific names in the package of processed fish products, except for species of Salmonidae and Gadidae.
Considering the findings of this current analysis, the unambiguous identification of the species, should not be limited simply to morphology, which is a requirement to prevent possible causes of replacement practices. Thus, our results suggest a need to include other methodologies to identify the products at species level, and that the tools based on DNA have proved to be useful and efficient for this type of study, especially, the barcode region (COI gene), which has demonstrated to be very powerful and sensitive for authenticating and certification of processed fish products.
Concluding Remarks. Despite substantial relevance of the species L. purpureus for economic and fishing scenarios, it was the first study using the molecular tool (DNA barcoding) to authenticate fish fillets marketed as “pargo” in supermarkets along the north of Brazil. The rate of substitution was 22%, and some factors lead us to suggest intentional substitutions or commercial fraud, especially, when related to the R. aurorubens - species with little relevance from the fishery standpoint.
Regarding labeling issues, companies that supply food products in general, and the fillets sampled herein, they must keep their informative labels handy, toward ensuring consumer rights. Thus, the present findings are warning about these unwanted activities, as they could justify regulatory and surveillance measures by the competent agencies.
Acknowledgments
The authors are grateful for the financial assistance received from the Brazilian National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq), which funded this study through a MSc Scholarship and bench fees (Grants 306233/2009-6 to IS, 306233/2009-6 to HS).
References
- Allen G-R. Snappers of the World. An annotated and illustrated catalogue of Lutjanid species known to date. Rome: FAO; 1985.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25 (17):3389-402.
- Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv. 2010; 143 (6):1438-43.
- Brito MA, Schneider H, Sampaio I, Santos S. DNA barcoding reveals high substitution rate and mislabeling in croaker fillets (Sciaenidae) marketed in Brazil: The case of “pescada branca” (Cynoscion leiarchus and Plagioscion squamosissimus). Food Res Int. 2015; 70:40-46.
- Carvalho DC, Guedes D, Trindade MG, Coelho RMS, Araujo PHL. Nationwide Brazilian governmental forensic programme reveals seafood mislabelling trends and rates using DNA barcoding. Fisheries Research. 2017; 191:30-35.
- Cawthorn DM, Steinman HA, Witthuhn RC. DNA barcoding reveals a high incidence of fish species misrepresentation and substitution on the South African Market. Food Res Int . 2012; 46(1):30-40.
- Cervigón F, Cipriani R, Fischer W, Garibaldi L, Hendrickx M Lemus AJ. et al FAO species identification sheets for fishery purpose. Field guide to the comercial marine and brackish-water resources of the northern coast of South America. Rome: FAO ; 1993.
- Cohen J, Deeds JE, Wong E, Hanner RH, Yanci HF, White KD et al Public health response to puffer fish (Tetrodotoxin) poisoning from mislabeled product. J Food Protect. 2009; 72(4):810-17.
- Cutarelli A, Amoroso MG, De Roma A, Girardi S, Galiero G, Guarino A et al Italian market fish species identification and commercial frauds revealing by DNA sequencing. Food Control. 2014; 37:46-50.
- Di Pinto A, Di Pinto P, Terio V, Bozzo G, Bonerba E, Ceci E et al DNA barcoding for detecting market substitution in salted cod fillets and battered cod chunks. Food Chem. 2013; 141(3):1757-62.
- Dias Neto J, Dornelles LDC. Diagnóstico da pesca marítima do Brasil. Brasília: IBAMA; 1996.
- Filonzi L, Chiesa S, Vaghi M, Marzano FN. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res Int . 2010; 43(5):1383-88.
- Food and Agriculture Organization FAO. (2016). The state of world fisheries and aquaculture. Rome: FAO ; 2016.
- Guallar E, Sanz-Gallardo I, Veer PV, Bode P, Aro A, Gómez-Aracena J et al Mercury, fish oils, and the risk of myocardial infarction. N Engl J Med. 2002; 347 (22):1747-54.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symp ser. 1999; 41:95-98.
- Hellberg RS, Morrisey MT. Advances in DNA-based techniques for the detection of seafood species substitution on the commercial market. Journal Lab Autom. 2011; 16(4):308-21.
- Jacquet JL Pauly D. Trade secrets: Renaming and mislabeling of seafood. Mar Policy. 2008; 32:309-18.
- Kimura MA. Simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111-20.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016; 33:1870-74.
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25(11):1451-52.
- Logan CA, Alter E, Haupt AJ, Tomalty K, Palumbi SR. An impediment to consumer choice: Overfished species are sold as Pacific red snapper. Biol Conserv . 2008; 141(6):1591-1599.
- Maralit BA, Aguila RD, Ventolero MFH, Perez SKL, Willette DA, Santos MD. Detection of mislabeled commercial fishery by-products in the Philippines using DNA barcodes and its implications to food traceability and safety. Food Control . 2013; 33(1):119-25.
- Marko PB, Lee SC, Rice AM, Gramling JM, Fitzhenry TM, McAlister JS et al Mislabelling of a depleted reef fish. Nature. 2004; 430:309-310.
- Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura Brasil 2008-2009; 2010.
- Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2010; 2012.
- Ministério da Pesca e Aquicultura MPA. Boletim estatístico da pesca e aquicultura, Brasil 2011; 2013.
- Moura RL, Lindeman KC. A new species of snapper (Perciformes: Lutjanidae) from Brazil, with comments on the distribution of Lutjanus griseus and L. apodus Zootaxa. 2007; 1422(3):31-43.
- Nelson JS, Grande TC, Wilson MVH. Fishes of the World. 5th ed. New Jersey: John Wiley & Sons; 2016.
- Paiva MP. Recursos pesqueiros estuarinos e marinhos do Brasil. Fortaleza: UFC Edições; 1997.
- Palmeira CAM, Rodrigues-Filho LFS, Sales JBL, Vallinoto M, Schneider H, Sampaio I. Commercialization of a critically endangered species large tooth sawfish, Pristis perotteti in fish markets of northern Brazil: authenticity by DNA analysis. Food Control . 2013; 34(1):249-252.
- Ratnasingham S, Hebert PDN. BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecology Notes. 2007; 7:355-64.
- Sanger F, Nichlen S, Coulson AR. DNA Sequencing with Chain-Termination Inhibitors. Proceeding of the National Academy of Science of the USA. 1977; 74(12):5463-68.
- Souza RFC. Dinâmica Populacional do pargo, Lutjanus purpureus Poey, 1875 (Pisces: Lutjanidae) na Plataforma Norte do Brasil. [Dissertação de mestrado]. Belém, PA: Universidade Federal do Pará; 2002.
- Stiles ML, Kagan A, Lahr HJ, Pullekines E, Walsh A. Seafood Sticker: Shock Why you may be paying too much for your fish. Oceana; 2013.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994; 22(22):4673-80.
- Van Leeuwen SPJ, Van Velzen MJM, Swart CP, Van Der Veen I, Traag WA, De Boer J. Halogenated contaminants in farmed salmon, trout, tilapia, pangasius, and shrimp. Environmental science & technology. 2009; 43(11):4009-15.
- Veneza I, Felipe B, Oliveira J, Silva R, Sampaio I, Schneider H et al A barcode for the authentication of the snappers (Lutjanidae) of the western Atlantic: rDNA 5S or mitochondrial COI? Food Control . 2014; 38:116-23.
- Ward RD. Genetics in fisheries management. Hydrobiologia. 2000; 420:191-201.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005; 360(1462):1847-57.
- Warner K, Timme W, Lowell B, Hirshfield M. Oceana Study Reveals Seafood Fraud Nationwide. Oceana; 2013.
- Wong EHK, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Research International. 2008; 41(8):828-37.
Publication Dates
-
Publication in this collection
2018
History
-
Received
28 May 2017 -
Accepted
03 Jan 2018