ABSTRACT
Characiformes is an order of freshwater fishes that includes many commercially important and emblematic species from South America and Africa, such as the popular piranhas, hatchetfishes, African tiger fishes and tetras. The order is split into two suborders with a total of 24 families, 282 genera and ca. 2,100 species. Here, we present an expanded phylogeny of characiform fishes, including data for 520 species and three genes (12S, 16S and RAG1), and the recently described family Tarumaniidae, which has not been examined by previous molecular analysis. Although our genetic coverage is limited to three gene fragments, the tree inferred based on maximum likelihood and Bayesian inference supports the monophyly of all characiform families and is largely congruent with results from recent studies that sampled less taxa but more genes. Also in agreement with a morphological hypothesis, our results strongly support the sister-group relationships between the family Tarumaniidae and Erythrinidae. Based on our results and that of the other molecular analyses, we propose a revised circumscription of the superfamily Erythrinoidea to include the families Tarumaniidae and Erythrinidae only.
Keywords:
Central Amazon; Erythrinoidea; Freshwater fishes; Ostariophysi; Tarumania
RESUMEN
Characiformes es un orden de peces de agua dulce que incluye un gran numero de especies emblemáticas y de importancia comercial en Sur América y África como lo son las populares pirañas, los peces voladores, los peces tigre de África y los tetras. El orden se divide en dos subórdenes con un total de 24 familias, 282 géneros y cerca de 2100 especies. Aquí, presentamos una filogenia expandida de Characiformes, que incluye datos de 520 especies, tres genes (12S, 16S y RAG1) y la recientemente descrita familia Tarumaniidae, la cual no ha sido examinada en previos análisis moleculares. Aunque nuestra cobertura genética esta limitada a tres genes, el árbol inferido basado en máxima verosimilitud e inferencia bayesiana apoya la monófila de todas las familias de Characiformes y es en gran medida congruente con los resultados de estudios recientes que examinaron menos especies pero más genes. También de acuerdo con una hipótesis morfológica, nuestros resultados apoyan firmemente las relaciones de grupos hermanos entre las familias Tarumaniidae y Erythrinidae. Con base en nuestros resultados y el de otros estudios moleculares, proponemos una circunscripción revisada de la superfamilia Erythrinoidea que incluye solo a las familias Tarumaniidae y Erythrinidae.
Palabras claves:
Amazonia Central; Erythrinoidea; Peces de agua dulce; Ostariophysi; Tarumania
Introduction
Characiformes are confined to freshwater basins in the Neotropical (South to Central America) and Paleotropical (Africa) regions (Eschmeyer et al., 2017Eschmeyer WN, Fricke R, van der Laan R. Catalog of fishes: genera, species, references [Internet]. San Francisco: California Academy of Science; 2017. Available from: http://researcharchive.calacademy.org
http://researcharchive.calacademy.org...
). The highest diversity of characiforms occurs in South America, with ca. 1,600 species in 20 families, compared to 500 species in four families that live in Africa. The past 10 years have seen considerable progress in resolving the phylogeny of characiforms using molecular data. Oliveira et al. (2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
) presented the relationships of 211 species based on three nuclear and two mitochondrial genes, proposing a revised classification for the family Characidae. Arcila et al. (2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.) inferred a phylogenomic tree based on 1,051 exon markers obtained from 110 characiform species along with 115 otophysan outgroups, supporting the monophyly of the order. Despite substantial differences in the datasets examined, these recent studies produced largely congruent results (Oliveira et al., 2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
; Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.).
The discovery of the so-called “mystery fish” took the ichthyology community by surprise when a juvenile specimen was first collected along a tributary of the Amazon over 15 years ago. Because of its distinct morphology with an anguilliform body shape, the apparent capacity for aerial breathing, and its multi-chambered swimbladder, early assessments concluded that the species was undescribed and its placement among Neotropical taxa remained unclear (de Pinna et al., 2017de Pinna M, Zuanon J, Rapp Py-Daniel L, Petry P. A new family of neotropical freshwater fishes from deep fossorial Amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool J Linnean Soc [serial on the Internet]. 2017; 182(1):76-106. Available from: https://doi.org/10.1093/zoolinnean/zlx028
https://doi.org/10.1093/zoolinnean/zlx02...
). Since its discovery, ca. 85 additional individuals have been collected and last year the species was formally described (Tarumania walkerae de Pinna, Zuanon, Rapp Py-Daniel & Petry, 2017) in a new monotypic family (Tarumaniidae). The original description also includes a first attempt to resolve its phylogenetic placement based on the examination of 128 morphological characters and 35 species, supporting its sister group relationships with the characiform family Erythrinidae, in the superfamily Erythrinoidea (de Pinna et al., 2017de Pinna M, Zuanon J, Rapp Py-Daniel L, Petry P. A new family of neotropical freshwater fishes from deep fossorial Amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool J Linnean Soc [serial on the Internet]. 2017; 182(1):76-106. Available from: https://doi.org/10.1093/zoolinnean/zlx028
https://doi.org/10.1093/zoolinnean/zlx02...
). Three undisputed morphological synapomorphies based on the round caudal fin, reduced coracoid and reduction of the caudal fin rays along with other circumstantial characters strongly supported the closest relationships between Tarumaniidae and Erythrinidae (de Pinna et al., 2017de Pinna M, Zuanon J, Rapp Py-Daniel L, Petry P. A new family of neotropical freshwater fishes from deep fossorial Amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool J Linnean Soc [serial on the Internet]. 2017; 182(1):76-106. Available from: https://doi.org/10.1093/zoolinnean/zlx028
https://doi.org/10.1093/zoolinnean/zlx02...
). The restricted distribution of Tarumaniidae to tributaries of the Rio Negro, Central Amazon has made collecting tissue samples for molecular analyses a major challenge, being among the few fish families without sequence data available in any public database. Assessing the phylogenetic placement of Tarumaniidae in a comprehensive phylogenetic context is critical to develop a framework for studying adaptations of fishes that survive in extreme environments such as temporary freshwater isolated pools and to shed light on the evolution of highly derived morphological adaptations such as the anguilliform body shape, aerial breathing and multi-chambered swimbladder (apparently unique among fishes).
In this study, we provide a first attempt to place the family Tarumaniidae in a phylogenetic framework based on analyses of DNA data (two mitochondrial and one nuclear gene fragments) and a dense taxonomic sampling. We analyze its placement in a large data matrix including 520 species of characiforms, using both maximum likelihood and Bayesian analyses. Although a few aspects of characiform phylogeny remain poorly resolved, the new phylogenetic tree is consistent with previous hypothesis and strongly supports Tarumaniidae as the sister group of Erythrinidae.
Material and Methods
Taxon sampling and DNA extractions. The diversity of characiform fishes is represented in this study by 520 species including all 24 valid characiform families and 202 (out of 284) genera (S1 - Available only as online supplementary file accessed with the online version of the article at http://www.scielo.br/ni). Tissue samples of Tarumania were obtained from an adult specimen deposited at Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil (INPA 21603, Paraná do Prato, Anavilhanas, Rio Negro, Municipality Novo Airão, AM, 20 Aug 2001). DNA was extracted using the DNesay tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Double-stranded DNA was synthesized via PCR using universal primers for two mitochondrial (ribosomal 16S and 12S rRNA subunits; Kocher et al., 1989Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989; 86(16):6196-200.; Palumbi et al., 1991Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The Simple Fool’s Guide to PCR, Version 2. Honolulu: University of Hawaii Zoology Department; 1991.) and a nuclear marker (recombination-activation gene 1 or RAG-1; Lopez et al., 2004Lopez JA, Chen WJ, Orti G. Esociform phylogeny. Copeia. 2004; 2004(3):449-64.). The PCR products were electrophoresed in 1.5% low melting point agarose gels using a tris-acetate buffer for quality control and submitted for purification and sequencing in both directions to High Throughput Sequencing Solutions (HTSeq.org), University of Washington, Seattle, Washington. Sequences were assembled using Sequencer v4.8 (Gene Codes Corporation, Ann Arbor, MI, USA) and aligned manually with Geneious v8.1.8. All sequences produced by this study have been deposited [GenBank: MH667495 (12S), MH667496 (16S), MH667497 (RAG-1)]. Additional sequence data examined herein were previously published in other studies (e.g., Oliveira et al., 2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
; Melo et al., 2014Melo BF, Sidlauskas B, Hoekzema K, Vari RP, Oliveira C. The first molecular phylogeny of Chilodontidae (Teleostei: Ostariophysi: Characiformes) reveals cryptic biodiversity and taxonomic uncertainty. Mol Phylogenet Evol [serial on the Internet]. 2014; 70:286-95. Available from: http://dx.doi.org/10.1016/j.ympev.2013.09.025
http://dx.doi.org/10.1016/j.ympev.2013.0...
; Thomaz et al., 2015Thomaz AT, Arcila D, Orti G, Malabarba L. Molecular phylogeny of the subfamily Stevardiinae Gill, 1858 (Characiformes: Characidae): classification and the evolution of reproductive traits. BMC Evol Biol [serial on the Internet]. 2015; 15(146):1-25. Available from: https://doi.org/10.1186/s12862-015-0403-4
https://doi.org/10.1186/s12862-015-0403-...
; Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.; Lavoue et al., 2017Lavoue S, Arnegard M, Rabosky D, McIntyre P, Arcila D, Vari R, Nishida M. Trophic evolution in African citharinoid fishes (Teleostei: Characiformes) and the origin of intraordinal pterygophagy. Mol Phylogenet Evol [serial on the Internet]. 2017; 113:23-32. Available from: http://dx.doi.org/10.1016/j.ympev.2017.05.001
http://dx.doi.org/10.1016/j.ympev.2017.0...
) and retrieved from GenBank (S1 - Availa ble only as online supplementary file accessed with the online version of the article at http://www.scielo.br/ni).
Phylogenetic analyses. Maximum likelihood tree searches were conducted with RAxML v8.2.10 (Stamatakis, 2006Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics [serial on the Internet]. 2006; 22(21):2688-90. Available from: https://doi.org/10.1093/bioinformatics/btl446
https://doi.org/10.1093/bioinformatics/b...
) executing ten separate searches with different starting trees under the GTRGAMMA model. Bootstrap analyses using 1000 pseudo replicates were performed to assess the statistical support of clades resulting from the phylogeny. For each gene, preliminary analyses were conducted using RAxML to detect possible cases of GenBank data contamination or species misidentification. When a representative of one species had an identical sequence to another, that sequence was excluded for downstream analyses. For the concatenation analyses, we used a partitioning scheme with four blocks: one block for the mitochondrial 12S and 16S markers, and three partitions for each of the codon position of RAG1. Following recent results based on morphology (Fink, Fink, 1981Fink SV, Fink WL. Interrelationships of the ostariophysan fishes. Zool J Linnean Soc. 1981; 72:297-353.) and phylogenomics (Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.), all trees were rooted with the African citharinoids.
Bayesian analysis were carried out using MrBayes v3.2.5 (Ronquist et al., 2012Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol [serial on the Internet]. 2012; 61(3):539-42. Available from: https://doi.org/10.1093/sysbio/sys029
https://doi.org/10.1093/sysbio/sys029...
) using two independent runs, with 20 Markov Chain Monte Carlo (MCMC) chains for each run. The same partitioned model implemented in RAxML was used in MrBayes. The heating parameters were set as “temp = 0.1” and “nswaps = 5” to improve the mixing of the MCMC chains. The runs were terminated after 10 million generations and checked for stationarity based on ESS values for all parameters being greater than 200. Parameter states were sampled every 1,000 generations (10,000 trees saved for each run). The majority rule 50% consensus tree was summarized from the sampled trees after discarding the burn-in samples before reaching stationarity. All trees obtained in this study are available at Figshare repository (doi: 10.6084/m9.figshare.5852844).
Topology tests were conducted to assess whether the monophyly of the traditionally recognized superfamily Erythrinoidea proposed by Buckup (1998Buckup PA. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleoste: Ostariophysi). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of Neotropical fishes. Porto Alegre: Edipucrs; 1998. p.123-144.) is consistent with the new data. We used RAxML to obtain maximum likelihood phylogenies constrained to enforce Buckup’s Erythrinoidea hypothesis. The unconstrained ML tree (supporting the non-monophyly of Erythrinoidea), was compared to the ML topology obtained by enforcing the monophyly of each circumscription of Erythrinoidea challenged by our results. For each of the analyses, the best trees of 10 independent searches were selected. To evaluate the differences in likelihood scores between constrained and unconstrained tree topologies, the site likelihood scores were extracted using RAxML and compared with the AU (approximately unbiased) test implemented in the program Consel (Shimodaira, 2002Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol . 2002; 51(3):492-508.).
Results and Discussion
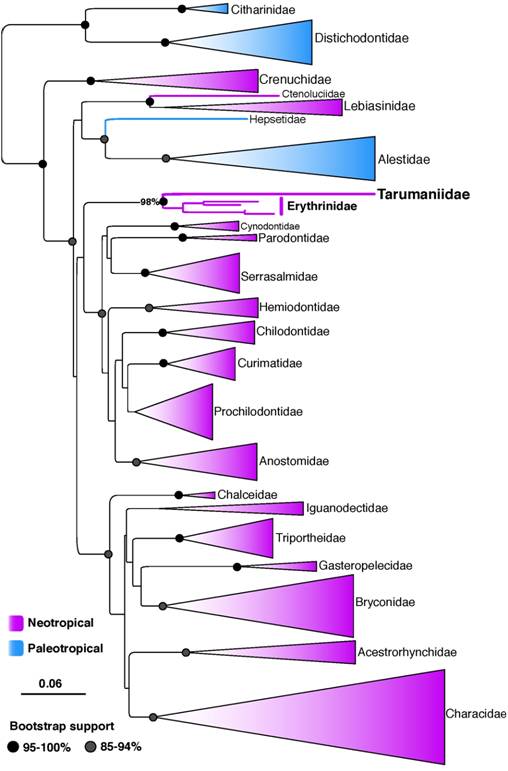
In this study, we present an expanded phylogeny for characiform fishes based on maximum likelihood and Bayesian analyses of 520 characiform species - including all valid families - to assess the phylogenetic placement of Tarumaniidae. Despite the modest amount of markers examined, higher-level relationships among major lineages of Characiformes resolved by our analyses (Fig. 1) are similar to those reported by previous multi-locus or phylogenomic studies (Oliveira et al., 2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
; Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.). We find strong support for the monophyly of all characiform families. The major discrepancy between our results and previous ones is the placement of the families Alestidae + Hepsetidae more closely related to Ctenoluciidae + Lebiasinidae (Fig. 1), but this placement was supported by low bootstrap values (45%) in our study.
Maximum likelihood tree resolving the phylogenetic placement of the family Tarumaniidae among 520 characiform species. Scale bar = 0.06 expected substitutions per position as estimated by RAxML.
The multi-locus trees as well as the individual gene trees inferred with mitochondrial (concatenated 12S and 16S) and nuclear (RAG1) data resolve Tarumaniidae as the sister group to Erythrinidae with strong bootstrap support (98 to 100%, Fig. 1; see also online supplementaries files S1, S2 and S3 - Availa ble only as online supplementary file accessed with the online version of the article at http://www.scielo.br/ni). However, our results do not support the monophyly of the superfamily Erythrinoidea following the circumscription proposed by Buckup (1998Buckup PA. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleoste: Ostariophysi). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of Neotropical fishes. Porto Alegre: Edipucrs; 1998. p.123-144.), which include the families Lebiasinidae, Erythrinidae, Hepsetidae and Ctenoluciidae, and more recently Tarumaniidae (de Pinna et al., 2017de Pinna M, Zuanon J, Rapp Py-Daniel L, Petry P. A new family of neotropical freshwater fishes from deep fossorial Amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool J Linnean Soc [serial on the Internet]. 2017; 182(1):76-106. Available from: https://doi.org/10.1093/zoolinnean/zlx028
https://doi.org/10.1093/zoolinnean/zlx02...
). This circumscription of Erythrinoidea proposed by Buckup (1998Buckup PA. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleoste: Ostariophysi). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of Neotropical fishes. Porto Alegre: Edipucrs; 1998. p.123-144.) based on 80 morphological characters and 27 species also was strongly rejected by the AU test (P-value = 0.00003).
Other studies suggested a sister group relationship between Lebiasinidae and Ctenoluciidae and a clade containing Chalceidae and Characidae (among other families) but excluding Erythrinidae (Oliveira et al., 2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
; Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.). Hepsetidae, on the other hand has been shown to be closely related to Alestidae. The placement of the Erythrinidae + Tarumaniidae clade among other families of characiforms is poorly resolved, but our study and three recent comprehensive molecular phylogenies suggest a close relationship of these two families with a big clade containing Anostomoidea, Serrasalmidae, Hemiodontidae, Parodontidae, and Cynodontidae (Oliveira et al., 2011Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
https://doi.org/10.1186/1471-2148-11-275...
; Arcila et al., 2017Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.; Mirande, 2018Mirande JM. Morphology, molecules and the phylogeny of Characidae (Teleostei, Characiformes). Cladistics; 2018; 34(2018):1-19. Available from: https://doi.org/10.1111/cla.12345
https://doi.org/10.1111/cla.12345...
). Further analyses with genomic datasets and deeper taxonomic sampling will be needed to settle this question. Based on our results and those of previous studies we suggest that the superfamily Erythrinoidea should be restricted to include the families Tarumaniidae and Erythrinidae.
The phylogenetic hypothesis supported in this study implies that air breathing evolved independently at least three times in characiform fishes that are able to survive low oxygen levels in the water (Graham, 1997Graham JB. Air-breathing fishes evolution, diversity, and adaptation. San Diego: Elsevier Inc; 1997.). In addition to Tarumaniidae, other families with air-breathing species are Lebiasinidae and Erythrinidae. Lebiasinidae includes around 67 species and 7 genera, with two known air breathing species (Piabucina festae and Lebiasina bimaculata), while Erythrinidae has 18 species and three genera, also with only two known air-breather (Erythrinus erythrinus and Hoplerythrinus unitaeniatus). Hoplias, on the other hand, is the most speciose erythrinid genus with 13 species, none of which are air-breathers but all of them are otherwise well adapted to live in hypoxic environments (Moraes et al., 1996Moraes G, Oliveira MA, Rantin FT. The metabolic pattern changes of Hoplias malabaricus from normoxia to hypoxia conditions. Rev Bras Biol. 1996; 56(2):191-96.; Graham, 1997Graham JB. Air-breathing fishes evolution, diversity, and adaptation. San Diego: Elsevier Inc; 1997.). Further studies are needed to resolve the relationships among erythrinids and lebiasinids, and other taxa to improve our understanding of the evolution of air breathing and adaptation to hypoxic conditions among characiform fishes.
Much work is still needed to resolve characiform phylogeny in general. For example, interrelationships among the families of two major clades that include (i) Cynodontidae, Parodontidae, Serrasalmidae, Hemiodontidae and families of Anostomoidea (this clade may be sister to Erythrinoidea), and (ii) Chalceidae, Iguanodectidae, Triportheidae, Gasteropelecidae, Bryconidae, Acestrorhynchidae and Characidae remain uncertain.
Acknowledgments
We thank Ricardo Betancur-R. (UPR-RP) for his suggestions on an earlier version of this manuscript. We also thank members of the Ortí Lab (Adela Roa-Varón and Chad Brock) for helping with DNA extractions. Marcos Mirande and two anonymous reviewers provided helpful comments on the manuscript. This study was supported by the National Science Foundation (NSF) grants (DIB-1811748) to DA and (DEB-1457426) to GO. This work was also supported by the Institutional Development Award (IDeA) INBRE (P20GM103475) from the National Institute of General Medical Sciences (NIGMS).
References
- Arcila D, Orti G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat Ecol Evol. 2017; 1(2):20.
- Buckup PA. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleoste: Ostariophysi). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of Neotropical fishes. Porto Alegre: Edipucrs; 1998. p.123-144.
- Eschmeyer WN, Fricke R, van der Laan R. Catalog of fishes: genera, species, references [Internet]. San Francisco: California Academy of Science; 2017. Available from: http://researcharchive.calacademy.org
» http://researcharchive.calacademy.org - Fink SV, Fink WL. Interrelationships of the ostariophysan fishes. Zool J Linnean Soc. 1981; 72:297-353.
- Graham JB. Air-breathing fishes evolution, diversity, and adaptation. San Diego: Elsevier Inc; 1997.
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989; 86(16):6196-200.
- Lavoue S, Arnegard M, Rabosky D, McIntyre P, Arcila D, Vari R, Nishida M. Trophic evolution in African citharinoid fishes (Teleostei: Characiformes) and the origin of intraordinal pterygophagy. Mol Phylogenet Evol [serial on the Internet]. 2017; 113:23-32. Available from: http://dx.doi.org/10.1016/j.ympev.2017.05.001
» http://dx.doi.org/10.1016/j.ympev.2017.05.001 - Lopez JA, Chen WJ, Orti G. Esociform phylogeny. Copeia. 2004; 2004(3):449-64.
- Melo BF, Sidlauskas B, Hoekzema K, Vari RP, Oliveira C. The first molecular phylogeny of Chilodontidae (Teleostei: Ostariophysi: Characiformes) reveals cryptic biodiversity and taxonomic uncertainty. Mol Phylogenet Evol [serial on the Internet]. 2014; 70:286-95. Available from: http://dx.doi.org/10.1016/j.ympev.2013.09.025
» http://dx.doi.org/10.1016/j.ympev.2013.09.025 - Mirande JM. Morphology, molecules and the phylogeny of Characidae (Teleostei, Characiformes). Cladistics; 2018; 34(2018):1-19. Available from: https://doi.org/10.1111/cla.12345
» https://doi.org/10.1111/cla.12345 - Moraes G, Oliveira MA, Rantin FT. The metabolic pattern changes of Hoplias malabaricus from normoxia to hypoxia conditions. Rev Bras Biol. 1996; 56(2):191-96.
- Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Orti G, Vari RP, Correa e Castro RM. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol [serial on the Internet]. 2011; 11(275):1-25. Available from: https://doi.org/10.1186/1471-2148-11-275
» https://doi.org/10.1186/1471-2148-11-275 - Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The Simple Fool’s Guide to PCR, Version 2. Honolulu: University of Hawaii Zoology Department; 1991.
- de Pinna M, Zuanon J, Rapp Py-Daniel L, Petry P. A new family of neotropical freshwater fishes from deep fossorial Amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool J Linnean Soc [serial on the Internet]. 2017; 182(1):76-106. Available from: https://doi.org/10.1093/zoolinnean/zlx028
» https://doi.org/10.1093/zoolinnean/zlx028 - Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol [serial on the Internet]. 2012; 61(3):539-42. Available from: https://doi.org/10.1093/sysbio/sys029
» https://doi.org/10.1093/sysbio/sys029 - Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol . 2002; 51(3):492-508.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics [serial on the Internet]. 2006; 22(21):2688-90. Available from: https://doi.org/10.1093/bioinformatics/btl446
» https://doi.org/10.1093/bioinformatics/btl446 - Thomaz AT, Arcila D, Orti G, Malabarba L. Molecular phylogeny of the subfamily Stevardiinae Gill, 1858 (Characiformes: Characidae): classification and the evolution of reproductive traits. BMC Evol Biol [serial on the Internet]. 2015; 15(146):1-25. Available from: https://doi.org/10.1186/s12862-015-0403-4
» https://doi.org/10.1186/s12862-015-0403-4
Edited by
Data availability
Data citations
Eschmeyer WN, Fricke R, van der Laan R. Catalog of fishes: genera, species, references [Internet]. San Francisco: California Academy of Science; 2017. Available from: http://researcharchive.calacademy.org
Publication Dates
-
Publication in this collection
11 Oct 2018 -
Date of issue
2018
History
-
Received
07 Feb 2018 -
Accepted
30 July 2018