ABSTRACT.
The distinction among Conyza canadensis, C. bonariensis, and C. sumatrensis is a challenge for weed science. In the current study, primers for microsatellite (SSR) loci were used to investigate the molecular divergence among the three species, the genetic structure of the populations at the molecular level and the level of genetic admixture among Conyza plants from southern Brazil. Twelve primers amplified well-defined DNA segments for all 88 samples of the three Conyza species. The estimated proportion of SSR polymorphic loci, number of alleles, and mean expected heterozygosity were higher in samples of C. bonariensis than in samples of C. sumatrensis or C. canadensis. Conyza canadensis was the species with the lowest molecular diversity. High genetic divergence was observed among the three species. The well-defined ancestral groups for each species led to the identification of samples of Conyza with ancestral genomes from the three species. Hybridization events between pairs of these species may have occurred in crop fields from southern Brazil. The high molecular diversity in resistant biotypes of C. sumatrensis indicated that these biotypes have a high potential to colonize new areas, which increases its potential as a weed.
Keywords:
horseweed; molecular divergence; SSR loci
RESUMO.
A identificação das espécies Conyza canadensis, C. bonariensis e C. sumatrensis tem sido desafiadora para a ciência das plantas daninhas. No presente estudo, primers para locos microssatélites (SSR) foram utilizados para investigar a divergência molecular entre as três espécies; mostrar como as populações estão geneticamente estruturadas em nível molecular e avaliar o nível de mistura genética entre as plantas de Conyza no Sul do Brazil. Doze primers amplificaram segmentos de DNA bem definidos em todas as 88 amostras das três espécies. A proporção estimada de locos SSR polimórficos, o número de alelos, e a heterozigosidade média esperada foram mais altos nas amostras de C. banariensis do que nas demais. C. canadensis foi a espécie com menor diversidade molecular. Divergência genética alta foi observada entre as três espécies. A formação de grupos ancestrais bem definidos para cada espécie levou à identificação de amostras de Conyza com genoma ancestral das três espécies. A ocorrência de hibridização entre as três espécies pode ter ocorrido nas lavouras do Sul do Brasil. A diversidade molecular alta em biótipos resistentes de C. sumatrensis indicou que estes biótipos têm um alto potencial para colonizar novas áreas, o que agrava seu potencial como planta daninha.
Palavras-chave:
buva; divergência molecular; locos SSR
Introduction
Some species of the genus Conyza are important weeds as they cause serious economic losses in agriculture (Thebaud & Abbott, 1995Thebaud, C., & Abbott, R. J. (1995). Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. American Journal of Botany, 82(3), 360-368.; Bossdorf et al., 2005Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E.,Siemann, E., & Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia, 144(1), 1-11.). The species C. canadensis (L.) Cronquist (horseweed), C. bonariensis (L.) Cronquist (hairy fleabane) and C. sumatrensis (Retz.) E. Walker (Sumatran fleabane) are examples of weeds that occur in orchards; vineyards; corn, soybean, cotton, and forage crops; pastures, and fallow areas (Lazaroto, Fleck, & Vidal, 2008Lazaroto, C. A., Fleck, N. G., & Vidal, R. A. (2008). Biologia e ecofisiologia de buva (Conyza bonariensis e Conyza canadensis). Ciência Rural, 38(3), 852-860.). Conyza canadensis is native to North America, whereas C. bonariensis and C. sumatrensis are native to South America. Among these three species, C. canadensis and C. sumatrensis are the most widespread worldwide (Thebaud & Abbott, 1995Thebaud, C., & Abbott, R. J. (1995). Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. American Journal of Botany, 82(3), 360-368.).
The increase in the economic relevance of these weeds is related to two main factors. One factor is the very efficient seed dispersal of these species and their prolific seed production, estimated at over 148,000 m-2 (Steckel & Gwathmey, 2009Steckel, L. E., & Gwathmey, C. O. (2009) Glyphosate-Resistant Horseweed (Conyza canadensis) Growth, Seed Production, and Interference in Cotton. Weed Science, 57(3), 346-350.). The second factor is the intense use of chemical options for their control, which has led to the selection of biotypes that are resistant to several herbicide mechanisms of action. To date, resistance to EPSPS inhibitors, ALS inhibitors and Photosystem I (PSI) and II (PSII) inhibitors individually, and multiple resistance to PSI + EPSPS, ALS + EPSPS or ALS + PSII have been reported (Matzraf, Lazar, Sibony, & Rubin, 2015Matzrafi, M., Lazar, T. W., Sibony, M., & Rubin, B. (2015). Conyza species: distribution and evolution of multiple target-site herbicide resistances. Planta, 242(1), 259-267.). Among all three species, cases of resistance to herbicides have been reported in 35 countries (Heap, 2016Heap, I. (2016). International survey of herbicide resistant weeds. Retrieved on July 24, 2016 from Retrieved on July 24, 2016 from http://www. weedscience.org
http://www. weedscience.org...
).
Conyza bonariensis and C. sumatrensis are common in Southern, Southeast and Midwestern of Brazil (Santos et al., 2014Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., Machado, M. F. P. S., Mangolin, C. A., & Nakajima, J. S. (2014b). Conyza sumatrensis: A new weed species resistant to glyphosate in the Americas. Weed Biology and Management, 14(2), 106-114.b). In contrast, C. canadensis occurs only in the Southern Region, particularly in Rio Grande do Sul State (Lazaroto et al., 2008Lazaroto, C. A., Fleck, N. G., & Vidal, R. A. (2008). Biologia e ecofisiologia de buva (Conyza bonariensis e Conyza canadensis). Ciência Rural, 38(3), 852-860.). Identification keys (Pruski & Sancho, 2006Pruski, J. F., & Sancho, G. (2006). Conyza sumatrensis var. leiotheca (Compositae: Astereae), a new combination for a common neotropical weed. Novon: A Journal for Botanical Nomenclature, 16(1), 96-101.) typically define characteristics that can be used to differentiate the three Conyza species. In C. canadensis, the leaves are yellowish green and glabrous, whereas in C. bonariensis and C. sumatrensis, they are greyish green and very hairy. The species vary in height and branching habit. Conyza canadensis branches from the middle of the main stem, C. bonariensis has branches that are taller than the main stem, and C. sumatrensis has branching towards the top of the main stem (Sansom, Saborido, & Dubois, 2013Sansom, M., Saborido, A. A., & Dubois, M. (2013). Control of Conyza spp. with glyphosate - a review of the situation in Europe. Plant Protection Science, 49(1), 44-53.). However, in South America, definite identification of and distinction among the three species of important weeds is difficult due to morphological variability within species, the occurrence of varieties within some species, and hybridization between species (Thebaud & Abbott, 1995Thebaud, C., & Abbott, R. J. (1995). Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. American Journal of Botany, 82(3), 360-368.).
Herbicide susceptibility varies among these species (Gonzalez-Torralva et al., 2010Gonzalez-Torralva, F., Cruz-Hipolito, H., Bastida, F., Malleder, N., Smeda, R. J., & De Prado, R. (2010). Differential susceptibility to glyphosate among the Conyza weed species in Spain. Journal of Agricultural and Food Chemistry, 58(7), 4361-4366.; Zheng et al., 2011Zheng, Q. A., Tai, C., Hu, J., Lin, H., Zhang, R., Su, F., & Yang, X. (2011). Satellite altimeter observations of nonlinear Rossby eddy-Kuroshio interaction at the Luzon Strait. Journal of Oceanography, 67(4), 365-376.); however, due to the difficulty in their identification, reports of unsuccessful control might involve weed misidentification. Developing a deeper understanding of the genetic structure of these species may help scientists to develop more precise identification tools and effective strategies for control.
In contrast to the several studies on the characteristics of resistant biotypes in other weeds (Yamauti et al., 2010Yamauti, M. S., Barroso, A. A. M., Souza, M. C., & Alves, P. L. C. A. (2010). Chemical control of glyphosate-resistant horseweed (Conyza canadensis) and hairy fleabane (Conyza bonariensis) biotypes. Revista Ciência Agronômica, 41(3), 495-500.; Xiao-Ling et al., 2011Xiao-Ling, S., Wu, J., Zhang, H., & Qiang, S. (2011). Occurrence of glyphosate-resistant horseweed (Conyza canadensis) population in China. Agricultural Sciences in China, 10(7), 1049-1055.; Santos et al., 2014Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., Machado, M. F. P. S., Mangolin, C. A., & Nakajima, J. S. (2014b). Conyza sumatrensis: A new weed species resistant to glyphosate in the Americas. Weed Biology and Management, 14(2), 106-114.a), few studies have addressed how the populations of C. canadensis, C. bonariensis and C. sumatrensis are genetically structured at the molecular level. Molecular analysis techniques allow us to estimate the genetic variability within and between species. They can also be used to search for specific unique molecular markers (alleles) of one or more species of Conyza; if found, such markers could be used to support the identification of and distinction among species. Differential allele frequencies could also be useful for species identification.
Simple sequences repeated of genomic DNA (SSR loci, also known as microsatellite loci) are efficient molecular markers to estimate genetic divergence and to reveal population structure in many plant species. In Conyza species, SSR loci have been used as molecular markers to investigate the evolution and spread of glyphosate resistance in C. canadensis in California (Yuan et al., 2010Yuan, J., Abercrombie, L., Cao, Y., Halfhill, M., Zhou, X., Peng, Y., & Stewart, C. (2010). Functional genomics analysis of horseweed (Conyza canadensis) with special reference to the evolution of non-target-site glyphosate resistance. Weed Science, 58(2), 109-117.; Okada et al., 2013Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart Jr., C. N., ... Jasienuk, M. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evolutionary Applications, 6(5), 761-777.).
Invasive species that grow under strong selection pressure from herbicide application usually show significant genetic differentiation. Moreover, less genetic diversity is expected in samples of resistant biotypes that have been under selection due to the recurrent use of herbicides. In the current study, SSR loci were used to investigate the molecular divergence among the three species (C. canadensis, C. bonariensis, and C. sumatrensis) and to evaluate the level of genetic admixture among Conyza plants from southern Brazil.
Material and methods
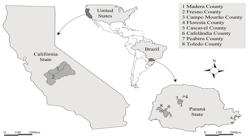
The seeds of C. sumatrensis were collected from several plants in Campo Mourão, Floresta, Cascavel, Cafelândia, Peabiru, and Toledo in Paraná State, Brazil, in January 2011 (Figure 1; Table 1). Seeds were collected from plants in fields of glyphosate-resistant (GR) soybeans with a history of glyphosate applications of at least four years and for which farmers reported decreasing levels of control over repeated glyphosate applications. The seeds from each collection site were individually placed in paper bags to prevent the mixture of seeds from different collection sites.
Seeds from each site were randomly distributed for germination in separated 500-mL pots containing sterile soil. Plants obtained from germinated seeds were maintained at room temperature in the greenhouse at the State University of Maringá (latitude 23°23'44.91"S and longitude 51°57'3.13"W, altitude 510 m), irrigated daily, and used for the experiments. A sample consisting of young leaves collected from 43 plants of C. sumatrensis was used for DNA extraction. The young leaves were collected from plants 15-30 days after plant emergence. Samples of these plants in full bloom were then sent to the Institute of Biology (Uberlandense Herbarium) at the Federal University of Uberlândia (Minas Gerais State, Brazil), where they were cataloged and classified as Conyza sumatrensis (Retz.) E. Walker. The plants of C. sumatrensis were analyzed for possible resistance to glyphosate, which was applied at different stages of development according to procedures described by Santos et al. (2014Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., Machado, M. F. P. S., Mangolin, C. A., & Nakajima, J. S. (2014b). Conyza sumatrensis: A new weed species resistant to glyphosate in the Americas. Weed Biology and Management, 14(2), 106-114.b). The samples from Cascavel and Toledo were recorded as glyphosate resistant (SFR 6 and SFR 9, respectively; Table 1), whereas the remaining samples of C. sumatrensis (SFS) were recorded as sensitive to glyphosate.
Distribution of the Conyza plants from which seeds were germinated to form samples representative of C. sumatrensis from Campo Mourão, Floresta, Cascavel, Cafelândia, Peabiru, and Toledo in Paraná State, Brazil, and of C. canadensis and C. bonariensis from the Central Valley of the State of California, United States.
The seeds of C. canadensis and C. bonariensis were collected in the Central Valley of the state of California, United States (Table 1; Figure 1). Since C. canadensis and C. bonariensis are well-differentiated species in North America, samples of the two species were collected in California and used as referential genomes to evaluate the level of genetic admixture among Conyza plants of southern Brazil. The broad morphological variability in Conyza plants from southern Brazil makes it difficult to obtain a secure identification of C. canadensis or C. bonariensis plants. Then, the seeds of resistant plants of C. canadensis (HWR) were collected in Madera County (Dinuba, CA), and the seeds of susceptible plants of C. canadensis (HWS 156) were collected in Fresno County. The seeds of C. bonariensis were collected from plants with multiple resistance (BH 51) to glyphosate and ALS in Fresno County (Parlier, CA) and from plants resistant to glyphosate (BH 53) in Parlier. Seeds were germinated in plastic trays (52 x 27 x 6 cm) in the greenhouse at the University of California (Davis, CA), using the substrate Sunshine Mix (Sungro Horticulture Canada, Ltd., Vancouver, British Columbia, Canada) for the establishment and growth of the plants. The plants were grown in greenhouse at the experimental field of the university (38°32'33.52"N and 121°45'47.92"W, elevation 17 meters). Samples consisting of young leaves collected from 45 plants of each species (C. canadensis and C. bonariensis) were used for DNA extraction.
Samples of Conyza sumatrensis (SF) obtained from several sites (Campo Mourão, Floresta, Cascavel, Cafelândia, Peabiru, and Toledo) in Paraná State, Brazil, and samples of Conyza canadensis (HW) and Conyza bonariensis (BH) obtained from Dinuba, Fresno, and Parlier in the Central Valley of the State of California, USA.
For DNA extraction, leaf pieces (300 mg) of each C. sumatrensis plant were separately ground in liquid nitrogen and homogenized with a glass rod in an Eppendorf microcentrifuge tube with the use of 800 μL extraction solution prepared with Tris-HCl 100 mM / EDTA 20 mM containing NaCl 1.4 M, CTAB (Cetyl Trimethyl Ammonium Bromide) 2%, PVP-40 (Polyvinylpyrrolidone-40) 2%, and β-mercaptoethanol 0.2% and maintained in an ice chamber. After homogenization, the microcentrifuge tubes were shaken gently and incubated at 65°C for 30 min. DNA was extracted according to the protocol by Doyle and Doyle (1990Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12(1), 13-15.).
DNA extraction from plants of C. canadensis and C. bonariensis was performed using the Soltis Lab CTAB DNA Extraction Protocol (2002) developed from Doyle and Doyle (1990Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12(1), 13-15.) and Cullings (1992Cullings, K. W. (1992). Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Molecular Ecology, 1(4), 233-240.) protocols (http://www.flmnh.ufl.edu/museum-voices/soltis-lab/files/2014/02/CTAB-DNA-Extraction.pdf). The young leaves were collected from plants with six weeks after plant emergence. For DNA extraction, leaf pieces (10-20 mg) were separately ground in liquid nitrogen and homogenized with a glass rod in an Eppendorf microcentrifuge tube with the use of 500 μL CTAB buffer extraction prepared according to the Soltis Lab CTAB DNA Extraction Protocol (2002).
After DNA extraction, DNA quantity and quality were determined by 0.8% agarose gel electrophoresis buffered with 1 x TAE (0.04 M Tris-Acetate and 0.001 M EDTA). A standard DNA (λ phage; 50, 100, and 150 ng) was used as a marker of concentration. The gel was stained with 0.5 μg/mL ethidium bromide, and the image was visualized with a Molecular Image Loccus L-PIX - HE (Loccus do Brasil Ltda., São Paulo, São Paulo State, Brazil) and the Picasa 3 program. The samples were also quantified using UV-visible Picodrop® spectrophotometer to verify the concentration of DNA per µL of each sample for dilution to be used in the PCR reactions.
Sixteen SSR primers previously developed for C. canadensis, i.e., HW01, HW02, HW06, HW07, HW14, HW27, HW29 (Abercrombie, Anderson, & Baldwin, 2009Abercrombie, L. G., Anderson, C. M., & Baldwin, B. G. (2009). Permanent genetic resources added to Molecular Ecology Resources database 1 January 2009-30 April 2009. Molecular Ecology Resources, 9(5), 1375-1379.), HW17, HWSSR01, HWSSR03, HWSSR04, HWSSR06, HWSSR07, HWSSR09, HWSSR11, and HWSSR12 (Okada et al., 2013Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart Jr., C. N., ... Jasienuk, M. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evolutionary Applications, 6(5), 761-777.), were used with eight DNA samples to define the DNA quantity used for polymerase chain reaction (PCR). After initial screening, a 1.0 µL aliquot of DNA was defined for further analysis. PCR was performed using a PTC-200 Peltier thermal cycler. The reaction mixtures were prepared in microtubes (1.5 mL) and then loaded on a multiplex plate as described by Okada et al. (2013), with a final volume of 10 µL per reaction: dNTP 125 mM, 0.375 units of Taq polymerase (QIAGEN, Valencia, CA, USA), 1 x PCR buffer (QIAGEN), 10 ng DNA, 10 mM of the reverse and forward primers, and Milli-Q water to make up to 10 µL.
Microsatellite amplification was initially performed with initial denaturation at 94°C for 5 min. followed by 34 cycles at 94°C for 1 min.; annealing was carried out at 55 °C for 1 min, and extension was at 72°C for 1 min. and 30 seconds. Electrophoresis was performed in a 2% agarose gel using 0.5 x TBE buffer (44.5 mmol·L-1 Tris, 44.5 mmol·L-1 boric acid, and 1 mmol·L-1 EDTA) at 109 V for 60 min. Each gel was stained with ethidium bromide at 0.5 μg·mL-1, and the image was captured using a Kodak EDAS 290 digital camera. The sizes of the amplified DNA segments (alleles) were determined using a 100-bp DNA Ladder (Invitrogen).
The polymorphic SSR loci in the C. sumatrensis, C. canadensis, and C. bonariensis samples were used to estimate the average number of alleles per locus, the average observed heterozygosity (Ho), and the expected heterozygosity (He). The genetic diversity among the six samples of C. sumatrensis (FST) was estimated using the POPGENE 1.32 program (Yeh, Boyle, & Xiyan, 1999Yeh, F. C., Boyle, T. Y. Z., & Xiyan, J. M. (1999). Popgene Version 1.31: Microsoft Window-based freeware for population genetic analysis. Edmonton, AL: University of Alberta.). The genetic distance matrix was calculated by the UPGMA clustering method (Unweighted Pair-Group Mean Average) using the values of Nei genetic distances (Nei, 1978Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a number of individuals. Genetics, 89(3), 538-590.).
Polymorphism in the SSR loci was also analyzed using the software Structure 2.0 (Pritchard, Wen, & Falush, 2010Pritchard, K. J., Wen, W., & Falush, D. (2010). Documentation for Structure software: Version 2.3. Chicago, IL: University of Chicago. ) and Instruct (Gao, Williamson, & Bustamante, 2007Gao, H., Williamson, S., & Bustamante, C. D. (2007). An MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics, 176(3), 1635-1651. ), which evaluate the level of genetic admixture among the three species of Conyza. The genotypes were clustered, with the number of clusters (K) ranging from 2 to 14, and were tested using the admixture model with a burn-in period of 200,000 repeats followed by 100,000 Markov Chain Monte Carlo (MCMC) repeats, considering the presence and absence of alleles across the sample. The true number of populations (K) is often identified using the maximal value of ∆ (K) returned by the software. The most likely number (K) of subpopulations was identified as described by Evanno, Regnaut, and Goudet (2005Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology, 14(8), 2611-2620.). The graphical output display of the Structure results was taken as input data using the Structure Harvester, a website and software that are used to visualize Structure output and to implement the Evanno method (Earl & Von Holdt, 2012Earl, D., & Vonholdt, B. M. (2012). Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conservation Genetic Resources, 4(2), 359-361.) to display a graphical representation. The graphical output display of the Instruct results was taken using the Distruct software (Rosenberg et al., 2002Rosenberg, N. A., Pritchard, J. K., Weber, J. L., Cann, H. M., Kidd, K. K., Zhivotovsky, L. A., & Feldman, M. W. (2002). Genetic structure of human populations. Science, 298(5602), 2381-2385.) to draw a bar plot of individual genome assignments. To explore the hierarchical partitioning of genetic variation within and among the samples of Conyza, we performed an Analysis of Molecular Variance (AMOVA, GenAlEx 6.2; Peakall & Smouse, 2006Peakall, R., & Smouse, P. E. (2006). GenAIEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Resources, 6(1), 288-295.).
Results and discussion
Four primers (HW01, HW27, HWSSR06, and HWSSR12) did not produce well-defined amplified DNA segments for all 88 samples of C. sumatrensis, C. canadensis, and C. bonariensis, and only 12 (HW02, HW06, HW07, HW14, HW29, HW17, HWSSR01, HWSSR03, HWSSR04, HWSSR07, HWSSR09, and HWSSR11) amplified two or more alleles per polymorphic locus (Table 2). The estimated proportion of SSR polymorphic loci (%P) was higher in samples of C. bonariensis (91.67%) than in samples of C. sumatrensis (83.33%) or C. canadensis (75%). However, the number of alleles (Na), effective number of alleles (Ne), and mean expected heterozygosity (He) were highest in samples of C. sumatrensis, indicating the highest molecular diversity. Samples of C. canadensis showed the lowest molecular diversity (Table 3).
The Nei identity (Nei, 1978Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a number of individuals. Genetics, 89(3), 538-590.) values calculated from analysis of the 12 microsatellite loci of the three species of Conyza showed that the highest value of genetic identity (I = 0.8293) was observed between C. canadensis and C. sumatrensis and that the most divergent species (I = 0.6303) were C. canadensis and C. bonariensis. The similarity between C. canadensis and C. bonariensis was lower than that between C. canadensis and C. sumatrensis and that between C. bonariensis and C. sumatrensis (0.7348) for number and frequencies of alleles at the 12 SSR loci. AMOVA showed higher genetic variation within (54%) than among (46%) the C. canadensis, C. bonariensis, and C. sumatrensis samples.
Simple Sequence Repeat (microsatellite) primers used for analysis of the Conyza sumatrensis, C. canadensis, and C. bonariensis plants, and the number of alleles at each locus.
Number of alleles (Na) and effective number of alleles (Ne) per polymorphic SSR locus, mean observed heterozygosity (Ho), expected heterozygosity (He), and percentage of polymorphic loci (%P) in the samples from the three Conyza species (C. canadensis, C. bonariensi s, and C. sumatrensis).
The clustering of the C. canadensis, C. bonariensis, and C. sumatrensis plants according to a model-based Bayesian algorithm is shown in Figure 2. Each bar in the graph represents a plant and its inferred proportion of genome admixture. The colors represent three different ancestral groups. The optimal K value determined by Bayesian analysis indicated that the plants were grouped into 3 clusters (∆K2 = 0.00; ∆K3 = 91.5046; ∆K4 = 0.3635; ∆K5 = 1.3088; ∆K6 = 0.9398; ∆K7 = 1.2130; ∆K8 = 0.00; ∆K9 = 1.4745; ∆K10 = 8.9503; ∆K11 = 0.1505; ∆K12 = 3.0456; ∆K13 = 0.2263; ∆K14 = 0.00). The bar plot obtained for the K value (K = 3; ΔK = 91.5046), and the results showed that 96.9% of the C. canadensis samples were in the red group, 96.7% of the C. bonariensis samples were in the blue group, and 79.7% and 15.1% of the C. sumatrensis samples were in the green and red groups, respectively (Table 4).
The bar plot graphic (Figure 2) shows that several plants identified as C. sumatrensis in the ancestral group that were defined as prevalent in samples of C. canadensis and C. bonariensis from the Northern Hemisphere (Central Valley of California, USA). Unlike the analysis using the Structure software, the analysis using Instruct identified ten clusters for the 88 samples of the three Conyza species. Plants sharing alleles from the ten groups are evident in samples of the species C. canadensis, C. bonariensis, and C. sumatrensis. The selfing rate was high, ranging from 0.655 ± 0.028 (cluster 1) to 0.803 ± 0.013 (cluster 10). In the plants of C. canadensis from Dinuba (HWR), the Instruct analysis identified the predominance of the ancestral genome from the pink group, which suggests the selection of one of the ancestral groups in the resistant biotypes of C. canadensis.
The bar plot shows that the plants of Campo Mourão represent all three groups but predominantly represent the red group (C. canadensis). Plants from the blue group (C. bonariensis) are also evident in samples of Campo Mourão. In the samples from Floresta and Cafelândia, high proportions of plants in the red group (C. canadensis) are evident. In the samples from Peabiru and Toledo, low proportions of plants of the red and blue groups are apparent.
Bar plot of descendants from Conyza canadensis obtained from Dinuba (HWR) and Fresno (HWS 156), C. bonariensis obtained from Parlier, and C. sumatrensis obtained from Campo Mourão (3), Floresta (4), Cascavel (5), Cafelândia (6), Peabiru (7), and Toledo (8) in three inferred groups based on ΔK values. Each plantlet is represented by a single vertical line broken into K colored segments (K = 3), with lengths proportional to each of the K inferred clusters. Each color represents the proportion of membership of each individual, represented by a vertical line.
Proportion of Conyza canadensis, C. bonariensis, and C. sumatrensis samples in each group (K = 3) and the number of sampled plants.
A lower proportion of polymorphic loci was observed for C. canadensis-R plants (HWR: 41.67%) than for C. canadensis-S plants (HWS 156: 66.67%), suggesting that resistant plants show a reduction of polymorphism in microsatellite loci. Lower polymorphism in C. bonariensis-2R (BH51: 50%) plants than in C. bonariensis-R (BH53: 91.67%) plants was also observed but not considered of significance since a relatively low number of C. bonariensis-2R plants were analyzed.
In the samples of C. sumatrensis distributed among the six sites from southern Brazil (Paraná State), the highest proportion of polymorphic loci (83.3%) was observed in samples from Campo Mourão and Cafelândia. However, the highest number of alleles (Na = 2.0833) and the highest effective number of alleles (Ne = 1.8016), as well as the highest mean expected heterozygosity value (He = 0.3632), were observed in the samples from Toledo-R identified by Santos et al. (2014Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., Machado, M. F. P. S., Mangolin, C. A., & Nakajima, J. S. (2014b). Conyza sumatrensis: A new weed species resistant to glyphosate in the Americas. Weed Biology and Management, 14(2), 106-114.b) as resistant to glyphosate (Table 5). A high number of alleles (Na = 1.9167) and a high effective number of alleles (Ne = 1.6171) were also observed in samples of resistant biotypes from Cascavel-R (Table 5). Values of Ho were lower than He in the samples characterized as susceptive to glyphosate and those characterized as resistant, indicating a deficit of heterozygosity in all of the samples of C. sumatrensis. However, the selection for resistant biotypes due to glyphosate application has not reduced the number of alleles (Na and Ne) in the resistant biotypes (from Toledo and Cascavel; Table 5).
Number of alleles (Na) and effective number of alleles (Ne) per polymorphic SSR locus, mean observed heterozygosity (Ho), expected heterozygosity (He), and percentage of polymorphic loci (%P), in the samples of Conyza sumatrensis from six sites in southern Brazil.
The differential allele frequency in samples of C. sumatrensis among the six sites in southern Brazil is very high (Fst = 0.31), indicating that the samples from the six sites are from structured sub-populations of C. sumatrensis. There are common alleles occurring at similar frequencies in all samples at the HW29 (HW291 ) and HWSSR11 (HWSSR111 ) loci, indicating an absence of genetic differentiation; however, the differential frequencies of alleles at other loci are sufficiently high to determine the genetic structure of the six samples of C. sumatrensis. The gene flow (Nm = 0.5535) was moderate among the samples of C. sumatrensis from the six sites, suggesting an exchange of alleles or dispersion of samples among sites.
The Nei identity (Nei, 1978Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a number of individuals. Genetics, 89(3), 538-590.) values calculated from the analysis of the 12 microsatellite loci of samples of C. sumatrensis from six sites in southern Brazil (Campo Mourão, Floresta, Cascavel, Cafelândia, Peabiru, and Toledo) showed that the highest value of genetic identity (I = 0.9275) was observed between samples from Cascavel and Peabiru and that the most divergent samples (I = 0.6931) were those from Floresta and Peabiru (Table 6). The high genetic similarity between the samples from Cascavel and Peabiru suggest that the implementation of similar control strategies could be effective in the two areas since plants with high genetic similarity can be expected to respond similarly to such strategies.
Although the functional significance of the SSR loci (HW02, HW06, HW07, HW14, HW29, HW17, HWSSR01, HWSSR03, HWSSR04, HWSSR07, HWSSR09, HWSSR11) in Conyza species is unknown, the high number of alleles and the high levels of observed and expected heterozygosity in C. sumatrensis suggest that this species a higher potential than does C. canadensis or C. bonariensis to colonize new areas. High heterozygosity has been considered to indicate that the plant population has a substantial amount of adaptive genetic variation since there is a greater chance of finding plants that respond differently to changes or pressures exerted by the environment. SSR loci that occur in tandem are abundantly distributed in coding and non-coding regions of plant genomes (see review by Kalia, Rai, Kalia, Singh, & Dhawan, 2011Kalia, R. K., Rai, M. K., Kalia, S., Singh, R., & Dhawan, A. K. (2011). Microsatellite markers: an overview of the recent progress in plants. Euphytica, 177(3), 309-334.) such that alterations in SSR loci that are located in a coding region or intron have the potential to regulate the differential expression of genes related to the environmental adaptive potential of the plant. The high molecular diversity estimated for C. sumatrensis appears to be consistent with the high morphological diversity that is reported by farmers and agronomists in the cropping fields in southern Brazil.
The molecular differentiation in SSR loci of the three Conyza species is consistent with the value of genetic identity found between the samples of C. canadensis and C. bonariensis (0.6303), C. canadensis and C. sumatrensis (0.8293), and C. bonariensis and C. sumatrensis (0.7348). Levels of genetic identity lower than 0.85 are frequently reported between populations from different species (Thorpe & Solé-Cava, 1994Thorpe, J. P., & Solé-Cava, A. M. (1994). The use of allozyme electrophoresis in invertebrate systematics. Zoologica Scripta, 23(1), 3-18.). The highest value of genetic identity reported in our study (I = 0.8293) was between C. canadensis and C. sumatrensis. The highest value of molecular similarity between C. canadensis and C. sumatrensis is in agreement with the highest similarity reported based on morphological parameters (Pruski & Sancho, 2006Pruski, J. F., & Sancho, G. (2006). Conyza sumatrensis var. leiotheca (Compositae: Astereae), a new combination for a common neotropical weed. Novon: A Journal for Botanical Nomenclature, 16(1), 96-101.; Vladimirov, 2009Vladimirov, V. (2009). Erigeron sumatrensis (Asteraceae): a recently recognized alien species in the Bulgarian flora. Phytologia Balcanica, 15(3), 361-365.).
AMOVA showed higher genetic variation within (54%) than among (46%) the samples of the three species, and the grouping of the C. canadensis, C. bonariensis, and C. sumatrensis plants according to Bayesian statistics revealed that the higher molecular diversity within the samples of the three species of Conyza is due to the high diversity within C. sumatrensis. The plants of Campo Mourão, Cafelândia, and Floresta represent all three ancestral groups (red, green, and blue), with genomes predominantly of the red group (C. canadensis) and the blue group (C. bonariensis), whereas in the samples from Peabiru and Toledo, low proportions of the genomes of the red and blue groups was observed.
The mixture of genomes, which were defined as ancestral groups prevalent in samples of C. canadensis and C. bonariensis from the Central Valley of California, in samples from sites in Paraná State suggest that all three species Conyza are present in these areas and that hybridization may be occurring. The presence of different Conyza species in an area can lead to difficulty in species identification. Seeds of a particular species can be carried to other areas by wind dispersal or via the movements of agricultural machinery. The potential exchange of alleles between C. canadensis and C. bonariensis has been noted in areas of São Paulo State, Brazil, based on analyses of loci for isozymes of esterase, malate dehydrogenase and acid phosphatase (Soares et al., 2015Soares, A. A. F., Fregonezi, A. M. D. T., Bassi, D., Mangolin, C. A., Collet, S. A. O., Oliveira Jr., R. S. O., & Machado, M. F. P. S. (2015). Evidence of high gene flow between samples of Conyza canadensis and C. bonariensis as revealed by isozyme polymorphisms. Weed Science, 63(3), 604-612.). Genomic admixture of Conyza species could explain why plants of one species (e.g., C. sumatrensis) can often exhibit the characteristics of another (e.g., C. bonariensis or C. canadensis). The hybridization between C. canadensis and C. ramosissima that was induced by Zelaya, Owen, and Vangessel (2007Zelaya, I. A., Owen, M. D. K., & Vangessel, M. J. (2007). Transfer of glyphosate resistance: evidence of hybridization in Conyza (Asteraceae). American Journal of Botany, 94(4), 660-673.) produced interspecific hybrids of Conyza that exhibited intermediate phenotypes between the parents.
Despite the genomic admixture of the three ancestral groups in some plants in southern Brazil (evident in the bar plot graphic; Figure 2), the selfing rate revealed by the Instruct analysis for the three species of Conyza was high (0.655-0.803). Self-compatibility in Conyza species might explain the low values of Ho (Table 3) in the three Conyza species. Conyza canadensis is described as a self-compatible species in which the pollen is released before the capitula have fully opened, suggesting that it is primarily self-pollinated (Weaver, 2001Weaver, S. E. (2001). The biology of Canadian weeds: Conyza canadensis. Canadian Journal of Plant Science, 81(4), 867-875.; Okada et al., 2013Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart Jr., C. N., ... Jasienuk, M. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evolutionary Applications, 6(5), 761-777.). High inbreeding coefficients (FIS = 0.850 to 0.966) in a large sample of C. canadensis were recently reported by Okada et al. (2013) in a microsatellite loci analysis. Self-pollination has also been described in C. bonariensis (Ferrer & Good-Avila, 2007Ferrer, M. M., & Good-Avila, S. V. (2007). Macrophylogenetic analyses of the gain and loss of self-incompatibility in the Asteraceae. New Phytologyst, 173(2), 401-414.) but has not been described to date in C. sumatrensis. In C. sumatrensis, the analysis of SSR polymorphism indicated moderate gene flow (Nm = 0.5535) among the samples from different sites (Campo Mourão, Floresta, Cascavel, Cafelândia, Peabiru, and Toledo), suggesting out-crossing among plants from these sites. Values of Nm ranging from 0.25 to 0.99 indicate intermediate gene flow among populations (Govindaraju, 1989Govindaraju, D. R. (1989). Variation in gene flow levels among predominantly self-pollinated plants. Journal of Evolutionary Biology, 2(3), 173-181.).
Despite the indication of moderate gene flow among the C. sumatrensis samples, the value of Fst (0.31) was high, indicating that the frequency of alleles at the SSR loci varied among the six samples. Values of FST ranging from 0.01 to 0.05 indicate minimal divergence among populations; those from 0.05 to 0.15 indicate moderate divergence, whereas those ranging from 0.15 to 0.25 indicate high divergence. The observed FST was ˃ 0.25, indicate very high divergence among the populations. Conyza sumatrensis species forms genetically well-structured populations according to both Wright's F-statistic (Wright, 1965Wright, S. (1965). The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19(3), 395-420.) and Bayesian statistics. Highly differentiated populations with Fst values ranging from 0.034 to 0.7530 have been reported by Okada et al. (2013Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart Jr., C. N., ... Jasienuk, M. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evolutionary Applications, 6(5), 761-777.) for 42 populations of C. canadensis from different geographical regions across the Central Valley of California. Most populations were highly differentiated, as expected for a highly selfing species. Since the establishment of a population of an invasive species often involves a small number of plants that contain only part of the variation present in the origin population, the "founder effect" in self-fertilization species can result in populations with different numbers and proportions of alleles. Moreover, invasive species that have evolved under strong selection pressure by herbicide application are expected to show high genetic differentiation.
The high proportion of polymorphic loci and the high values of observed and expected heterozygosity (Table 6) in samples of C. sumatrensis that were identified as resistant to glyphosate as well as those identified as susceptive to glyphosate conflicts with our hypothesis that lower genetic diversity is expected in samples of resistant biotypes that have been selected for by the use of herbicides. The molecular diversity estimated in the samples C. sumatrensis that were resistant to glyphosate indicates that the selection of resistant biotypes has not reduced the number of alleles or the observed and expected heterozygosity in the resistant biotypes.
Conclusion
The high genetic similarity at the molecular level observed between C. canadensis and C. sumatrensis is consistent with the high morphological similarity between the two species. The well-defined groups of ancestral genomes in the microsatellite loci for each species led to the identification of plants with the ancestral genome of C. canadensis in southern Brazil, where C. sumatrensis is the prevalent species, and to the identification of samples with admixture of ancestral genomes of two or three species. Moreover, the high molecular diversity in the resistant biotypes of C. sumatrensis indicated that these biotypes have a high potential to colonize new areas, which will facilitate their persistence and increase the difficulty of their control. The high molecular diversity of the resistant biotypes is consistent with the hypothesis of multiple mechanisms that are involved in herbicide resistance in Conyza species. The high and low values of genetic similarity between the C. sumatrensis samples may be used to investigate the efficiency of similar and different strategies, respectively, to control infestation in different areas.
Acknowledgements
We would like to thank CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for their financial support. We would also like to thank the researchers, technicians and students of the Jasieniuk Lab (Department of Plant Science at the University of California, Davis, CA), who offered to give in part of the biological material and allowed the execution of part of the lab work in this work.
References
- Abercrombie, L. G., Anderson, C. M., & Baldwin, B. G. (2009). Permanent genetic resources added to Molecular Ecology Resources database 1 January 2009-30 April 2009. Molecular Ecology Resources, 9(5), 1375-1379.
- Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E.,Siemann, E., & Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia, 144(1), 1-11.
- Cullings, K. W. (1992). Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Molecular Ecology, 1(4), 233-240.
- Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus, 12(1), 13-15.
- Earl, D., & Vonholdt, B. M. (2012). Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conservation Genetic Resources, 4(2), 359-361.
- Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology, 14(8), 2611-2620.
- Ferrer, M. M., & Good-Avila, S. V. (2007). Macrophylogenetic analyses of the gain and loss of self-incompatibility in the Asteraceae. New Phytologyst, 173(2), 401-414.
- Gao, H., Williamson, S., & Bustamante, C. D. (2007). An MCMC approach for joint inference of population structure and inbreeding rates from multi-locus genotype data. Genetics, 176(3), 1635-1651.
- Gonzalez-Torralva, F., Cruz-Hipolito, H., Bastida, F., Malleder, N., Smeda, R. J., & De Prado, R. (2010). Differential susceptibility to glyphosate among the Conyza weed species in Spain. Journal of Agricultural and Food Chemistry, 58(7), 4361-4366.
- Govindaraju, D. R. (1989). Variation in gene flow levels among predominantly self-pollinated plants. Journal of Evolutionary Biology, 2(3), 173-181.
- Heap, I. (2016). International survey of herbicide resistant weeds Retrieved on July 24, 2016 from Retrieved on July 24, 2016 from http://www. weedscience.org
» http://www. weedscience.org - Kalia, R. K., Rai, M. K., Kalia, S., Singh, R., & Dhawan, A. K. (2011). Microsatellite markers: an overview of the recent progress in plants. Euphytica, 177(3), 309-334.
- Lazaroto, C. A., Fleck, N. G., & Vidal, R. A. (2008). Biologia e ecofisiologia de buva (Conyza bonariensis e Conyza canadensis). Ciência Rural, 38(3), 852-860.
- Matzrafi, M., Lazar, T. W., Sibony, M., & Rubin, B. (2015). Conyza species: distribution and evolution of multiple target-site herbicide resistances. Planta, 242(1), 259-267.
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a number of individuals. Genetics, 89(3), 538-590.
- Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart Jr., C. N., ... Jasienuk, M. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evolutionary Applications, 6(5), 761-777.
- Peakall, R., & Smouse, P. E. (2006). GenAIEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Resources, 6(1), 288-295.
- Pritchard, K. J., Wen, W., & Falush, D. (2010). Documentation for Structure software: Version 2.3. Chicago, IL: University of Chicago.
- Pruski, J. F., & Sancho, G. (2006). Conyza sumatrensis var. leiotheca (Compositae: Astereae), a new combination for a common neotropical weed. Novon: A Journal for Botanical Nomenclature, 16(1), 96-101.
- Rosenberg, N. A., Pritchard, J. K., Weber, J. L., Cann, H. M., Kidd, K. K., Zhivotovsky, L. A., & Feldman, M. W. (2002). Genetic structure of human populations. Science, 298(5602), 2381-2385.
- Sansom, M., Saborido, A. A., & Dubois, M. (2013). Control of Conyza spp. with glyphosate - a review of the situation in Europe. Plant Protection Science, 49(1), 44-53.
- Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., & Osipe, J. B. (2014a). Multiple resistance of Conyza sumatrensis to chlorimuron-ethyl and to glyphosate. Planta Daninha, 32(2), 409-416.
- Santos, G., Oliveira Jr., R. S., Constantin, J., Francischini, A. C., Machado, M. F. P. S., Mangolin, C. A., & Nakajima, J. S. (2014b). Conyza sumatrensis: A new weed species resistant to glyphosate in the Americas. Weed Biology and Management, 14(2), 106-114.
- Soares, A. A. F., Fregonezi, A. M. D. T., Bassi, D., Mangolin, C. A., Collet, S. A. O., Oliveira Jr., R. S. O., & Machado, M. F. P. S. (2015). Evidence of high gene flow between samples of Conyza canadensis and C. bonariensis as revealed by isozyme polymorphisms. Weed Science, 63(3), 604-612.
- Steckel, L. E., & Gwathmey, C. O. (2009) Glyphosate-Resistant Horseweed (Conyza canadensis) Growth, Seed Production, and Interference in Cotton. Weed Science, 57(3), 346-350.
- Thebaud, C., & Abbott, R. J. (1995). Characterization of invasive Conyza species (Asteraceae) in Europe: quantitative trait and isozyme analysis. American Journal of Botany, 82(3), 360-368.
- Thorpe, J. P., & Solé-Cava, A. M. (1994). The use of allozyme electrophoresis in invertebrate systematics. Zoologica Scripta, 23(1), 3-18.
- Vladimirov, V. (2009). Erigeron sumatrensis (Asteraceae): a recently recognized alien species in the Bulgarian flora. Phytologia Balcanica, 15(3), 361-365.
- Weaver, S. E. (2001). The biology of Canadian weeds: Conyza canadensis Canadian Journal of Plant Science, 81(4), 867-875.
- Wright, S. (1965). The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19(3), 395-420.
- Xiao-Ling, S., Wu, J., Zhang, H., & Qiang, S. (2011). Occurrence of glyphosate-resistant horseweed (Conyza canadensis) population in China. Agricultural Sciences in China, 10(7), 1049-1055.
- Yamauti, M. S., Barroso, A. A. M., Souza, M. C., & Alves, P. L. C. A. (2010). Chemical control of glyphosate-resistant horseweed (Conyza canadensis) and hairy fleabane (Conyza bonariensis) biotypes. Revista Ciência Agronômica, 41(3), 495-500.
- Yeh, F. C., Boyle, T. Y. Z., & Xiyan, J. M. (1999). Popgene Version 1.31: Microsoft Window-based freeware for population genetic analysis Edmonton, AL: University of Alberta.
- Yuan, J., Abercrombie, L., Cao, Y., Halfhill, M., Zhou, X., Peng, Y., & Stewart, C. (2010). Functional genomics analysis of horseweed (Conyza canadensis) with special reference to the evolution of non-target-site glyphosate resistance. Weed Science, 58(2), 109-117.
- Zelaya, I. A., Owen, M. D. K., & Vangessel, M. J. (2007). Transfer of glyphosate resistance: evidence of hybridization in Conyza (Asteraceae). American Journal of Botany, 94(4), 660-673.
- Zheng, Q. A., Tai, C., Hu, J., Lin, H., Zhang, R., Su, F., & Yang, X. (2011). Satellite altimeter observations of nonlinear Rossby eddy-Kuroshio interaction at the Luzon Strait. Journal of Oceanography, 67(4), 365-376.
Publication Dates
-
Publication in this collection
Oct-Dec 2017
History
-
Received
01 Aug 2016 -
Accepted
23 Nov 2016