Abstracts
Tamarillo is one of the main fruit crops in the Andean region of Colombia. However, due to expansion of viral diseases, the cultivated area has undergone a rapid decline during recent years. In this work, we report the taxonomical identity of some of the viruses present in the main tamarillo producing regions in Colombia. The presence of AMV, CMV, PLRV, Potyvirus, ToMV, ToRSV and TSWV was evaluated by ELISA in the provinces of Antioquia, Boyacá, Cundinamarca, Nariño and Putumayo. These results were complemented with RT-PCR and cDNA sequencing of the corresponding coat regions. Potyvirus, CMV and PLRV are the most predominant viruses, each virus being detected in at least one sample. Sequencing results revealed high levels of identity of PVY and PLRV from tamarillo with virus strains from potato crops, suggesting the possibility of cross infection. Our findings confirm that a virus complex is responsible for the decline of tamarillo productivity in Colombia.
Potyvirus; Polerovirus; ELISA; mixed infection; RT-PCR; tamarillo; tree tomato
Tomate de árvore (Solanum betaceum) é uma das principais fruteiras da região dos Andes da Colômbia. No entanto, devido à expansão de doenças virais, a área cultivada diminuiu-se rapidamente nos últimos anos. Neste trabalho, nós relatamos a identificação taxonômica de alguns dos vírus presentes nas principais regiões produtoras tomate de árvore na Colômbia. A presença de AMV, CMV, PLRV, Potyvirus, ToMV, TSWV e ToRSV foi avaliada pelo ELISA, nos estados de Antioquia, Boyacá, Cundinamarca, Nariño e Putumayo. Estes resultados foram complementados com RT-PCR e seqüenciamento do DNA das regiões correspondentes ao gene da proteína capsidial. Potyvirus, CMV e PLRV foram os vírus mais prevalentes, tendo sido cada um deles detectado em pelo menos uma amostra. Resultados do seqüenciamento mostram altos níveis de identidade dos isolado de PVY e PLRV obtidos de S. betaceum com isolados do vírus da batata, sugerindo a possibilidade de infecção cruzada. Os resultados confirmam a proposta de um complexo de vírus como responsável pela redução na produção do tomate de árvore na Colômbia.
Potyvirus; Polerovirus; ELISA; infecção multi-viral; RT-PCR; tamarillo; tomate de árvore
RESEARCH ARTICLE ARTIGO
Margarita JaramilloI; Pablo Andrés GutiérrezII; Luz Estela LagosIII; José Miguel CotesIV; Mauricio MarínII
IFaculty of Administrative and Agricultural Sciences, Corporación Universitaria Lasallista, Caldas, Antioquia, Colombia
IILaboratories of Molecular Biology and Industrial Microbiology, Faculty of Sciences, National University of Colombia, Medellín, Colombia, AA 3840
IIIDepartment of Biology, Faculty of Natural Sciences and Mathematics, University of Nariño, Pasto, Colombia
IVDepartment of Agronomy, Faculty of Agricultural Sciences, National University of Colombia, Medellín, Colombia, AA 3840
ABSTRACT
Tamarillo is one of the main fruit crops in the Andean region of Colombia. However, due to expansion of viral diseases, the cultivated area has undergone a rapid decline during recent years. In this work, we report the taxonomical identity of some of the viruses present in the main tamarillo producing regions in Colombia. The presence of AMV, CMV, PLRV, Potyvirus, ToMV, ToRSV and TSWV was evaluated by ELISA in the provinces of Antioquia, Boyacá, Cundinamarca, Nariño and Putumayo. These results were complemented with RT-PCR and cDNA sequencing of the corresponding coat regions. Potyvirus, CMV and PLRV are the most predominant viruses, each virus being detected in at least one sample. Sequencing results revealed high levels of identity of PVY and PLRV from tamarillo with virus strains from potato crops, suggesting the possibility of cross infection. Our findings confirm that a virus complex is responsible for the decline of tamarillo productivity in Colombia.
Key words:Potyvirus, Polerovirus, ELISA, mixed infection, RT-PCR, tamarillo, tree tomato.
RESUMO
Tomate de árvore (Solanum betaceum) é uma das principais fruteiras da região dos Andes da Colômbia. No entanto, devido à expansão de doenças virais, a área cultivada diminuiu-se rapidamente nos últimos anos. Neste trabalho, nós relatamos a identificação taxonômica de alguns dos vírus presentes nas principais regiões produtoras tomate de árvore na Colômbia. A presença de AMV, CMV, PLRV, Potyvirus, ToMV, TSWV e ToRSV foi avaliada pelo ELISA, nos estados de Antioquia, Boyacá, Cundinamarca, Nariño e Putumayo. Estes resultados foram complementados com RT-PCR e seqüenciamento do DNA das regiões correspondentes ao gene da proteína capsidial. Potyvirus, CMV e PLRV foram os vírus mais prevalentes, tendo sido cada um deles detectado em pelo menos uma amostra. Resultados do seqüenciamento mostram altos níveis de identidade dos isolado de PVY e PLRV obtidos de S. betaceum com isolados do vírus da batata, sugerindo a possibilidade de infecção cruzada. Os resultados confirmam a proposta de um complexo de vírus como responsável pela redução na produção do tomate de árvore na Colômbia.
Palavras-chave:Potyvirus, Polerovirus, ELISA, infecção multi-viral, RT-PCR, tamarillo, tomate de árvore.
INTRODUCTION
Tamarillo (Solanum betaceum Bosh) is a fruit tree of Andean origin mainly cultivated in Colombia and Ecuador for use in the fresh fruit market and the food processing industry. Tamarillo is also produced less widely in Zambia, Zimbabwe, Uganda, Sri Lanka, India and New Zealand (Ministerio de Agricultura y Desarrollo Rural 2006; Stangeland et al., 2009). In Colombia, tamarillo orchards cover a total cultivated area of 7,646 ha distributed in 18 provinces. However, Antioquia and Cundinamarca alone account for two-thirds of the total production (50 and 14%, respectively) (Ministerio de Agricultura y Desarrollo Rural 2006). In Colombia, the production of tamarillo has declined dramatically due to several phytosanitary problems such as anthracnose (Colletotrichum acutatum), root-knot nematodes (Meloidogyne spp.) and a complex of viruses. The latter has caused a 50-80% reduction in the cultivated area of traditional tamarillo regions such as eastern and northern Antioquia (Mejía et al., 2009).
Several studies have reported viral infections of tamarillo in different Colombian regions caused by unknown species of potyvirus and isometric viruses (Sánchez de Luque et al., 1982a; Tamayo 1990, 1996; Tamayo et al., 1999; Betancourth et al., 2003; Cruz 2005; Cuspoca 2007; Gil et al., 2009a). Some of the symptoms associated with these diseases include mosaics, outgrowth of veins, dark-green blisters, severe leaf deformation, yellowing, ringspots, defoliation and drastic reduction in fruit production and plant longevity. Immature fruits show color breaking ranging from yellow to pale red. In mature fruits the pulp is hard and the skin presents purple patches (Tamayo 1996; Betancourth et al., 2003; Gil et al., 2009a).
Studies in different countries have shown that viral infections of tamarillo are caused by a mixture of viruses that can include members of genera Alfamovirus (Alfalfa mosaic virus, AMV), Cucumovirus (Cucumber mosaic virus, CMV), Nepovirus (Tomato ringspot virus, ToRSV), Polerovirus (Potato leafroll virus, PLRV), Potexvirus (Potato aucuba mosaic virus, PAMV), Potyvirus (Potato virus Y, PVY; Tamarillo mosaic virus, TaMV; Potato virus A, PVA), Tobamovirus (Tomato mosaic virus, ToMV) and Tospovirus (Tomato spotted wilt virus, TSWV) (Vizuete et al., 1990; Eagles et al., 1994). Due to the complex mixture of viruses associated with viral infections of tamarillo and the lack of knowledge of the predominant species in Colombia, characterization studies are required for the implementation of appropriate control strategies. In this paper we investigated the presence of AMV, CMV, PLRV, Potyvirus, ToMV, ToRSV and TSWV viruses in five tamarillo producing regions in Colombia using a combination of ELISA and RT-PCR.
MATERIALS AND METHODS
Sources of virus-infected plant material
Tamarillo samples were collected in the Colombian provinces of Antioquia, Boyacá, Cundinamarca, Nariño and Putumayo. A total of twelve orchards per province were sampled. Each sample consisted of four young leaves from plants with symptoms of virus diseases. A detailed characterization of plant symptoms observed in leaves, fruits and flowers was also carried out for each orchard.
ELISA tests
Each sample was tested for the presence of AMV, CMV, PLRV, Potyvirus, ToMV, ToRSV and TSWV viruses using ELISA. Detection antibodies were purchased from Agdia (Indiana, USA). Potyvirus antibodies were specific for all aphid-transmitted members of the Potyvirus Group. Each test was run in parallel with a positive control provided by the manufacturer and a negative control. Absorbance values were recorded using a Multiscan Reader (Labsystem, Finland). A sample was considered positive if the absorbance reading at 405 nm was at least twice the value of the negative control (Matthews, 1993).
Sequencing
Total RNA was isolated from positive ELISA samples using the RNeasy plant mini kit (Qiagen, CA, USA). 100 mg of leaf tissue was macerated in 450 µL of RLT buffer and 10 µL β-mercaptoetanol following manufacturer's recommendations. Total RNA was resuspended in 40 µL of DEPC-treated water. RT was performed in a reaction mixture (20 µL) containing 1X buffer, 2.5 mM MgCl2, 0.5 mM of each dNTP, 2.5 µM of reverse primer (Table 1), 100 U M-MuLV Reverse transcriptase (Fermentas, Lithuania), 10 U RNasin (Fermentas) and 5 µL of total RNA at 37°C for 30 min. PCR was performed in 2.5 µL of the synthesized cDNA, 1X PCR buffer, 2.5 mM MgCl2, 2 mM dNTPs, 0.5 U DNA Taq polymerase (Fermentas) and 1 µM of forward and reverse primers (Table 1). Conditions for PCR were as follows: 94°C for 5 min and 35 cycles of 94°C for 1 min, 50-58°C for 1 min, 72°C for 2 min, followed by 10 min at 72°C, performed on a Biometra thermal cycler (Göttingen, Germany). Due to the lack of sequence data for tamarillo viruses in the Andean region, RT-PCR primers were chosen based on previous reports of viruses infecting this crop in other countries (Table 1). RT-PCR amplifications were run in 1.5% agarose gels containing ethidium bromide (10 mg/mL) and were visualized using a Biometra UV transilluminator. Amplicon size was verified by comparison with the Generuler 100 pb DNA ladder (Fermentas). RT-PCR products were purified using QIAquick PCR Purification kit (Qiagen) and gel purified using the QIAquick Gel Extraction Kit (Qiagen) when required. Sequencing reactions were performed using the primers used in the RT-PCR. The reactions were run on an ABI Prism 3730xl sequencer (PE Applied Biosystems) at Macrogen (South Korea).
Phylogenetic analysis
DNA sequences were edited with BioEdit 6.0.6 and Chromas 2.01 and analyzed with Blastn (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Sequences for other closely related virus species were obtained from GenBank and analyzed using PAUP 4.0b10* (Swofford, 2002). Sequences were aligned with the program Clustal X and results were checked manually. Trees were obtained by the stepwise addition of 1,000 replicates using the heuristic search option with stepwise addition of PAUP. Confidence intervals using 1,000 bootstrap replicates were calculated (Felsenstein, 1985).
RESULTS
Symptoms
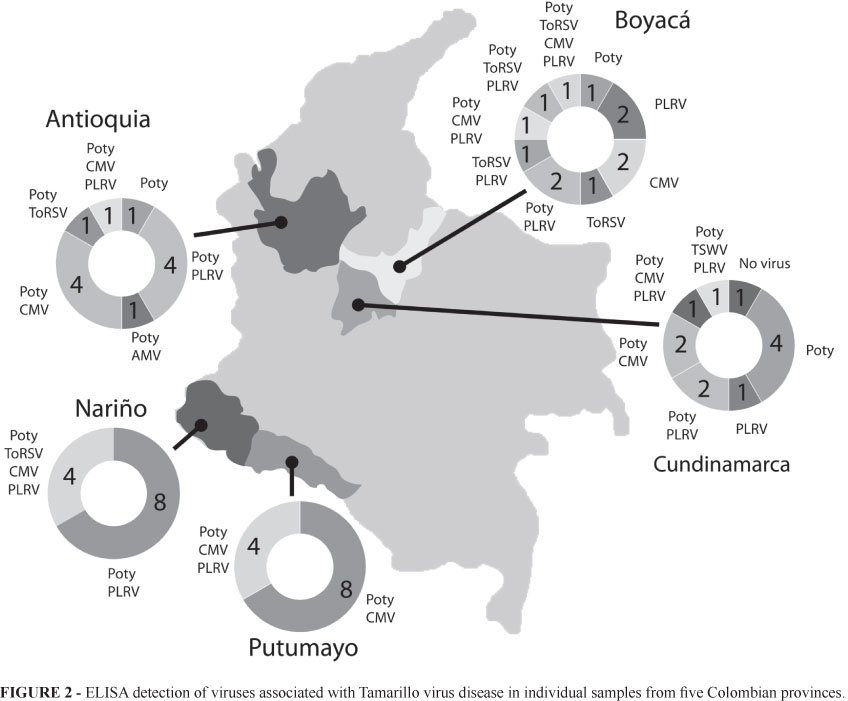
In spite of being infected by a wide variety of viruses, samples shared a similar symptomatology. The most common were general mosaics with green blisters in leaves, vein outgrowth, shoot deformation, flower and fruit breaking, hardness of fruit pulp, purple patches on fruit skin and severe defoliation. Some plants also presented more specific symptoms such as vein banding, etching, oily and concentric ringspots and bronzing (Figure 1).
ELISA tests
Each virus was detected by ELISA in at least one sample out of 60 (Table 2). The most prevalent viruses were Potyvirus and CMV, found in all provinces, and PLRV, only absent in Putumayo. ToMV was only found in south Colombia (Nariño and Putumayo) while AMV, ToRSV and TSWV were found marginally in some provinces. Co-infection was common, especially in Antioquia, Boyacá and Cundinamarca. Four samples from Nariño and one from the municipality of Saboyá (Boyacá) revealed the presence of four viruses: Potyvirus, CMV, PLRV, ToMV (in Nariño) and ToRSV (in Boyacá). In the southern provinces, four out of 12 samples contained three or more different viruses (Figure 2). It is important to note that all samples tested, with the exception of one from San Bernardo, Cundinamarca, were positive for virus infection.
Sequencing and phylogenetic analysis
A total of 26 amplicons with the expected sized of 330 bp were obtained with primers PLRV F-R. These samples were collected in Antioquia (12 samples), Boyacá (3 samples), Cundinamarca (7 samples) and Nariño (4 samples). DNA sequencing of 10 amplicons confirmed the presence of PLRV. Comparison with other PLRV sequences isolated from different potato strains showed a percent identity higher than 97%. Phylogenetic analysis grouped all PLRV strains in a single cluster with a bootstrap value of 100%. CYDV-RPV was used as outgroup (Figure 3).
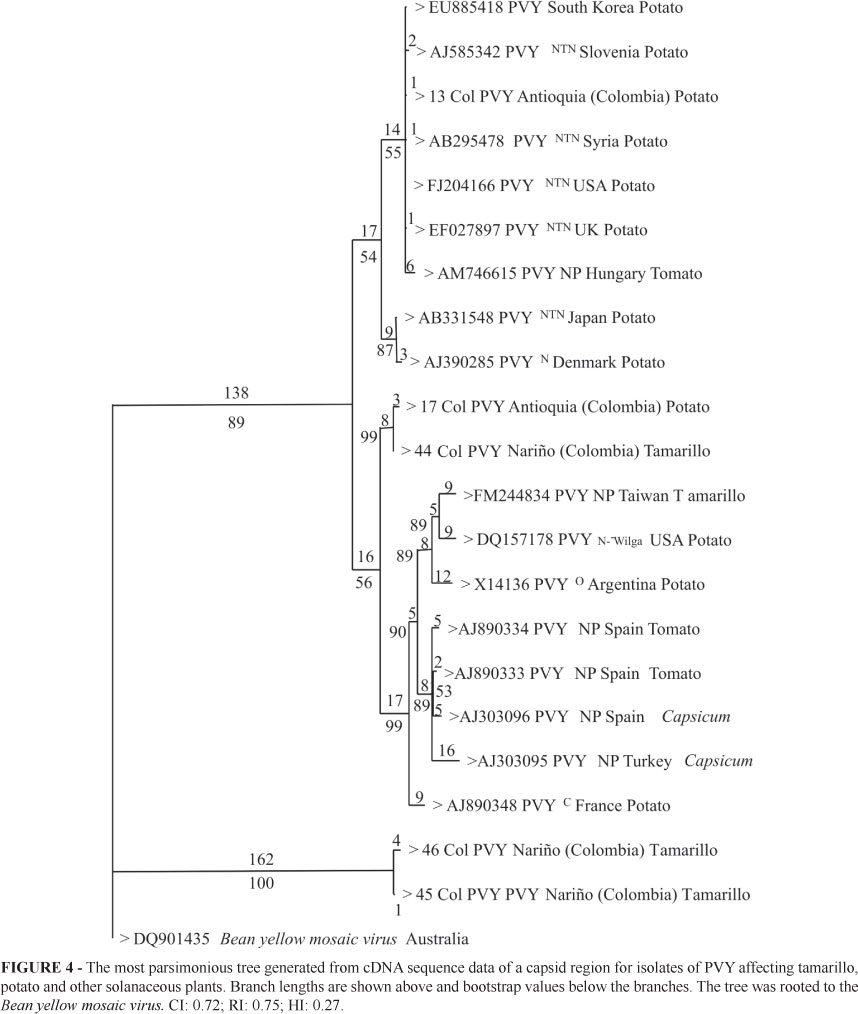
For PVY, a total of eight amplicons of 480 bp were obtained using primers PVY F-R. Samples were collected in Antioquia (2 samples), Cundinamarca (3 samples) and Nariño (3 samples). Only samples from Nariño gave high quality sequences and their comparison with PVYº, PVYC, PVYN, PVYNTN and PVYN-Wilga, PVY-NP showed a percent identity higher than 87%. This analysis revealed a close relationship between strain 44Col from tamarillo samples collected in the municipality of Córdoba (Nariño) and PVYNTN isolates from different countries. Percent identity between these two strains was higher than 96%. Strain 44Col was identical to strain 17Col reported by Gil et al. (2009b) in potato crops in La Unión (Antioquia). The remaining sequences are less than 93% identical to strain 17Col and the reference sequences (Gil et al., 2009b). Phylogenetic analysis segregated PVY strains into two clusters with high bootstrap values (89% and 100% for groups I and II, respectively). Group I was divided into three subgroups as follows: the first subgroup was composed of strains closely related to PVYNT, PVYN and PVY isolate 13Col. The second subgroup included Colombian PVY isolates 44Col from tamarillo (Nariño) and 17Col from potato (La Unión, Antioquia). Finally, the third subgroup was composed of PVY races O, C and N-Wilga as well as NP strains (Non-Potato type, Aramburu et al., 2006) isolated from Capsicum spp. and S. lycopersicum. Group II is composed of two PVY isolates from Nariño not related to any of the PVY reference strains included in the analysis (Figure 4). The BYMV sequence, with an approximate percent identity of 65 % to the PVY isolates, clustered outside Groups I and II. RT-PCR did not amplify fragments of the correct size for AMV, CMV, ToRSV, ToMV and TSMW. This might be due to variations in the annealing regions of the generic primers used to detect these viruses.
DISCUSSION
Virus diseases are the most limiting phytosanitary problem in the production of tamarillo in Colombia (Mejía et al., 2009). Serological and electron microscopy studies demonstrated the association of this disease with potyvirus infection (Saldarriaga et al., 1997; Tamayo et al., 1999; Betancourth et al., 2003). Our serological and molecular studies confirmed those results and identified Potato virus Y as one of the potyviruses infecting tamarillo plants in Colombia. Preliminary work by Tamayo (1996) and Tamayo et al. (1999), also suggest the presence of at least an isometric virus in tamarillo plants exhibiting mosaic symptoms. Using ELISA, we also detected the presence of isometric viruses AMV, CMV, PLRV and ToRSV. Some studies have postulated the occurrence of viral complexes in tamarillo orchards. This hypothesis was supported by the wide variety of symptoms associated with this crop in New Zealand (Chamberlain, 1954) and further confirmed by ELISA tests in Ecuador (Vizuete et al., 1990) and New Zealand (Eagles et al., 1994).
Potyviruses are commonly found in tamarillo. In Ecuador the most common potyvirus is PVY while in New Zealand TaMV has been reported with an incidence as high as 100% in some regions of this country. PLRV, ToRSV and AMV have been frequently found in the valleys of the province of Pichincha, Ecuador. In New Zealand CMV, PaMV, AMV, TSWV and ArMV have also been detected (Eagles 1994; Eagles et al., 1994). In this work, serological tests confirmed the presence of all the viruses under study in at least one sample out of a total of 60. CMV and PLRV were the most common in contrast to AMV and TSWV, which were only detected in one sample. ToMV and ToRSV were found in eight and five samples, respectively. It is interesting to note that five samples were simultaneously infected with four viruses. Simultaneous infection with two or three viruses, one of them a potyvirus, was also common. These results confirm the observation of flexuous rods about 750 to 800 nm long in tamarillo plants from Colombia with symptoms of viral infection (Maldonado & Sánchez de Luque 1984; Saldarriaga & Bernal, 1995; Betancourth et al., 2003). However, the identity of these viruses could not be determined using the universal antibodies for the potyvirus group designed by Jordan & Hammond (1991) and distributed by Agdia (USA). In the literature, the most common potyviruses infecting tamarillo are PVY and TaMV. PVY was first reported by Chamberlain (1954) in New Zealand and by Bhargava & Joshi (1959) in India. The impact of PVY on tamarillo was later confirmed by Vizuete et al. (1990) in Ecuador. They found that this virus is the most common in tamarillo orchards in the valleys of Pichincha and responsible for symptoms such as yellowing and leaf deformation. RT-PCR detection of PVY confirmed the presence of these viruses in samples from Antioquia, Cundinamarca and Nariño (Colombia). This was later verified by sequencing results from samples collected in Nariño. PVY is commonly divided into races based on biological, serological and molecular evaluations (Singh et al., 2008).
Phylogenetic analysis shows that PVY strains from tamarillo in Nariño cluster in two clades. The first clade is composed of strain 44Col and strains of the PVYNTN race from different countries. The second clade separated strains 45Col and 46Col from all the other PVY strains used in this study (Bootstrap of 100%), including strains NP from Capsicum sp. and S. lycopersicum. Even though this subgroup is clearly related to PVY, it was not possible to find a close association with any particular race and this probably represents a new variant of PVY. PVY isolate from tamarillo plants in Taiwan clustered in a branch that includes PVYW, O type strains from potato. These strains belong to a well supported clade with NP strains of PVY (Bootstrap of 92%). Isolate PVY 44Col from tomato tree in Nariño shares 99% identity with the capsid sequence of strain 17 Col from potato (Gil et al., 2009b) and it is possible that both plants can serve as host to similar PVY strains. This result suggests that phytosanitary measures should probably consider both crops as a source of PVY inoculum, especially as potato and tamarillo crops are commonly found in the same Andean agroecosystems. This hypothesis should be verified experimentally in greenhouse and field cross-pathogenicity tests and complemented with the entire genome sequencing of both viral strains.
On the other hand, the grouping of strains 45Col and 46Col in a clade different from NP and the PVY (N, O, C) races supports the postulate of Singh et al. (2008) which claims that the PVY species is composed of a large number of variants that arose by multiple mutation, recombination and gene rearrangement events. The characterization of novel PVY strains from different hosts will improve our knowledge of the degree of variability in this species.
Serological detection of PLRV in all four provinces is surprising as this virus has only been marginally detected in tomato tree in Ecuador (Vizuete et al., 1990), has not been detected in New Zealand and Solanum betaceum is not considered a natural host by the International Committee on Taxonomy of Viruses. PLRV can infect plants from Solanaceae, Amaranthaceae, Cruciferae and Portulacaceae families but S. tuberosum and S. lycopersicum are believed to be its natural hosts. Using ELISA, Vizuete et al. (1990) correlated the presence of PLRV with symptoms such as vein yellowing, mosaics and oily ringspots. However, the presence of other viruses such as AMV, ToRSV and PVY made it difficult to attribute these symptoms exclusively to PLRV. Ochoa & Insuasti (2005) demonstrated the appearance of vein outgrowth, leaf rolling, yellowing and necrosis in older leaves after transmission of PLRV in tamarillo by M. persicae. In potato, symptoms of primary infection consist typically of chlorosis and leaf rolling of young leaves, which have a reddish appearance. Plants with secondary infection can develop chlorosis of upper leaves, rolling of older leaves and a severe reduction of plant growth (Salazar, 1995).
Detection of PLRV in tamarillo plants in this study raises several issues that should be addressed in future investigations. First of all the specific symptoms caused by PLRV and its effect on fruit production and plant longevity should be determined. Secondly, given the fact that commercial propagation of tamarillo is very different to potato (tubers vs sexual seed), it is important to establish whether PRLV transmission is similar in both hosts. It is worth mentioning that tamarillo and potato are visited by some of the same aphid species in Colombia (Martínez et al., 2009).
Analysis of partial PLRV coat sequences isolated from tamarillo reveal a high percent identity (> 98%) between Antioquia, Cundinamarca and Nariño strains and strain 4Col isolated from a potato crop in the municipality of la Unión (Antioquia) (Gil et al., 2009b). Strain 4Col is also more than 99% identical to isolates 25Col and 28Col identified in tamarillo in the municipality of Córdoba (Nariño). It is then reasonable to propose that potato and tomato tree share the same PLRV variants, as might also be the case with PVY. This hypothesis could be verified by cross pathogenicity tests in future studies.
The dendrogram grouped all tomato tree PLRV sequences with potato reference strains from all over the world. Percent identity inside this group was higher than 97.5%. The high levels of identity between the PLRV strains found in this work are compatible with previous studies done in different countries and continents (Guyader & Ducray, 2002; Taliansky et al., 2003; Mukherjee et al., 2003; Plchova et al., 2009). The level of variation for the whole PLRV genome does not exceed 6%, with ORF 3, coding for the coat protein, being the most conserved with percent identities above 96.5%. The most variable regions of the PLRV genome correspond to ORF 0 and the 5' of ORF 1, but their percent identity exceeds 94.5% (Guyader & Ducray, 2002; Plchova et al., 2009). These ORFs have been used to classify PLRV into three lineages. Lineage II corresponds exclusively to Australian strains. Lineages I and III are composed of strains from different geographical regions with no difference in their degree of virulence or aphid transmission efficiency (Guyader & Ducray, 2002). It would be interesting to sequence these regions in Colombian PRLV strains and determine their phylogenetic relationship to the aforementioned lineages. Additionally, pathogenicity tests of tamarillo strains on Physalis floridana, Datura stramonium, Capsicum spp. and S. lycopersicum will make it possible to determine if these variants belong to races that can infect hosts other than potato such as Tobacco yellow top virus, Tomato yellow top strain (Thomas, 1984) and Capsicum yellows virus (Gunn & Pares, 1990).
CMV, detected in all five provinces and 29 out of 60 samples, was another frequently found virus. CMV belongs to the Bromoviridae family and has a single-stranded RNA trisegmented genome encapsidated in isometric capsids. This virus is characterized by an extremely broad host range that includes more than 1000 species from at least 65 different plant families and is transmitted by more than 60 aphid species (García-Arenal et al., 2000). CMV can act as helper virus of satellite RNAs between 300-400 nt which can affect symptom expression as shown with systemic necrosis on tomato (García-Arenal & Palukaitis, 1999). CMV was first reported on tamarillo by Chamberlain (1954) in New Zealand where its prevalence was confirmed in at least two provinces (Eagles et al., 1994). CMV induces bootlacing and is commonly found in co-infection with the TaMV potyvirus. On the other hand, Vizuete et al. (1990), using serological tests, did not detect CMV in Ecuador; however Ochoa & Insuasti (2005) showed that this virus is distributed in all producing regions in that country.
Failures in the molecular diagnosis of CMV in serological positive samples could be explained on the grounds of sequence variability and silent mutations. Several authors have reported that CMV consists of a wide range of variants with marked differences in their hosts and phathogenicity (Rizos et al., 1992; De Blas et al., 1994; García-Arenal et al., 2000; Yu et al., 2005) and that gene rearrangements has played an important role in its evolution. Future work should address the molecular characterization of CMV strains infecting tamarillo in Colombia and their phylogentic relationship to other strains. As mentioned before, AMV, ToMV, ToRSV and TSWV were only marginally found in some Colombian provinces. ToMV was only found in the south while ToRSV was mainly detected in province of Boyacá. These viruses have been detected in tamarillo producing regions in New Zealand; however, their specific symptomatology and effect on tamarillo orchards are not well established (Vizuete et al., 1990; Eagles et al., 1994; Ochoa & Unsuasti, 2005). The effect of each of these viruses and their combinations on the symptom development and production of tamarillo in Colombia should be studied in the future. The possibility that other viruses than those assayed are infecting tamarillo cannot be precluded and must also be investigated.
This study is a step towards the understanding of viral diseases affecting tamarillo in Colombia and the world. The presence of a viral complex makes it necessary to deepen the understanding of the epidemiological variables involved in the establishment and expansion of this disease in the Andean regions of Colombia and South America. Future studies should address the transmission mechanism for each virus, susceptibility of different tamarillo varieties, the existence of alternative hosts and characterization of the specific symptoms caused by each virus.
ACKNOWLEDGEMENTS
This work was financed by COLCIENCIAS grant: 1118-405-20317 (408-2007) and DIME-BICENTENARIO 2008-2009 grant: 20101007745. We thank tamarillo producers that allowed sample collection and members of the Cell and Molecular Biology Laboratory at the National University of Colombia, Medellin campus.
Received 13 January 2011
Accepted 21 June 2011
Author for correspondence: Mauricio Marín, e-mail: mamarinm@unal.edu.co
TPP235
Section Editor: F. Murilo Zerbini
- Aramburu J, Galipienso L, Matas M (2006) Characterization of potato virus Y isolates from tomato crops in northeast Spain. European Journal of Plant Pathology 115:247-258.
- Betancourth C, Goyes R, Bravo DA (2003) Caracterización biológica de un virus del tomate de árbol (Solanum betaceum Send) en el departamento de Nariño. Fitopatología Colombiana 27:7-10.
- Bhargava KS, Joshi RD (1959) A virus disease of tree tomato, Cyphomandra betacea Sendt. Due to Potato virus Y American Journal of Potato Research 36:8.
- De Blas C, Borja MJ, Saiz M, Romero J (1994) Broad spectrum detection of Cucumber Mosaic Virus (CMV) using the Polymerase Chain Reaction. Journal of Phytopathology 141:323-329.
- Chamberlain EE (1954) Plant Viruses in New Zealand. Bulletin No.108. DSIR. Department of Scientific and Industrial Research.
- Chu FH., Chao CH, Chung MH, Chen CC, Yeh SD (2001) Completion of the genome sequence of Watermelon silver mottle virus and utilization of degenerate primers for detecting tospoviruses in five serogroups. Phytopathology 91:361-368.
- Cruz LF (2005) Identificación de virus en Solanum betaceum. Tesis de Ingeniería Agronómica. Universidad Nacional de Colombia, sede Bogotá
- Cuspoca J (2007) Evaluación de virus de tomate de árbol (Solanum betaceum) en plantas indicadoras y su detección por PCR. Tesis de Ingeniería Agronómica. Universidad Nacional de Colombia sede Bogotá
- Eagles R (1994) Tamarillo mosaic potyvirus: characterization and resistance. Thesis PhD, Plant Sciences, School of Biological Sciences. University of Auckland.
- Eagles R, Gardner R, Forster R (1994) Incidence and distribution of six virus infecting Tamarillo (Cyphomandra betacea) in New Zealand. New Zealand Journal of Crop Horticultural Science 22:453-458.
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791.
- García-Arenal F, Palukaitis P (1999) Structure and functional relationships of satellite RNAs of Cucumber mosaic virus In: Vogt PK, Jackson AO (Eds.) Satellites and defective viral RNAs. Berlin. Springer-Verlag. pp. 37-63.
- García-Arenal F, Escriu F, Aranda MA, Alonso-Prados JL, Malpica JM,, Fraile A (2000) Molecular epidemiology of Cucumber mosaic virus and its satellite RNA. Virus Research 71:1-8.
- Gil JF, Ayala ML, Marín M, González EP (2009a) Identificación de Potyvirus en cultivos de tomate de árbol (Solanum betaceum Cav.) en Antioquia mediante detección serológica. Revista Politécnica 5:112-120.
- Gil JF, Quintero M, González P, Cotes JM, Marín M (2009b) Detección serológica y molecular de virus en cultivos de papa de tres regiones de Antioquia. In: 29 Congreso Nacional de Fitopatología y Ciencias Afines. Medellín, Colombia.
- Gunn LV, Pares RD (1990) Capsicum yellows a disease induced by a Luteovirus in glasshouse peppers (Capsicum annuum) in Australia Journal Phytopathology 129:210.
- Guyader S, Ducray DG (2002) Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. Journal of General Virology 83:1799-1807.
- Hsu HT, Barzuna L, Hsu YH, Bliss W, Perry KL (2000) Identification and Subgrouping of Cucumber mosaic virus with Mouse Monoclonal Antibodies. Phytopathology 90:615-620.
- Jacobi V, Bachand GD, Hamelin RC, Castello JD (1998) Development of a multiplex immunocapture PCR assay for detection and differentiation of tomato and tobacco mosaic tobamoviruses. Journal of Virological Methods 74:167-178.
- Jordan R, Hammond J (1991) Comparison and differentiation of potyvirus isolates and identification of strain, virus, subgroup-specific and potyvirus group-common epitopes using monoclonal antibodies. Journal of General Virology 72:25-36.
- Maldonado C, Sánchez de Luque C (1984) Caracterización, purificación y serología del virus de la necrosis anular del tomate de árbol (Cyphomandra betacea sendt). In: VI Congreso ASCOLFI. Santa Marta, Colombia. p. 54.
- Martínez JE, Zuluaga C, Álvarez JA, Lagos LE, Marín M (2009) Detección serológica y molecular de virus en afidos asociados a cultivos de tomate de árbol con síntomas de virosis en Antioquia, Cundinamarca y Nariño. In: 29 Congreso Nacional de Fitopatología y Ciencias Afines. Medellín. p. 123.
- Mason G, Roggero P, Tavella L (2003). Detection of Tomato spotted wilt virus in its vector Frankliniella occidentalis by reverse transcription-polymerase chain reaction. Journal of Virological Methods 109:69-73.
- Matthews REF (1993) Diagnosis of plant virus diseases. New York NY.CRC Press.
- Mejía DM, Rodas EI, Patiño LF, González EP (2009) Efecto del acibenzolar-s-metil sobre el desarrollo de la virosis causada por potyvirus en tomate de árbol. Agronomía Colombiana 27:87-93.
- Ministerio de Agricultura y Desarrollo Rural (2006) Observatorio agrocadenas Colombia. Disponível em: <http://www.agrocadenas.gov.co>. Acceso em: Fev 2008.
- Murkherjee K, Verma Y, Chakrabarti S, Singh MN, Khurana SMP (2003) Cloning and sequencing of coat protein gene of an Indian Potato leafroll virus (PLRV) isolate and its similarity with other members of Luteoviridae. Virus genes 26:247-253.
- Nie X, Singh RP (2001) A novel usage of random primers for multiplex RT-PCR detection of virus and viroid in aphids, leaves, and tubers. Journal of Virological Methods 91:37-49.
- Ochoa J, Insuasti AM (2005) Etiología de las enfermedades virales del tomate de árbol en Ecuador. Instituto Nacional de Investigaciones Agropecuarias, Quito (Ecuador). Informe Técnico Anual - INIAP. Ecuador.
- Plchova H, Cerovska N (2009) Short communication: Molecular analysis of Potato leafroll virus isolates from the Czech Republic. Virus Genes 39:153-155.
- Rizos H, Gunn L, Pares RD, Gillings MR (1992) Differentiation of Cucumber mosaic virus isolates using the polymerase chain reaction. Journal of General Virology 73:2099-2103.
- Salazar LF (1995) Los virus de la papa y su control. Centro Internacional de la Papa. Lima, Perú
- Saldarriaga A, Bernal JA (1995) Virus en tomate de árbol (Cyphomandra betacea Sendt). In: XV Congreso ASCOLFI. Bogotá, Colombia.
- Saldarriaga A, Bernal JA, Tamayo P (1997) Virosis del tomate de árbol. In: Enfermedades del cultivo de árbol en Antioquia: Guía de reconocimiento y control. Boletín técnico CORPOICA, Colombia.
- Samuitiene M, Zitikaite I, Navalinskiene M, Valiunas D (2003) Identification of tomato ringspot nepovirus by RT-PCR. Biologija (Lithuania) 4:35-38.
- Sánchez de Luque C (1982) Estudios de hospedantes de un nuevo virus en el tomate de árbol (Cythomandra betacea Sendt). In: V Congreso ASCOLFI, XXII Reunión Anual de la Sociedad Americana de Fitopatología. División Caribe, ASP- CD, Cali, Colombia. pp 41-42.
- Singh R, Kurz J, Boiteau G, Bernard G (1995) Detection of Potato leafroll virus in single aphids by the reverse transcription polymerase chain reaction and its potential epidemiological application. Journal of Virological Methods 55:133-143.
- Singh R, Valkonen T, Gray SM, Boonham N, Jones RAC, Kerlan C, Schubert J (2008) Brief Review Discussion paper: The naming of Potato virus Y strains infecting potato. Archives of Virology 153:1-13.
- Stangeland T, Remberg SV, Lye KA (2009) Total antioxidant activity in 35 Ugandan fruits and vegetables. Food Chemistry 113:85-91.
- Swofford DL (1998) PAUP: Phylogenetic analysis using parsimony (* and other methods). Version: 4. Sunderland, EEUU. Sinauer Associates.
- Taliansky M, Mayo M, Barker H (2003). Pathogen profile Potato leafroll virus: a classic pathogen shows some new tricks. Molecular Plant Pathology 4:81-89.
- Tamayo PJ (1990) Mosaico del tomate de árbol. ASCOLFI Informa 16:54-55.
- Tamayo PJ (1996) Enfermedades virales del tomate de árbol (Cyphomandra betacea (Cav).Sendt.) en Colombia. ASCOLFI Informa 22:26-29.
- Tamayo PJ, Zapata JL, Salazar LF (1999) El mosaico y la virosis del tomate de árbol en el altiplano norte de Antioquia. Revista Facultad Nacional de Agronomía Medellín 52:781-785.
- Thomas JE (1984) Characterisation of an Australian isolate of tomato yellow top virus. Annals of Applied Biology 104:79-86.
- Vizuete B, Insuasti ML, Ochoa J, Ellis M (1990) Biological and serological characterization of tree tomato virus diseases in Ecuador. INIAP, Ohio State University.
- Xu H, Nie J (2006) Identification, characterization, and molecular detection of Alfalfa mosaic virus in potato. Phytopathology 96:1237-1242.
- Yu C, Jianxiang Wu, Xueping Zhou (2005) Detection and subgrouping of Cucumber mosaic virus isolates by TAS-ELISA and immunocapture RT-PCR. Journal of Virological Methods 123:155-161.
Detection of a complex of viruses in tamarillo (Solanum betaceum) orchards in the Andean region of Colombia
Publication Dates
-
Publication in this collection
24 Aug 2011 -
Date of issue
June 2011
History
-
Received
13 Jan 2011 -
Accepted
21 June 2011