Abstracts
The genus Babesia comprises protozoa that cause diseases known as babesiosis. Dogs are commonly affected by Babesia canis or Babesia gibsoni. Babesia canis is divided into the subspecies Babesia canis canis, Babesia canis vogeli and Babesia canis rossi. Among these, Babesia canis vogeli predominates in Brazil. The objective of this study was to conduct a phylogenetic analysis on Babesia isolates from dogs in Goiânia, Goiás. Blood samples were obtained from 890 dogs presenting clinical signs suggestive of canine babesiosis that were attended at a veterinary hospital of Goiás. Only samples presenting typical intraerythrocytic parasites were used in the study. These were subjected to DNA extraction and amplification of a fragment of the 18S rRNA, by means of PCR. The PCR products were purified and sequenced. Sequences were obtained from 35 samples but only 17 of these were kept after quality assessment. Similarity analysis using BLASTn demonstrated that all 17 sequences corresponded to B. canis vogeli. Analysis using the Mega4 software showed that the isolates of B. canis vogeli from dogs in Goiânia present a high degree of molecular similarity (99.2 to 100%) in comparison with other reference isolates from other regions of Brazil and worldwide, deposited in GenBank.
Babesia canis vogeli; canine babesiosis; molecular characterization; phylogeny
O gênero Babesia compreende protozoários causadores de enfermidades denominadas babesioses. Cães geralmente são acometidos por Babesia canis ou Babesia gibsoni, sendo a primeira classificada em subespécies Babesia canis canis, Babesia canis vogeli e Babesia canis rossi. Entre essas, Babesia canis vogeli predomina no Brasil. O objetivo desse trabalho foi realizar estudo filogenético de amostras de Babesia em cães, em Goiânia, Goiás. Amostras de sangue foram obtidas de 890 cães atendidos no Hospital Veterinário de Goiás, apresentando sinais clínicos de babesiose. Somente amostras com presença de parasitos intraeritrocitários típicos foram utilizadas. Estas foram submetidas a extração de DNA e amplificação de fragmento do gene 18S rRNA pela PCR. Os produtos de PCR foram purificados e sequenciados. Foram sequenciadas 35 amostras, das quais apenas 17 foram mantidas após avaliação de qualidade. A análise de similaridade fornecida pelo BLASTn demonstrou que as 17 sequências deste estudo eram correspondentes a Babesia canis vogeli. Pela utilização do programa Mega4, foi possível verificar que as amostras de Babesia canis vogeli, provenientes de cães da cidade de Goiânia, apresentam, alto grau de similaridade molecular (99,2 a 100%) com isolados de referência de outras regiões do Brasil e do mundo, depositados em GenBank.
Babesia canis vogeli; babesiose canina; caracterização molecular; filogenia
FULL ARTICLE
Phylogenetic characterization of Babesia canis vogeli in dogs in the state of Goiás, Brazil
Caracterização filogenética de Babesia canis vogeli em cães do estado de Goiás, Brasil
Sabrina Castilho DuarteI; Juliana Alves ParenteII; Maristela PereiraII; Célia Maria de Almeida SoaresII; Guido Fontgalland Coelho LinharesI
IParasitic Diseases Laboratory, Veterinary School, Federal University of Goiás UFG
IIMolecular Biology Laboratory, Biological Sciences Institute, Federal University of Goiás UFG
Corresponding author Corresponding author: Sabrina Castilho Duarte Escola de Veterinária, Universidade Federal de Goiás UFG, Campus II, CP 131 CEP 74001-970, Goiás, GO, Brasil e-mail: sabrinacd@gmail.com
ABSTRACT
The genus Babesia comprises protozoa that cause diseases known as babesiosis. Dogs are commonly affected by Babesia canis or Babesia gibsoni. Babesia canis is divided into the subspecies Babesia canis canis, Babesia canis vogeli and Babesia canis rossi. Among these, Babesia canis vogeli predominates in Brazil. The objective of this study was to conduct a phylogenetic analysis on Babesia isolates from dogs in Goiânia, Goiás. Blood samples were obtained from 890 dogs presenting clinical signs suggestive of canine babesiosis that were attended at a veterinary hospital of Goiás. Only samples presenting typical intraerythrocytic parasites were used in the study. These were subjected to DNA extraction and amplification of a fragment of the 18S rRNA, by means of PCR. The PCR products were purified and sequenced. Sequences were obtained from 35 samples but only 17 of these were kept after quality assessment. Similarity analysis using BLASTn demonstrated that all 17 sequences corresponded to B. canis vogeli. Analysis using the Mega4 software showed that the isolates of B. canis vogeli from dogs in Goiânia present a high degree of molecular similarity (99.2 to 100%) in comparison with other reference isolates from other regions of Brazil and worldwide, deposited in GenBank.
Keywords:Babesia canis vogeli, canine babesiosis, molecular characterization, phylogeny.
RESUMO
O gênero Babesia compreende protozoários causadores de enfermidades denominadas babesioses. Cães geralmente são acometidos por Babesia canis ou Babesia gibsoni, sendo a primeira classificada em subespécies Babesia canis canis, Babesia canis vogeli e Babesia canis rossi. Entre essas, Babesia canis vogeli predomina no Brasil. O objetivo desse trabalho foi realizar estudo filogenético de amostras de Babesia em cães, em Goiânia, Goiás. Amostras de sangue foram obtidas de 890 cães atendidos no Hospital Veterinário de Goiás, apresentando sinais clínicos de babesiose. Somente amostras com presença de parasitos intraeritrocitários típicos foram utilizadas. Estas foram submetidas a extração de DNA e amplificação de fragmento do gene 18S rRNA pela PCR. Os produtos de PCR foram purificados e sequenciados. Foram sequenciadas 35 amostras, das quais apenas 17 foram mantidas após avaliação de qualidade. A análise de similaridade fornecida pelo BLASTn demonstrou que as 17 sequências deste estudo eram correspondentes a Babesia canis vogeli. Pela utilização do programa Mega4, foi possível verificar que as amostras de Babesia canis vogeli, provenientes de cães da cidade de Goiânia, apresentam, alto grau de similaridade molecular (99,2 a 100%) com isolados de referência de outras regiões do Brasil e do mundo, depositados em GenBank.
Palavras-chave:Babesia canis vogeli, babesiose canina, caracterização molecular, filogenia.
Introduction
The natural hosts of the protozoa Babesia canis and Babesia gibsoni are canids, and these protozoa present extensive geographic distribution. On the basis of vector specificity and antigenic properties, B. canis has been subdivided into three subspecies: B. canis canis, B. canis vogeli and B. canis rossi (UILENBERG et al., 1989). Nevertheless, other researchers have adopted phylogenetic criteria to support the taxonomic classification for these organisms into three distinct species (ZAHLER et al., 1998). The red dog tick Rhipicephalus sanguineus, which is recognized as the tick vector of B. gibsoni and B. canis vogeli, is widespread in Brazil, especially in urban areas (SZABÓ et al., 2007).
Babesia canis rossi occurs in South Africa and Sudan, whereas B. canis. canis is endemic in Europe and B. canis vogeli in northern Africa, North America, Europe, Australia, Sudan, Turkey and South America. Babesia gibsoni is predominantly found in Asia, Africa, North America, Europe and Australia (EIRAS et al., 2008; IRWIN, 2009). In Brazil, the most common piroplasm in dogs is B. canis vogeli (PASSOS et al., 2005; DUARTE et al., 2008a; RAMOS et al., 2010). However, there have been two reports on the presence of B. gibsoni in southern Brazil (TRAPP et al., 2006).
Direct microscopic examination is the conventional method for detecting Babesia spp. in animal blood samples. This is a conclusive, feasible and low cost diagnostic method, but not necessarily reliable for differentiation among species or subspecies (CACCIO et al., 2002).
Recent approaches within the field of molecular biology have increased the use of molecular techniques such that they have become suitable tools for taxonomic studies (UILENBERG, 2006). Consequently, genetic analysis has improved and this has promoted advanced phylogenetic characterization of microorganisms, thereby resulting in the emergence of new groups and also proposals for taxonomic changes (KJEMTRUP; CONRAD, 2006; HUNFELD et al., 2008).
Based on analysis of the 18S rRNA gene, Zahler et al. (2000) performed genotypic characterization of a small piroplasm isolated from dogs with babesiosis in Spain and concluded that it was more closely related to B. microti, Babesia rodhaini and Theileria equi than to B. gibsoni. These authors regarded it as a new species and named it Theileria annae. Later on, Camacho et al. (2001) demonstrated that this species was widespread in northwestern Spain.
Molecular techniques have also been used to confirm the uncommon presence of T. equi in blood samples from asymptomatic dogs in Spain (CRIADO-FORNELIO et al., 2004) and, concerning B. gibsoni, to demonstrate that indeed it represents at least three phylogenetically distinct species (IRWIN, 2009).
This present study had the aim of phylogenetically investigating piroplasms present in dogs with symptoms compatible with canine babesiosis, in the city of Goiânia, state of Goiás, Brazil (16º 40' 58" S and 49º 15' 65" W).
Material and Methods
1. Sample collection
Venous blood samples were collected into EDTA from 890 dogs referred to the Veterinary Hospital of the Veterinary School, Federal University of Goiás (UFG) showing clinical signs compatible with canine babesiosis, between January and August, 2009. The samples were screened by means of direct microscopic examination to detect piroplasmid-like parasites in Giemsa-stained blood smears. The morphological features of the intraerythrocytic stages of parasites were evaluated based on Hoskins (1991). Positive samples were immediately subjected to DNA extraction and maintained at 20 ºC for PCR tests.
2. DNA extraction
Total DNA was extracted from blood samples that presented positive results in the parasitological examination (n = 35). A commercial kit (IllustraTM, GE Healthcare) was used for this procedure, in accordance with the manufacturer's instructions, and was adjusted to a volume of 200 µL of blood. The eluted DNA was kept at 20 ºC before PCR amplification.
3. Design of primers for targeting 18S rDNA amplification
A pair of oligonucleotides was designed for targeting the 18S rRNA gene amplification by means of PCR. Initially, the following sequences (with respective access numbers) were retrieved from the GenBank database: B. canis vogeli (AY072925), B. canis canis (AY072926), B. canis rossi (L19079), B. gibsoni (AF231350), B. bovis (L31922), B. bigemina (X59604) and B. equi (Z15105). These sequences were subjected to multiple alignment using the ClustalW method (THOMPSON et al., 1999). The pair of primers was then picked out from conserved regions of the sequences.
The selected primers Bab7 (5'-GGC TAC CAC ATC TAA GGA AG-3') and Bab9 (5'-CTA AGA ATT TCA CCT CTG ACA G-3) were then evaluated for similarities against the GenBank database by means of the BLASTn® algorithm (ALTSCHUL et al., 1990).
4. PCR assay
Total DNA samples extracted from the piroplasm-like positive dog blood samples were subjected to PCR in order to amplify the genus-specific 18S rRNA target fragment.
The PCR mix was prepared for a final volume of 50 µL, as described by Duarte et al. (2008a), as follows: 1× PCR buffer (Invitrogen, Carlsbad, USA); 2.0 mM magnesium chloride (Invitrogen); 0.2 mM dNTP (GE Healthcare, Buckinghamshire, England); 10 pM forward primer Bab7; 10 pM reverse primer Bab9; 1.25 U of Taq DNA polymerase (Invitrogen) and 5 µL of DNA sample. Amplification was performed in a thermocycler (Eppdendorf Mastercycler Personal, Hamburg, Germany), programmed for 35 cycles of denaturation at 94 ºC for 30 seconds, annealing at 56 ºC for 30 seconds and extension at 72 ºC, preceded by an initial denaturation for 2 minutes and followed by a final elongation at 72 ºC. The positive-control DNA used in the PCR test consisted of an isolate of B. canis vogeli that had been obtained from the Parasitic Diseases Laboratory of the Veterinary School of UFG. Ultra-pure water (Invitrogen, Carlsbad, USA) was used as the negative control.
The specificities of the primer sequences were compared for similarities with the data available in GenBank, by means of the BLAST algorithm tool (http://www.ncbi.nlm.nih.gov/BLAST/) (ALTSCHUL et al., 1990). Genomic DNA extracted from 10-fold serial dilution (101 to 1012) of B. canis vogeli was used to assess the analytical sensitivity of the PCR assay, as described by Read; Hyde (1993).
The amplified products (10 µL of each sample) were subjected to electrophoresis in 1.2% agarose gel (GE Healthcare, Buckinghamshire, England) in 1× TBE buffer. The gels were stained in 0.4 µg.mL1 ethidium bromide solution for 10 minutes and then photographed using an imaging system (Vilber Lourmat, Torcy, France) under UV transillumination.
5. 18S rRNA fragment sequencing
The Babesia genus-specific PCR products were purified with QIAquick Gel Extraction Kit (Qiagen, Düsseldorf, Germany), in accordance with the manufacturer's protocol. The purified amplicons were then directly sequenced using the same primers and the DYEnamic™ ET dye terminator kit MegaBACE™ (GE Healthcare, Buckinghamshire, England), in an automatic sequencing apparatus (MegaBACE1000 - GE Healthcare, Buckinghamshire, England).
The sequences were finally analyzed by means of the Phred software (EWING, 1998), and only those with quality values over 20 were accepted for dendrogram construction.
6. Similarity and phylogenetic analysis
Sequences of the corresponding PCR products obtained with the Bab7/Bab9 primer set from the piroplasm-like organisms that had been isolated were analyzed using BLASTn to confirm their identities. They were then compared with the 18S rRNA Babesia sequences from GenBank by means of multiple alignments using the ClustalW algorithm (THOMPSON et al., 1999). Phylogenetic trees were inferred using neighbor-joining analysis, by means of the MEGA4 software with a bootstrap of 1,000 replications and evolutionary distances adjusted through Kimura-2 parameter nucleotide substitution (TAMURA et al., 2007).
7. Nucleotide sequences
The 18S rRNA sequences from GenBank included in this study consisted of B. canis vogeli strains reported from: Egypt (AY371297), Australia (AY102163), Japan (AB083374), Brazil (AY371196), Venezuela (DQ297390), Argentina (EU362993), USA (AY371198), France (AY072925), Spain (AY150061) and Turkey (AM183215). In addition to B. canis vogeli, the following closely related species were included for comparison: B. canis rossi (L19079, DQ111760), B. canis canis (AY072926, AY611729), B. gibsoni Asia-1 (AF175300) and Asia-2 (AF175301), B. canis presentii (AY649326), T. annae (AF188001), B. equi (DQ287951), B. microti (U09833) and B. conradae (AF158072). The sequences of Theileria annulata (EU083801) and Plasmodium falciparum (M19172) were used as distant phylogenetic organisms in order to root the tree.
Results
1. Samples
Direct microscopic screening of the dog blood smears detected 35 samples with intraerythrocytic piroplasm-like stages that most closely resembled babesid trophozoites or merozoites.
2. Primer specificity and sensitivity
Babesia genus-specific primers named Bab7 (forward, with 21 nucleotides, located at the position 619-639) and Bab9 (reverse, with 22 bases, at the position 1091-1112) were constructed from the 18S rRNA sequence alignments. The pair was designed to flank a DNA target fragment of approximately 490 bp.
Similarity analysis using BLASTn confirmed that the primer set Bab7/Bab9 is specific for all Babesia species, as well as for T. annae. The annealing temperature (Ta) for both primers was calculated as 56 ºC (Table 1).
Evaluation of the PCR assay specificity showed that the primer pair Bab7/Bab9 was suitable for detecting 1 Babesia-infected erythrocytes.
3. Amplification of the 18S rRNA gene fragments
Fragments of around 490 bp were obtained from all the 35 DNA samples from dogs that had previously been identified as positive by means of parasitological examination (Figure 1).
Six samples with less intensive staining signal bands were discarded in order to avoid low-quality DNA for sequencing. The remaining 29 samples generated clear bands of the expected size, without any nonspecific amplification.
Sequencing of the 29 PCR products only produced 17 with Phred values over 20. These 17 sequences shared at least 269 straight nucleotides and were selected for phylogenetic analysis.
Based on similarity analysis using BLASTn, all 17 sequences were identified as B. canis vogeli. The sequences were named as B. canis vogeli GO followed by the numbers 1 to 17, and were then deposited in the GenBank database under accession numbers GU386266-GU386282.
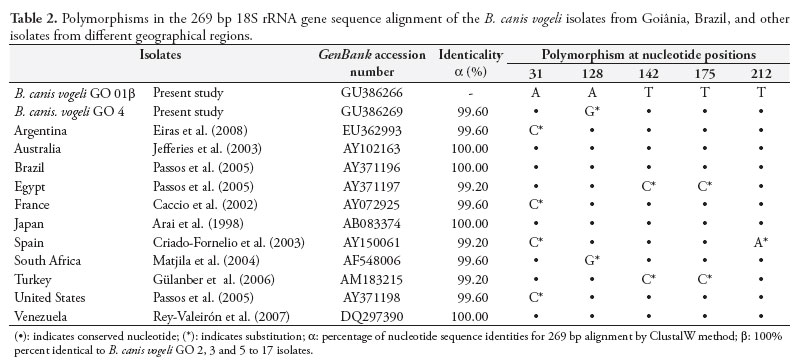
Analysis on nucleotide pleomorphism demonstrated that all the sequences in this study except for B. canis vogeli GO 4 were 100% identical to one another. The isolate B. canis vogeli GO 4 differed from any other in a single nucleotide (99.6%), consisting of a substitution (A for G) at position 128 of the alignment (Table 2). With the exception of B. canis vogeli isolate GO 4, all other isolates in this study were 100% identical to isolates from Australia, Brazil, Japan and Venezuela. A similarity of 99.6% was found, in comparison with isolates from South Africa, Argentina, the United States and France, with a unique nucleotide substitution, while a similarity of 99.2% was found, in relation to isolates from Spain, Egypt and Turkey, with two substitutions (Table 2).
4. Phylogenetic analysis
The phylogenetic tree generated from the alignment, including the partial sequences of 18S rRNA of the autochthonous B. canis vogeli GO 1 and B. canis vogeli GO 4 isolates, and the corresponding sequences of other piroplasmids used in this analysis, revealed that: 1) the sequences clustered into two distinct clades. One clade (bootstrap = 99%) was composed of B. gibsoni, B. canis vogeli, B. canis canis and B. c. rossi. The other clade (bootstrap = 74%) included the babesid species B. microti and B. conradae and the theilerids T. annae, T. equi and T. annulata; 2) B. canis vogeli GO 1 and GO 4 were clustered together to form a well-defined group with other B. canis vogeli strains from different geographical regions. This close phylogenetic relationship was sustained by a high bootstrap value for the neighbor-joining algorithm; 3) the isolate B. canis vogeli GO 4 was characterized as a branch linked to the B. canis vogeli from South Africa (bootstrap = 64%); 4) among the babesid sequences, B. canis presentii and B. canis canis presented the most distant phylogenetic relationship with the isolates B. canis vogeli GO 1 and B. canis vogeli GO 4 (bootstrap = 92%), followed by B. gibsoni (bootstrap = 68%) and B. canis rossi (bootstrap = 99%); and 5) B. conradae, T. equi, T. annulata, B. microti and T. annae generated an independent clade that was demonstrated to be the most distantly related group of species in this molecular analysis (Figure 1).
With the exception of B. canis vogeli GO isolate 4, the results obtained for the other 15 isolates in this study were the same as observed for B. canis vogeli GO 1, since they shared 100% identicality in the corresponding sequences.
Discussion
The direct parasitological examination was performed as a screening test in order to separate out piroplasmid-infected blood samples to be used in the molecular study. This procedure was restricted to identifying typical parasite stages that could undoubtedly be recognized as piroplasms, since the morphological feature may fail to discriminate among piroplasmid species, as reported elsewhere (BIRKENHEUER et al., 2004; PASSOS et al., 2005; ALLSOPP; ALLSOPP, 2006; DUARTE et al., 2008b).
The molecular similarity analysis with the 17 sequences obtained in this study revealed that they shared in common at least 99.2% similarity with the B. canis vogeli reference strains from the GenBank database. The similarity ranged from 99.2 to 100%. This result, in addition to what was inferred from the phylogenetic tree, confirmed that the present isolates presented an affiliation with other B. canis vogeli isolates from different geographical regions.
The results from this study are comparable with those presented by Passos et al. (2005), concerning similarities between B. canis vogeli from Brazil and Japan. Studies conducted in Turkey by Gülanber et al. (2006) demonstrated that their regional B. canis vogeli isolate was 100% similar to an isolate from Egypt and 99% to isolates from Brazil, Japan, France and Spain. Therefore, the results presented in this paper corroborate other studies conducted in Brazil and worldwide.
Eiras et al. (2008) concluded that B. canis vogeli isolates from Argentina were phylogenetically distant to isolates from Brazil and Venezuela. However, these authors emphasized that the sample size used was insufficient to reach any definitive conclusion. On the other hand, the present study demonstrated that the EU362993 GenBank sequence from Argentina (EIRAS et al., 2008) clustered in a branch together with isolates from Spain, France and the USA, which were linked to other B. canis vogeli from different regions including Brazil.
Occurrences of B. canis vogeli have previously been confirmed in Brazil by means of molecular techniques. Similarities of between 99.4 and 100% were reported on the basis of B. canis vogeli 18S rRNA gene analyses on isolates from five dogs in the states of Minas Gerais and São Paulo (PASSOS et al., 2005). B. canis vogeli has also been identified in samples from Rio de Janeiro by means of RFLP-PCR (DE SÁ et al., 2006) and from Goiás by means of a novel subspecies-specific PCR assay (DUARTE et al., 2008a).
The subspecies B. canis canis and B. canis rossi, which occur in other continents (IRWIN, 2009), were not found in any sample in the present study. Nor have they been found in any other study conducted in South America so far (PASSOS et al., 2005; DUARTE et al., 2008a).
Morphological examination of blood smears revealed small piroplasms similar to B. gibsoni in one dog in southern Brazil (BRACCINI et al., 1992) but this finding was not confirmed by means of any molecular technique. Based on sequence analysis on a partial fragment of the 18S rRNA gene, B. gibsoni genotype Asia 1 was characterized from four dogs in southern Brazil (TRAPP et al., 2006). This species was not detected in any of the isolates of the present study but, nevertheless, other further research is needed in order to evaluate a larger number of samples that could promote a more accurate screening in dog populations.
Other piroplasmid species have also been recognized as agents of hemoparasitosis in dogs, such as T. annae (ZAHLER et al., 2000), B. microti (ZAHLER et al., 2000, CAMACHO et al., 2001), B. conradae (BIRKENHEUER et al., 2004, KJEMTRUP et al., 2006) and T. equi (CRIADO-FORNELIO et al., 2004). However, none of these species was found in the present study.
Most studies conducted in Brazil on canine babesiosis have been based on parasitological or serological diagnostic methods. Molecular biology techniques are now emerging within this research field in this country, which may indeed reinforce the reliability and scientific consistency of further studies. There is an increasing demand for more studies in Brazil, in order to enhance knowledge about the etiological spectrum of canine babesiosis in this country.
Conclusion
Similarity evaluation and phylogenetic analysis on partial fragments of the 18S rRNA gene endorse the molecular identity of B. canis vogeli in isolates from dogs in the city of Goiânia, state of Goiás, Brazil.
Isolates of B. canis vogeli from Goiânia exhibit high percentages of molecular similarity and participate in the well-defined phylogenetic group composed by reference strains for B. canis vogeli from other parts of Brazil and from other regions of the world.
Acknowledgements
The authors thank the National Council for Technological and Scientific Development (CNPq) for its financial support.
Received February 11, 2011
Accepted April 19, 2011
- ALLSOPP, M. T. E.; ALLSOPP, B. Molecular sequence evidence for the reclassification of the some Babesia species. Annals of New York Academy of Sciences, v. 1081, n. 1, p. 509-517, 2006. PMid:17135560. http://dx.doi.org/10.1196/annals.1373.076
- ALTSCHUL, S. F. et al. Basic local alignment search tool. Journal of Molecular Biology, v. 215, n. 3, p. 403-410, 1990.
- ARAI, S. et al. Babesia canis infection in canine-red blood cell-substituted SCID mice. International Journal for Parasitology, v. 28, n. 9, p. 1429-1435, 1998. http://dx.doi.org/10.1016/S0020-7519(98)00094-0
- BIRKENHEUER, A. J. et al. Detection and molecular characterization of a novel large Babesia species in a dog. Veterinary Parasitology, v. 124, n. 3-4, p. 151-160, 2004. PMid:15381295. http://dx.doi.org/10.1016/j.vetpar.2004.07.008
- BRACCINI, G. L. et al. Resultados de exames laboratoriais realizados no setor de Protozoologia da Faculdade de Veterinária da Universidade Federal do Rio Grande do Sul, Porto Alegre, nos anos 1986 a 1990. Arquivos da Faculdade de Veterinária da UFRGS, v. 20, p. 134-149, 1992.
- CACCIO, S. M. et al. Molecular characterization of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Veterinary Parasitology, v. 106, n. 4, p. 285-292, 2002. http://dx.doi.org/10.1016/S0304-4017(02)00112-7
- CAMACHO, A. T. et al. Infection of dogs in north-west Spain with a Babesia microti-like agent. Veterinary Record, v. 149, n. 18, p. 552-555, 2001. PMid:11720208. http://dx.doi.org/10.1136/vr.149.18.552
- CRIADO-FORNELIO, A. et al. The "expanding universe" of piroplasms. Veterinary Parasitology, v. 119, n. 4, p. 337-345, 2004. PMid:15154598. http://dx.doi.org/10.1016/j.vetpar.2003.11.015
- DE SÁ, A. G. et al. Detection and Molecular Characterization of Babesia canis vogeli from naturally infected Brazilian dogs. International Journal of Applied Research in Veterinary Medicine, v. 4, n. 2, 2006.
- DUARTE, S. C. et al. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Veterinary Parasitology, v. 152, n. 1-2, p. 16-20, 2008a. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
- DUARTE, S. C. et al. Diagnóstico Parasitológico e Molecular da Babesiose Canina na cidade de Goiânia-GO. Revista de Patologia Tropical, v. 37, n. 3. p.229-236, 2008b.
- EIRAS, D. F. et al. First molecular characterization of Babesia vogeli in two naturally infected dogs of Buenos Aires, Argentina. Veterinary Parasitology, v. 157, n. 3-4, p. 294-298, 2008. PMid:18786765. http://dx.doi.org/10.1016/j.vetpar.2008.07.037
- EWING, B. et al. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research, v. 8, n. 3, p. 175-185, 1998. PMid:9521921.
- GÜLANBER, A. et al. First molecular diagnosis of Babesia vogeli in domestic dogs from Turkey. Veterinary Parasitology, v. 139, n. 1-3, p. 224-230, 2006. PMid:16584843. http://dx.doi.org/10.1016/j.vetpar.2006.02.035
- HOSKINS, J. D. Veterinary clinics of North America Philadelphia: Saunders Company, 1991. v. 21, n. 1, 201 p.
- HUNFELD, K. P., HILDEBRANDTH, A.; GRAY, J. S. Babesiosis: recent insights into an ancient disease. International Journal for Parasitology, v. 38, n. 11, p. 1219-1237, 2008. PMid:18440005. http://dx.doi.org/10.1016/j.ijpara.2008.03.001
- IRWIN, P. J. Canine babesiosis: from molecular taxonomy to control. Parasites & Vectors, v. 2, 2009. Suplemento 1.
- JEFFERIES, R. et al. Two Species of Canine Babesia in Australia: Detection and Characterization by PCR. Journal for Parasitology, v. 89, n.2, p. 409-412, 2003. http://dx.doi.org/10.1645/0022-3395(2003)089[0409:TSOCBI]2.0.CO;2
- KJEMTRUP, A. M. et al. Babesia conradae, sp. Nov., a small canine Babesia identified in California. Veterinary Parasitology, v. 138, n. 1-2, p. 103-111, 2006. PMid:16524663. http://dx.doi.org/10.1016/j.vetpar.2006.01.044
- KJEMTRUP, A. M., Conrad, P. A. A review of the small canine piroplasms from California: Babesia conradae in the literature. Veterinary Parasitology, v. 138, n. 1-2, p. 112-117, 2006. PMid:16522352. http://dx.doi.org/10.1016/j.vetpar.2006.01.045
- MATJILA, P. T. et al. Confirmation of occurrence of Babesia canis vogeli in domestic dogs in South Africa. Veterinary Parasitology, v. 122, n. 2, p. 119-125, 2004. PMid:15177716. http://dx.doi.org/10.1016/j.vetpar.2004.03.019
- PASSOS, L. M. F. et al. First molecular detection of Babesia vogeli in dogs from Brazil. Veterinary Parasitology, v. 127, n. 1, p. 81-85, 2005. PMid:15619377. http://dx.doi.org/10.1016/j.vetpar.2004.07.028
- RAMOS, R. et al. Molecular survey and genetic characterization of tick-borne pathogens in dogs in metropolitan Recife (north-eastern Brazil). Parasitology Research, v. 107, n. 5, p. 1115-1120, 2010. PMid:20680344. http://dx.doi.org/10.1007/s00436-010-1979-7
- READ, M., HYDE, J. E. Methods in Molecular Biology Totowa: Humana Press Inc., 1993. p. 43-55. PMid:8220733.
- REY VALEIRÓN, C. et al. Parasitological and molecular characterization of a Venezuelan isolate of Babesia canis Revista Científica Maracaibo, v. 17, n. 1, p. 21-27, 2007.
- SZABÓ, M. P. J.; OLEGÁRIO, M. M. M.; SANTOS, A. L. Q. Tick fauna from two locations in the Brazilian savannah. Experimental and Applied Acarology, v. 43, n. 1, p. 73-84, 2007. PMid:17828441. http://dx.doi.org/10.1007/s10493-007-9096-8
- TAMURA, K. et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, v. 24, n. 1, p. 1596-1599, 2007. PMid:17488738. http://dx.doi.org/10.1093/molbev/msm092
- THOMPSON, J. D., PLEWNIAK, F., POCH, O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Research, v. 27, n. 13, p. 2682-2690, 1999. PMid:10373585. PMCid:148477. http://dx.doi.org/10.1093/nar/27.13.2682
- TRAPP, S. M. et al. Babesia gibsoni genotype Ásia in dogs from Brazil. Amsterdam, Veterinary Parasitology, v. 141, n. 1-2, p. 177-180, 2006. PMid:16765518. http://dx.doi.org/10.1016/j.vetpar.2006.04.036
- UILENBERG, G. et al. Three groups of Babesia canis distinguished and a proposal for nomenclature. Veterinary Quarterly, v. 1, p. 33-40, 1989. PMid:2655263. http://dx.doi.org/10.1080/01652176.1989.9694194
- UILENBERG, G. Babesia - a historical overview. Veterinary Parasitology, v. 138, n. 1-2, p. 3-10, 2006. PMid:16513280. http://dx.doi.org/10.1016/j.vetpar.2006.01.035
- ZAHLER, M. et al. Characteristic genotypes discriminate between Babesia canis isolates of differing vector specificity and pathogenicity to dogs. Parasitology Research, v. 84, n. 7, p. 544-548, 1998. PMid:9694369. http://dx.doi.org/10.1007/s004360050445
- ZAHLER, M. et al. Detection of a new pathogenic Babesia microti-like species in dogs. Veterinary Parasitology, v. 89, n. 3, p. 241-248, 2000. http://dx.doi.org/10.1016/S0304-4017(00)00202-8
Corresponding author:
Publication Dates
-
Publication in this collection
21 Dec 2011 -
Date of issue
Dec 2011
History
-
Received
11 Feb 2011 -
Accepted
19 Apr 2011