Abstracts
Babesiosis is a hemolytic disease caused by protozoans of the genus Babesia (Apicomplexa). This disease occurs worldwide and is transmitted by ticks to a variety of mammals, including humans. The objective of the present study was to optimize a molecular approach for the detection of a fragment of 18S rDNA of Babesia canis, Babesia vogeli, Babesia rossi or Babesia gibsoni based on a single semi-nested Polymerase Chain Reaction (PCR), and compare the efficiency of this approach with that of a simple PCR protocol. To this end, 100 blood samples collected from dogs with suspected hemoparasite infections were analyzed. A comparison of the results of simple PCR and semi-nested PCR indicated a highly significant difference (p value = 0.0000). While only five (5%) of the samples tested positive using the simple protocol, 22 (22%) were positive using the snPCR technique. The results of this study reinforce the findings of previous studies, which have demonstrated the greater sensitivity of tests based on nested or semi-nested PCR. Therefore, to avoid false-negative results due to low levels of parasitemia, we suggest the preferential use of this protocol in epidemiological studies of canine babesiosis, particularly those that require reliable estimates of the prevalence of infection.
Babesia ; molecular diagnosis; semi-nested PCR; eastern Amazonia
A babesiose é uma doença hemolítica de ocorrência mundial, causada por protozoários do gênero Babesia(Apicomplexa), que são transmitidos por carrapatos a diversos mamíferos, incluindo o homem. O objetivo deste estudo foi otimizar um método molecular para a detecção de fragmento do 18S rDNA de Babesia canis, Babesia vogeli, Babesia rossi ou Babesia gibsoni com base em uma única semi-nested (snPCR), comparando sua eficiência com um protocolo de PCR simples. Para isso, 100 amostras de sangue de cães com suspeita de hemoparasitoses foram analisadas e, enquanto o protocolo de PCR simples indicou somente 5% (5/100) de amostras positivas, o protocolo de snPCR, com 22% (22/100) de amostras positivas, apresentou maior sensibilidade (pvalor = 0,0000). Este resultado está de acordo com outros estudos que mostram a maior sensibilidade de detecção dos testes baseado em nested ou snPCR. Assim, como uma forma de prevenir resultados falso-negativos devido à baixa parasitemia, sugere-se que este protocolo seja preferencialmente usado nos estudos epidemiológicos de babesiose canina, em especial naqueles que tratam da sua prevalência.
Babesia ; diagnóstico molecular; semi-nested PCR; Amazônia oriental
Introduction
Babesiosis is a hemolytic disease caused by protozoans of the genus Babesia (Apicomplexa). This disease occurs worldwide and is transmitted to a range of different mammal species (BOOZER; MACINTIRE, 2003Boozer AL, Macintire DK. Canine babesiosis. Vet Clin North Am Small Anim Pract 2003; 33(4): 885-904. PMid:12910748.). Typical clinical signs include fever, depression, and anemia (PASSOS et al., 2005Passos LMF, Geiger SM, Ribeiro MFB, Pfister K, Zahler-Rinder M. First molecular detection of Babesia vogeli in dogs from Brazil. Vet Parasitol 2005; 127(1): 81-85. PMid:15619377.). Four protozoan species – Babesia canis, Babesia vogeli, Babesia rossi and Babesia gibsoni – are known to cause canine babesiosis or piroplasmosis. The first three species are recognized based on the vector (tick) species, virulence, and geographic distribution. Babesia canis is transmitted by Dermacentor reticularis, a tick species found in Europe (BOOZER; MACINTIRE, 2003Boozer AL, Macintire DK. Canine babesiosis. Vet Clin North Am Small Anim Pract 2003; 33(4): 885-904. PMid:12910748.), while the vector of Babesia vogeli is the red tick, Rhipicephalus sanguineus, which is found in the United States and in tropical and subtropical regions around the world, including Brazil (PASSOS et al., 2005Passos LMF, Geiger SM, Ribeiro MFB, Pfister K, Zahler-Rinder M. First molecular detection of Babesia vogeli in dogs from Brazil. Vet Parasitol 2005; 127(1): 81-85. PMid:15619377.). Babesia rossi, the most virulent of the three species, is transmitted by Haemaphysalis leachi, a tick found in South Africa (LOBETTI, 1998Lobetti RG. Canine Babesiosis. Comp Cont Ed Vet Pract 1998; 20(4): 418-431.). In Brazil, canine babesiosis is typically caused by B. vogeli, although there are some reports of B. gibsoni in the south of the country (JOJIMA et al., 2008).
Procedures used for the diagnosis of babesiosis include direct detection, which is based on the presence of merozoites in the red cells of blood smears stained with Giemsa, Field's or Wright's solutions. This is a specific approach, which is adequate for the acute phase of the disease, although it has low sensitivity and certain limitations, given that the characteristics and duration of the parasitemia may vary considerably (GREENE, 2006Greene C. Ehrlichiosis, Neorickettsiosis, Anaplasmosis and Wolbachia Infection. In: Greene C. Infectious diseases of the dog and cat. 3rd ed. St. Louis: Saunders; 2006. p. 203-231.). Indirect diagnosis is based on serological tests such as the Indirect Immunofluorescence Reaction (IIR), which is relatively sensitive but of low specificity due to cross reactions among the different Babesia species (VIDOTTO; MANDUCA, 2004). Diagnosis by IIR may also be ineffective in the early phase of the infection, when antibody concentrations may be too low to be detected. Both direct and indirect approaches thus present certain limitations, which may hamper diagnosis and lead to either false-negative or false-positive results.
Molecular tests, particularly those based on the Polymerase Chain Reaction (PCR), appear to represent a promising tool for the diagnosis of many parasitic diseases (GASSER, 2006Gasser RB. Molecular tools – advances, opportunities and prospects. Vet Parasitol 2006; 136(2): 69-89. PMid:16457951.), given that they are both sensitive and specific (BÖSE et al., 1995Böse R, Jorgensen WK, Dalgliesh RJ, Friedhoff KT, de Vos AJ. Current state and future trends in the diagnosis of babesiosis. Vet Parasitol 1995; 57(1-3): 61-74. PMid:7597794.). The sensitivity of this approach can be further enhanced by the application of nested (nPCR) or semi-nested (snPCR) techniques, which can be especially useful when parasitemia is low. Birkenheuer et al. (2003)Birkenheuer AJ, Levy MG, Breitschwerdt EB. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J Clin Microbiol 2003; 41(9): 4172-4177. PMid:12958243. http://dx.doi.org/10.1128/JCM.41.9.4172-4177.2003
http://dx.doi.org/10.1128/JCM.41.9.4172-...
recommended a semi-nested PCR approach for the amplification of DNA fragments from B. gibsoni, B. canis, B. vogeli, and B. rossi, with a specific snPCR protocol for each taxon. The objective of the present study was to optimize a molecular method for the detection of the DNA of these four Babesia species based on a single snPCR.
Materials and Methods
One hundred samples were selected randomly from those collected from dogs with clinical signs of hemoparasitosis treated at the Veterinary Hospital of the Federal Rural University of Amazonia (HOVET-UFRA), Belém (Pará State) between August and October 2011. The blood samples were drawn into tubes containing EDTA.
The total DNA of each sample was extracted from a 300 µl aliquot of the blood using the standard phenol-chloroform procedure described by Sambrook et al. (1989)Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory; 1989.. DNA quality control was checked by electrophoresis in agarose gel, followed by quantification using a Qubit fluorometer (Invitrogen). The molecular diagnosis was based on the PCR protocol proposed by Duarte et al. (2008)Duarte SC, Linhares GFC, Romanowsky TN, Silveira OJ N°, Borges LMF. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Vet Parasitol 2008; 152(1-2): 16-20. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
http://dx.doi.org/10.1016/j.vetpar.2007....
, which involves the amplification of a segment of the 28S rDNA gene. During the present study, a snPCR protocol was optimized based on the simple PCR protocol developed by Kordick et al. (1999)Kordick SK, Breitschwerdt EB, Hegarty BC, Southwick KL, Colitz CM, Hancock, SI, et al. Coinfection with multiple tick-borne pathogens in a walker hound kennel in North Carolina. J Clin Microbiol 1999; 37(8): 2631-2638. PMid:10405413. and Martin et al. (2006)Martin AR, Dunstan RH, Roberts TK, Brown GK. Babesia canis vogeli: A novel PCR for its detection in dogs in Australia. Exp Parasitol 2006; 112(1): 63-65. PMid:16256109., which amplifies a fragment of the 18S rDNA gene of the Babesia species. This involved conducting an initial reaction in a total volume of 25 µL containing 10-20 ng of the DNA template, 1.5 mM of MgCl2, 2.5 mM of each dNTP, 10 mM of Tris-HCl, 50 mM of KCl, 5 µM of each primer (Bab-f [MARTIN et al., 2006Martin AR, Dunstan RH, Roberts TK, Brown GK. Babesia canis vogeli: A novel PCR for its detection in dogs in Australia. Exp Parasitol 2006; 112(1): 63-65. PMid:16256109.] + Babesiacommon [KORDICK et al., 1999Kordick SK, Breitschwerdt EB, Hegarty BC, Southwick KL, Colitz CM, Hancock, SI, et al. Coinfection with multiple tick-borne pathogens in a walker hound kennel in North Carolina. J Clin Microbiol 1999; 37(8): 2631-2638. PMid:10405413.]), and 1 U of Taq DNA polymerase (Invitrogen). The second reaction was also conducted in a total volume of 25 µL, containing 1 µL of the product of the first reaction, 1.5 mM of MgCl2, 2.5 mM of each dNTP, 10 mM of Tris-HCl, 50 mM of KCl, 5 µM of each primer (B. canis/Babesiacommon [KORDICK et al., 1999Kordick SK, Breitschwerdt EB, Hegarty BC, Southwick KL, Colitz CM, Hancock, SI, et al. Coinfection with multiple tick-borne pathogens in a walker hound kennel in North Carolina. J Clin Microbiol 1999; 37(8): 2631-2638. PMid:10405413.]), and 1 U of Taq DNA polymerase (Invitrogen). The amplification protocol of both reactions consisted of 35 cycles of 1 minute at 94 °C, 2 minutes at 65 °C, and 1 minute at 72 °C, preceded by 3 minutes at 94 °C and followed by 5 minutes at 72 °C. The DNA of peripheral blood sample from one dog (from Belém) treated at HOVET-UFRA and showing large intraerythrocytic piroplasms morphologically compatible with Babesia vogeli, which was examined by light microscopy after panoptic staining, was used as the positive control, while sterile bi-distilled water was used as the negative control.
All the PCR products were visualized after electrophoresis in 1.5% agarose gel in Tris-acetate-EDTA (TAE) buffer, using GelRed™ Nucleic Acid stain (Biotium) and an ultraviolet transilluminator. The samples that produced fragments of 590 bps for the BAB1/BAB4 primers (DUARTE et al., 2008Duarte SC, Linhares GFC, Romanowsky TN, Silveira OJ N°, Borges LMF. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Vet Parasitol 2008; 152(1-2): 16-20. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
http://dx.doi.org/10.1016/j.vetpar.2007....
) and 394 and 208 bps for the first and second reactions of the snPCR, respectively, were considered to be positive. A 100-bp molecular marker (Invitrogen DNA ladder) was used to estimate the size of each amplified fragment.
The amplicon from the positive control sample was purified with a GFX PCR DNA and gel purification kit (GE Healthcare) and ligated to the pGEM-T vector plasmid (Promega), which was then inserted into TOP 10 Escherichia coli (Invitrogen Life Technologies). The cloned fragment was obtained by PCR from the recombinant clones of the colonies using the M13F/M13R primers and sequenced automatically in a 3500xl Genetic Analyzer (Applied Biosystems), according to the manufacturer's specifications. The BioEdit program (HALL, 1999Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999; 41: 95-98.) was used to align the forward and reverse sequences with AY072925, obtained from Genbank (NCBI).
Sensitivities of simple and semi-nested PCR for the detection of Babesia vogeli were compared by McNemar's test using BioEstat 5.3 software (AYRES et al., 2007).
Results and Discussion
All the DNA samples showed high purity and integrity. The positivity of the control was confirmed based on a comparison of the nucleotide sequence of the fragment amplified with the Bab-f/Babesiacommon primers with sequence AY072925 obtained from Genbank (NCBI - www.ncbi.nlm.nih.gov).
Martin et al. (2006)Martin AR, Dunstan RH, Roberts TK, Brown GK. Babesia canis vogeli: A novel PCR for its detection in dogs in Australia. Exp Parasitol 2006; 112(1): 63-65. PMid:16256109.used the BLAST program and observed that the primer Bab-f is highly specific to Babesia genus and also observed 100% specificity and sensitivity with results that were reproducible in later experiments, involving both sets of primers individually. Therefore, we simply checked the specificity of the protocol proposed here by obtain the sequence (GenBank accession number KF753247) of the product amplified from the positive control in the first reaction.
An identity of 100% was recorded between the nucleotide sequence obtained in the present study (positive control) and that of B. vogeli obtained from Genbank, which confirms, for the first time, the occurrence of this protozoan species in the metropolitan area of the city of Belém.
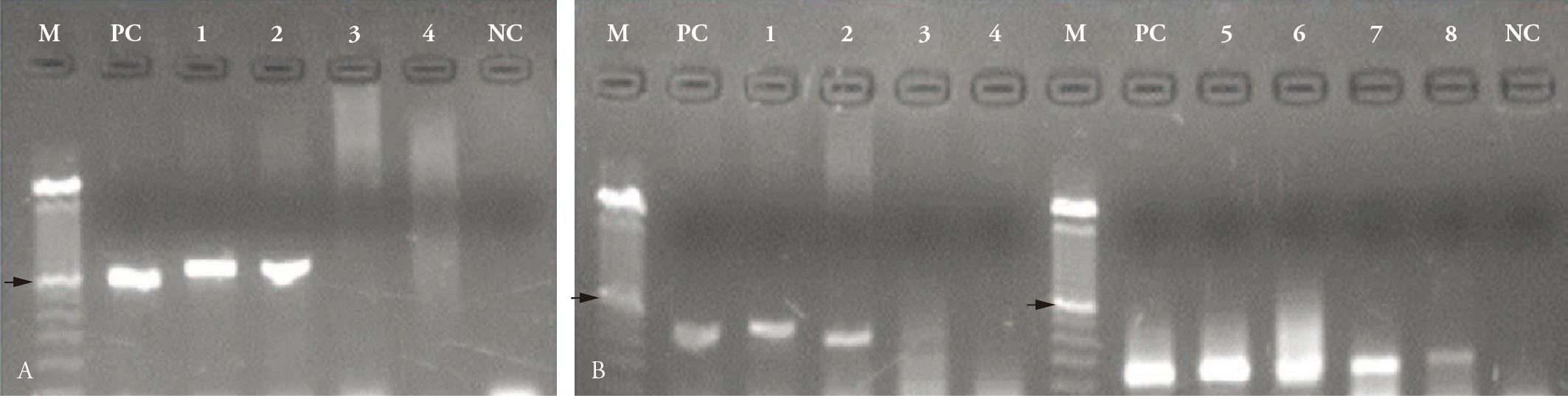
Both, the simple and the two semi-nested PCR, amplified fragments with lengths similar to those obtained for the positive control sample (Figure 1) were in agreement with the original descriptions (KORDICK et al., 1999Kordick SK, Breitschwerdt EB, Hegarty BC, Southwick KL, Colitz CM, Hancock, SI, et al. Coinfection with multiple tick-borne pathogens in a walker hound kennel in North Carolina. J Clin Microbiol 1999; 37(8): 2631-2638. PMid:10405413.; MARTIN et al., 2006Martin AR, Dunstan RH, Roberts TK, Brown GK. Babesia canis vogeli: A novel PCR for its detection in dogs in Australia. Exp Parasitol 2006; 112(1): 63-65. PMid:16256109.; DUARTE et al., 2008Duarte SC, Linhares GFC, Romanowsky TN, Silveira OJ N°, Borges LMF. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Vet Parasitol 2008; 152(1-2): 16-20. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
http://dx.doi.org/10.1016/j.vetpar.2007....
). A comparison of the results of simple PCR and semi-nested PCR revealed a highly significant difference (p value = 0.0000). While only five (5%) of the samples tested positive using the simple protocol, 22 (22%) proved positive using the snPCR technique.
Detection of Babesia vogeli by: A) simple PCR based on the Bab1/Bab4 primers; B) semi-nested PCR based on the Bab-f/Babesia common primers (lines 1-4) and B. canis/Babesiacommon (lines 5-8). M = 100 base-pair (bp) molecular marker, the arrows indicate the 600 bp fragments; PC = positive control; NC = negative control; reactions 1, 2, 3, and 4 included different samples.
Calder et al. (1996)Calder JA, Reddy GR, Chieves L, Courtney CH, Littell R, Livengood JR, et al. Monitoring Babesia bovis infections in cattle by using PCR-based tests. J Clin Microbiol 1996; 34(11): 2748-2755. PMid:8897177.compared the clinical sensitivity of a serological test with a PCR-based molecular approach by analyzing cattle infected experimentally with Babesia bovis, and found that 30% of the samples presented false-negative results in the molecular test, particularly in cases of chronic infection, which are characterized by low levels of parasitemia. The most probable explanation for the discrepancy between that study and the present one is that the snPCR is more sensitive than the single PCR technique since, depending of the level of the parasitemia of the host, the number of copies of the target DNA may not be sufficient to be detected in a single reaction. In fact, in comparison with other diagnostic procedures, a number of studies have shown that nPCR or snPCR is much more effective for the detection of pathogens, including hemoparasites such as Ehrlichia canis and Anaplasma platys(CHANG; PAN, 1996Chang WL, Pan MJ. Specific amplification of Ehrlichia platys DNA from blood specimens by two-step PCR. J Clin Microbiol 1996; 34(12): 3142-3146. PMid: 8940461.; FERREIRA et al., 2007Ferreira RF, Cerqueira AMF, Pereira AM, Guimarães CM, Sá AG, Abreu FS, et al. Anaplasma platys diagnosis in dogs: Comparison between morphological and molecular tests. Intern J Appl Res Vet Med 2007; 5(3): 113-119.; MARTIN et al., 2005Martin AR, Brown GK, Dunstan RH, Roberts TK. Anaplasma platys: an improved PCR for its detection in dogs. Exp Parasitol 2005; 109(3): 176-180. PMid:15713449.; McBRIDE et al., 1996McBride JW, Corstvet RE, Gaunt SD, Chinsangaram J, Akita GY, Osburn BI. PCR detection of acute Ehrlichia canis infection in dogs. J Vet Diagn Invest 1996; 8(4): 441-447. PMid:8953528.; RAMOS et al., 2009; RUFINO et al., 2013Rufino CP, Moraes PH, Reis T, Campos R, Aguiar DC, McCulloch JA, et al. Detection of Ehrlichia canis and Anaplasma platys DNA using multiplex PCR. Vector Borne Zoonotic Dis 2013; 13(12): 846-850. http://dx.doi.org/10.1089/vbz.2013.1303
http://dx.doi.org/10.1089/vbz.2013.1303...
).
Birkenheuer et al.'s study (2003) appears to be the only published report of the use of a semi-nested PCR protocol for the detection of Babesia protozoans. In this case, however, different primers were used for the detection of each of the following species B. gibsoni, B. canis, B. vogeli, and B. rossi. The protocol optimized in the present study thus represents a relatively cheap and effective test for epidemiological and prevalence studies, especially in areas where no information is available on the occurrence of the four different taxa.
A number of studies have reported the occurrence of canine babesiosis in Brazil and, according to them, B. vogeli is now known to occur in the south (JOJIMA et al., 2008; VIEIRA et al., 2013Vieira TSWJ, Vieira RFC, Nascimento DAG, Tamekuni K, Toledo RS, Chandrashekar R, et al. Serosurvey of tick-borne pathogens in dogs from urban and rural areas from Parana State, Brazil. Rev Bras Parasitol Vet 2013; 22(1): 104-109. PMid:24252955. http://dx.doi.org/10.1590/S1984-29612013000100019
http://dx.doi.org/10.1590/S1984-29612013...
), southeast (COSTA-JUNIOR et al., 2009; LEMOS et al., 2012Lemos DL, Cerqueira AMF, Toma HK, Silva AV, Corrêa RGB, Paludo GR, et al. Detection and molecular characterization of piroplasms species from naturally infected dogs in southeast Brazil. Rev Bras Parasitol Vet 2012; 21(2): 137-142. PMid:22832754.), mid-west (DUARTE et al., 2008Duarte SC, Linhares GFC, Romanowsky TN, Silveira OJ N°, Borges LMF. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Vet Parasitol 2008; 152(1-2): 16-20. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
http://dx.doi.org/10.1016/j.vetpar.2007....
; SPOLIDORIO et al., 2011Spolidorio MG, Torres MM, Campos WN, Melo ALT, Igarashi M, Amude AM, et al. Molecular detection of Hepatozoon canis and Babesia canis vogeli in domestic dogs from Cuiabá, Brazil. Rev Bras Parasitol Vet 2011; 20(3): 253-255. PMid: 21961759.), northeast (RAMOS et al., 2010Ramos R, Ramos C, Araújo F, Oliveira R, Souza I, Pimentel D, et al. Molecular survey and genetic characterization of tick-borne pathogens in dogs in metropolitan Recife (north-eastern Brazil). Parasitol Res 2010; 107(5): 1115-20. PMid:20680344. http://dx.doi.org/10.1007/s00436-010-1979-7.
http://dx.doi.org/10.1007/s00436-010-197...
; SILVA et al., 2012Silva AB, Costa AP, De Sá JC, Costa FB, Dos Santos ACG, Guerra RMS. Molecular detection of Babesia canis vogeli in dogs and in Rhipicephalus sanguineus from the middle-west region of Maranhão, northeast Brazil. Ciênc Anim Bras 2012; 13(3): 388-395. http://dx.doi.org/10.5216/cab.v13i3.18439
http://dx.doi.org/10.5216/cab.v13i3.1843...
) and north of Brazil (SPOLIDORIO et al., 2013Spolidorio MG, Minervino AHH, Valadas SYOB, Soares HS, Neves KAL, Labruna MB, et al. Serosurvey for tick-borne diseases in dogs from the Eastern Amazon, Brazil. Rev Bras Parasitol Vet 2013; 22(2): 214-219. PMid:23802236. http://dx.doi.org/10.1590/S1984-29612013005000023
http://dx.doi.org/10.1590/S1984-29612013...
), while B. gibsoni has only been confirmed in the south (TRAPP et al., 2006Trapp SM, Messick JB, Vidotto O, Jojima FS, De Morais HS. Babesia gibsoni genotype Asia in dogs from Brazil. Vet Parasitol 2006; 141(1-2): 177-180. PMid:16765518.). The present study is the first to identify the occurrence of B. vogeli in metropolitan Belém (eastern Amazonia).
Overall, given the zoonotic potential of this pathogen (KJEMTRUP; CONRAD, 2000Kjemtrup AM, Conrad PA. Human babesiosis: an emerging tick-borne disease. Int J Parasitol 2000; 30(12-13): 1323-1337. PMid:11113258.; VANNIER; KRAUSE, 2012Vannier E, Krause PJ. Human Babesiosis. N Engl J Med 2012; 366: 2397-2407. PMid:22716978. http://dx.doi.org/10.1056/NEJMra1202018
http://dx.doi.org/10.1056/NEJMra1202018...
), the protocol optimized in the present study, which is clearly far more sensitive than simple PCR procedures, should be used for all future epidemiological studies of babesiosis caused by B. gibsoni, B. canis, B. vogeli, and B. rossi, especially those that aim to evaluate the prevalence of these protozoans.
We are grateful to FAPESPA (ICAAF 012/2012) for its financial support and to CAPES for masters scholarships granted to Pablo Moraes and Claudia Pinheiro Rufino. We are also indebted to the anonymous reviewers for their comments on the original manuscript.
References
- Ayres M, Ayres JRM, Ayres DL, Santos AS. BioEstat 5.0 – Aplicações Estatísticas nas Áreas das Ciências Biológicas e Médicas: Sociedade Civil Mamirauá, Belém. Brasília: CNPq; 2007. 290 p.
- Birkenheuer AJ, Levy MG, Breitschwerdt EB. Development and evaluation of a seminested PCR for detection and differentiation of Babesia gibsoni (Asian genotype) and B. canis DNA in canine blood samples. J Clin Microbiol 2003; 41(9): 4172-4177. PMid:12958243. http://dx.doi.org/10.1128/JCM.41.9.4172-4177.2003
» http://dx.doi.org/10.1128/JCM.41.9.4172-4177.2003 - Boozer AL, Macintire DK. Canine babesiosis. Vet Clin North Am Small Anim Pract 2003; 33(4): 885-904. PMid:12910748.
- Böse R, Jorgensen WK, Dalgliesh RJ, Friedhoff KT, de Vos AJ. Current state and future trends in the diagnosis of babesiosis. Vet Parasitol 1995; 57(1-3): 61-74. PMid:7597794.
- Calder JA, Reddy GR, Chieves L, Courtney CH, Littell R, Livengood JR, et al. Monitoring Babesia bovis infections in cattle by using PCR-based tests. J Clin Microbiol 1996; 34(11): 2748-2755. PMid:8897177.
- Chang WL, Pan MJ. Specific amplification of Ehrlichia platys DNA from blood specimens by two-step PCR. J Clin Microbiol 1996; 34(12): 3142-3146. PMid: 8940461.
- Costa-Júnior LM, Ribeiro MFB, Rembeck K, Rabelo EML, Zahler-Rinder M, Hirzmann J, et al. Canine babesiosis caused by Babesia canis vogeli in rural areas of the State of Minas Gerais, Brazil and factors associated with its seroprevalence. Res Vet Sci 2009; 86(2): 257-260. PMid:18723199. http://dx.doi.org/10.1016/j.rvsc.2008.07.002
» http://dx.doi.org/10.1016/j.rvsc.2008.07.002 - Duarte SC, Linhares GFC, Romanowsky TN, Silveira OJ N°, Borges LMF. Assessment of primers designed for the subspecies-specific discrimination among Babesia canis canis, Babesia canis vogeli and Babesia canis rossi by PCR assay. Vet Parasitol 2008; 152(1-2): 16-20. PMid:18242863. http://dx.doi.org/10.1016/j.vetpar.2007.12.013
» http://dx.doi.org/10.1016/j.vetpar.2007.12.013 - Ferreira RF, Cerqueira AMF, Pereira AM, Guimarães CM, Sá AG, Abreu FS, et al. Anaplasma platys diagnosis in dogs: Comparison between morphological and molecular tests. Intern J Appl Res Vet Med 2007; 5(3): 113-119.
- Gasser RB. Molecular tools – advances, opportunities and prospects. Vet Parasitol 2006; 136(2): 69-89. PMid:16457951.
- Greene C. Ehrlichiosis, Neorickettsiosis, Anaplasmosis and Wolbachia Infection. In: Greene C. Infectious diseases of the dog and cat. 3rd ed. St. Louis: Saunders; 2006. p. 203-231.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999; 41: 95-98.
- Jojima FS, Garcia JL, Vidotto MC, Balarin MRS, Fabretti AK, Gasparini MR, et al. Ocorrência e caracterização molecular de espécies de Babesia em cães de uma população hospitalar da região de Londrina, PR. Rev Bras Parasitol Vet 2008; 17(S1): 277-283. PMid:20059862.
- Kjemtrup AM, Conrad PA. Human babesiosis: an emerging tick-borne disease. Int J Parasitol 2000; 30(12-13): 1323-1337. PMid:11113258.
- Kordick SK, Breitschwerdt EB, Hegarty BC, Southwick KL, Colitz CM, Hancock, SI, et al. Coinfection with multiple tick-borne pathogens in a walker hound kennel in North Carolina. J Clin Microbiol 1999; 37(8): 2631-2638. PMid:10405413.
- Lemos DL, Cerqueira AMF, Toma HK, Silva AV, Corrêa RGB, Paludo GR, et al. Detection and molecular characterization of piroplasms species from naturally infected dogs in southeast Brazil. Rev Bras Parasitol Vet 2012; 21(2): 137-142. PMid:22832754.
- Lobetti RG. Canine Babesiosis. Comp Cont Ed Vet Pract 1998; 20(4): 418-431.
- Martin AR, Brown GK, Dunstan RH, Roberts TK. Anaplasma platys: an improved PCR for its detection in dogs. Exp Parasitol 2005; 109(3): 176-180. PMid:15713449.
- Martin AR, Dunstan RH, Roberts TK, Brown GK. Babesia canis vogeli: A novel PCR for its detection in dogs in Australia. Exp Parasitol 2006; 112(1): 63-65. PMid:16256109.
- McBride JW, Corstvet RE, Gaunt SD, Chinsangaram J, Akita GY, Osburn BI. PCR detection of acute Ehrlichia canis infection in dogs. J Vet Diagn Invest 1996; 8(4): 441-447. PMid:8953528.
- Passos LMF, Geiger SM, Ribeiro MFB, Pfister K, Zahler-Rinder M. First molecular detection of Babesia vogeli in dogs from Brazil. Vet Parasitol 2005; 127(1): 81-85. PMid:15619377.
- Ramos CAN, Ramos RAN, Araújo FR, Guedes D Jr, Souza IIF, Ono TM et al. Comparação de nested-PCR com o diagnóstico direto na detecção de Ehrlichia canis e Anaplasma platys em cães. Rev Bras Parasit Vet 2009; 18(S1): 58-62. PMid:20040193.
- Ramos R, Ramos C, Araújo F, Oliveira R, Souza I, Pimentel D, et al. Molecular survey and genetic characterization of tick-borne pathogens in dogs in metropolitan Recife (north-eastern Brazil). Parasitol Res 2010; 107(5): 1115-20. PMid:20680344. http://dx.doi.org/10.1007/s00436-010-1979-7.
» http://dx.doi.org/10.1007/s00436-010-1979-7 - Rufino CP, Moraes PH, Reis T, Campos R, Aguiar DC, McCulloch JA, et al. Detection of Ehrlichia canis and Anaplasma platys DNA using multiplex PCR. Vector Borne Zoonotic Dis 2013; 13(12): 846-850. http://dx.doi.org/10.1089/vbz.2013.1303
» http://dx.doi.org/10.1089/vbz.2013.1303 - Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory; 1989.
- Silva AB, Costa AP, De Sá JC, Costa FB, Dos Santos ACG, Guerra RMS. Molecular detection of Babesia canis vogeli in dogs and in Rhipicephalus sanguineus from the middle-west region of Maranhão, northeast Brazil. Ciênc Anim Bras 2012; 13(3): 388-395. http://dx.doi.org/10.5216/cab.v13i3.18439
» http://dx.doi.org/10.5216/cab.v13i3.18439 - Spolidorio MG, Torres MM, Campos WN, Melo ALT, Igarashi M, Amude AM, et al. Molecular detection of Hepatozoon canis and Babesia canis vogeli in domestic dogs from Cuiabá, Brazil. Rev Bras Parasitol Vet 2011; 20(3): 253-255. PMid: 21961759.
- Spolidorio MG, Minervino AHH, Valadas SYOB, Soares HS, Neves KAL, Labruna MB, et al. Serosurvey for tick-borne diseases in dogs from the Eastern Amazon, Brazil. Rev Bras Parasitol Vet 2013; 22(2): 214-219. PMid:23802236. http://dx.doi.org/10.1590/S1984-29612013005000023
» http://dx.doi.org/10.1590/S1984-29612013005000023 - Trapp SM, Messick JB, Vidotto O, Jojima FS, De Morais HS. Babesia gibsoni genotype Asia in dogs from Brazil. Vet Parasitol 2006; 141(1-2): 177-180. PMid:16765518.
- Vannier E, Krause PJ. Human Babesiosis. N Engl J Med 2012; 366: 2397-2407. PMid:22716978. http://dx.doi.org/10.1056/NEJMra1202018
» http://dx.doi.org/10.1056/NEJMra1202018 - Vidotto O, Manduca S. Babesiose canina. Rev Bras Parasit Vet 2004; 13(S1): 58-61.
- Vieira TSWJ, Vieira RFC, Nascimento DAG, Tamekuni K, Toledo RS, Chandrashekar R, et al. Serosurvey of tick-borne pathogens in dogs from urban and rural areas from Parana State, Brazil. Rev Bras Parasitol Vet 2013; 22(1): 104-109. PMid:24252955. http://dx.doi.org/10.1590/S1984-29612013000100019
» http://dx.doi.org/10.1590/S1984-29612013000100019
Publication Dates
-
Publication in this collection
Jan-Mar 2014
History
-
Received
21 Aug 2013 -
Accepted
12 Nov 2013