Abstracts
Tick-borne infections can result in serious health problems for wild ruminants, and some of these infectious agents can be considered zoonosis. The aim of the present study was the post-mortem detection of hemoparasites in free-living Mazama gouazoubira from Minas Gerais state, Brazil. The deer samples consisted of free-living M. gouazoubira (n = 9) individuals that died after capture. Necropsy examinations of the carcasses were performed to search for macroscopic alterations. Organ samples were collected for subsequent imprint slides, and nested PCR assays were performed to detect hemoparasite species. Imprint slide assays from four deer showed erythrocytes infected with Piroplasmida small trophozoites, and A. marginale corpuscles were observed in erythrocytes from two animals. A. marginale and trophozoite co-infections occurred in two deer. A nested PCR analysis of the organs showed that six of the nine samples were positive for Theileria sp., five were positive for A. phagocytophilum and three were positive for A. marginale, with co-infection occurring in four deer. The results of the present study demonstrate that post-mortemdiagnostics using imprint slides and molecular assays are an effective method for detecting hemoparasites in organs.
Anaplasma spp.; Piroplasmida; molecular diagnostic; brown brocket deer; free-living; post-mortem detection; hemoparasites
Patógenos transmitidos por carrapatos podem resultar em sérios problemas de saúde para os ruminantes selvagens, e alguns podem ser zoonoses. O objetivo do presente estudo foi a detecção post mortem de hemoparasitos, em Mazama gouazoubira de vida livre, oriundos de Minas Gerais, através da análise de lâminas de impressão e nested PCR. Foram amostrados nove M. gouazoubira de vida livre, que morreram após a captura. Exames de necropsia foram realizados, e as carcaças foram examinadas para detectar alterações macroscópicas. Amostras dos órgãos foram coletadas para a realização de imprint em lâminas e para nested PCR à procura de hemoparasitos. A análise das lâminas mostrou pequenos trofozoítos de Piriplasmida nos eritrócitos de quatro dos oito animais examinados, e corpúsculos de Anaplasma marginale foram observados nos eritrócitos de dois dos cervídeos. A coinfecção com A. marginale e trofozoítos de piroplasmas ocorreu em dois animais. As análises de nPCR dos órgãos mostraram que seis dos nove animais estavam positivos para Theileria sp., cinco para A. phagocytophilum e três para A. marginale, sendo que a coinfecção ocorreu em quatro cervídeos. Os resultados do presente estudo demonstram que os diagnósticos post-mortem, pelas imprints em lâminas e ensaios moleculares, são métodos eficazes de detecção de hemoparasitos nos principais órgãos parasitados.
Anaplasma spp.; Piroplasmida; diagnostico molecular; veado catingueiro; vida livre; detecção post-mortem; hemoparasitos
Introduction
Tick-borne infections caused by protozoa and rickettsial organisms can result in serious problems for wild and domestic hosts, particularly when the animals have been weakened by other parasites or are undernourished or stressed (FOWLER, 1993Fowler ME. Zoo and Wild Animal Medicine Current Therapy, 3rd ed. WB Saunders, Philadelphia; 1993.). The brown brocket deer (Mazama gouazoubira) is a small forest cervid with solitary habits that can live in anthropogenic areas and in close proximity to farmers in neotropical forest areas. This behavior favors the interchange of infectious microorganisms between wild and domestic populations (DUARTE, 1997Duarte JMB. Biologia e conservação de cervídeos Sul-americanos: Blastocerus, Ozotoceros e Mazama, FUNEP, Jaboticabal; 1997.).
Babesia and Theileria species have been detected by molecular means in Brazilian cervids from Minas Gerais and Mato Grosso do Sul states (SILVEIRA et al., 2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66., 2013Silveira JAG, Rabelo EML, Lacerda ACL, Borges PAL, Tomás WM, Pellegrin AO, et al. Molecular detection and identification of hemoparasites in pampas deer (Ozotoceros bezoarticus Linnaeus, 1758) from the Pantanal Brazil. Ticks Tick Borne Dis 2013; 4(4): 341-345.). These species are known to cause clinical signs and hematological alterations in stressed animals (KREIER et al., 1962Kreier JP, Ristic M, Watrach AM. Theileria sp. in a deer in the United States. Am J Vet Res 1962; 23: 657-662.; BARKER et al., 1973Barker RW, Hoch AL, Buckner RG, Hair JA. Hematological changes in White-tailed-deer fawns, Odocoileus virginianus, infested with Theileria-infected lone star ticks. J Parasitol 1973; 59(6): 1091-1098.). On the other hand members of the Anaplasmataceae family can infect brown brocket deer, but the consequences of the infection on deer health are not well established.
In Brazil, the molecular detections of E. chaffeensis, A. marginale, A. bovis and Anaplasma spp. have been described in wild cervids (Blastocerus dichotomus, Ozotoceros bezoarticus and M. gouazoubira) in Mato Grosso do Sul and Minas Gerais states (MACHADO et al., 2006Machado RZ, Duarte JMB, Dagnone AS, Szabó MPJ. Detection of Ehrlichia chaffeensis in Brazilian marsh deer (Blastocerus dichotomus). Vet Parasitol 2006; 139(1-3): 262-266.; PICOLOTO et al., 2010Picoloto G, Ferreira de Lima R, Olegario LAO, Carvalho CME, Lacerda ACR, Tomas WM, et al. Real time polymerase chain reaction to diagnose Anaplasma marginale in cattle and deer (Ozotoceros bezoarticus leucogaster) of the Brazilian Pantanal. Rev Bras Parasitol Vet 2010; 19(3): 186-188.; SACCHI et al., 2012Sacchi ABV, Duarte JMB, Andre MR, Machado RZ. Prevalence and molecular characterization of Anaplasmataceae agents in free-ranging Brazilian marsh deer (Blastocerus dichotomus). Comp Immunol Microbiol Infect Dis 2012; 35(4): 325-334.; SILVEIRA et al., 2012, 2013).
Anaplasma marginale strains from deer exhibit low virulence in bovine hosts, and they have been studied as alternatives for controlling A. marginale infection in cattle (KUTTLER; ZAUG, 1988; KOCAN et al., 2003Kocan KM, de la Fuente J, Guglielmone AA, Meléndez RD.Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev 2003; 16(4): 698-712; SILVEIRA, 2012Silveira JAG. Ocorrência de hemoparasitos e ectoparasitos em veado-catingueiro (Mazama gouazoubira Fischer, 1814), veado-campeiro (Ozotocerus bezoarticus Linnaeus, 1758) e cervo-do-pantanal (Blastocerus dichotomus Illiger, 1815): utilização de métodos parasitológicos e moleculares. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2012.). Anaplasma phagocytophilum infects humans and a wide range of other animal hosts including rodents, ruminants, felids, horses, donkeys, dogs and birds (MADIGAN; PUSTERLA, 2000Madigan JE, Pusterla N. Ehrlichial diseases. Vet Clin North Am Equine Pract 2000; 16(3): 487-499.; STUEN, 2007). Capturing free-range deer is a fundamental concern for studies that require close access to the animals for sample collection and other ecological studies. However, cervid immobilization is a difficult process because these animals are easily frightened and very sensitive to capture myopathy (DUARTE, 2006Duarte JMB. Artiodactyla – Cervidae (Veado - catingueiro, Veado-campeiro, Cervo - do - pantanal). In: Cubas ZS, Silva JCR, Catão-Dias JL. Tratado de animais selvagens - Medicina Veterinária. Roca, São Paulo; 2006. p. 641-664.). In some cases, a restrained deer can die a few days after being captured. Therefore, an alternative way to study hemoparasites from these hosts is to harvest organs from free-living deer when these animals are found injured or bowled over. The aim of the present study was the post-mortem detection of hemoparasites in organs from free-living Mazama gouazoubira in Minas Gerais state.
Material and Methods
This study was approved by the Ethical Committee on Animal Experimentation (CETEA/UFMG, Belo Horizonte, MG, Brazil) under protocol no. 142/08 and by the Brazilian Institute for Environment and Natural Renewable Resources (IBAMA, Belo Horizonte, MG, Brazil) under license no. 16064-1.
Animal samples
This study was performed from June 2007 to April 2013. The deer samples came from free-living M. gouazoubira (n = 24) that were captured by the Forestry Police of Minas Gerais state and transported to IBAMA. Blood from these animals was collected by puncturing the jugular vein, and the samples were transferred immediately to vials containing EDTA. Blood smears were prepared, subjected to quick Romanowsky staining and examined under an optical microscope at 100X magnification. At least 40 microscopic fields were observed for each deer blood sample. Blood sample aliquots were frozen and stored for subsequent DNA extraction.
Nine (9/24) of these animals eventually died, and necropsy examinations were performed. The animals most likely died from capture myopathy or because of erroneous immobilization. Dog attacks were confirmed in three cases (MGM specimens). The deer were identified as follows: MGI8, MGI11, MGI12 and MGM1 (adult females), MGM2 (pregnant female) MGI13 and MGI19 (young females), MGM3 (adult male) and MGI14 (young male). Their ages were determined according to the following parameters: young (1-2 months), for animals presenting white hair spots; or adult, for animals presenting mature features.
Necropsy
Necropsy examinations were performed on nine dead deer, and the carcasses were evaluated for macroscopic alterations. Imprintslides of lymph node, spleen, liver and brain samples were prepared, and samples of these organs were collected and frozen for subsequent DNA extraction. The imprint slides were subjected to quick Romanowsky staining and examined under the optical microscope at 100× magnification.
DNA extraction and PCR amplification
DNA was extracted from the cervid organ and blood samples using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA), which was employed according to the manufacturer's instructions for animal tissue and whole blood, respectively.
Nested PCR assays were performed to detect Babesia/Theileria species (18S rRNA region) (SILVEIRA et al., 2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66.), monocytic (16S rRNA region) (KAWAHARA et al., 2009Kawahara M, Tajima T, Torii H, Yabutani M, Ishii J, Harasawa M, et al. Ehrlichia chaffeensis infection of Sika deer, Japan. Emerg Infect Dis 2009; 15(12): 1991-1993.) and granulocytic Anaplasmataceae agents (16S rRNA region) (MASSUNG et al., 1998Massung RF, Slater K, Owens JH, Nicholson WL, Mather TN, Solberg VB, et al. Nested PCR assay for the detection of granulocytic ehrlichiae. J Clin Microbiol 1998; 36(4): 1090-1095.) A. phagocytophilum(msp4 gene) (DE LA FUENTE et al. 2005; BOWN et al., 2007Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, et al. High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol 2007; 45(6): 1771-1776.) and A. marginale (msp4 gene) (DE LA FUENTE et al. 2008; SILVEIRA et al. 2012) (Table 1). Double-distilled water was used as the negative control (with no DNA). DNA was extracted from 300 µl of whole blood from a calf that had been experimentally infected with A. marginale (strain UFMG1) (BASTOS et al., 2010Bastos CV, Passos LMF, Facury-Filho EJ, Rabelo EM, de la Fuente J, Ribeiro MFB. Protection in the absence of exclusion between two Brazilian isolates of Anaplasma marginale in experimentally infected calves. Vet J 2010; 186(3): 374-378.) and another that was experimentally infected with both B. bovis (strain BbovMG) and B. bigemina (strain BbigMG) (COSTA et al., 2013Costa VMM, Ribeiro MFB, Duarte ALL, Mangueira JM, Pessoa AFA, Azevedo SS, et al. Seroprevalence and risk factors for cattle anaplasmosis, babesiosis, and trypanosomiasis in a Brazilian semiarid region. Rev Bras Parasitol Vet 2013; 22(2): 207-213.) for the positive control. DNA from IDE8 cell cultures infected with A. phagocytophilum (as isolated from a German dog and kindly donated by Dr. Erich Zweygarth-Institut für Vergleichende Tropenmedizin und Parasitologie, Ludwig Maximilians Universität München), was used for granulocytic Anaplasmataceae agents and A. phagocytophilumtests, or a dog infected with E. canis (Jaboticabal strain) (MOREIRA, 2001Moreira SM. Estudo retrospectivo (1998–2001) da erliquiose canina em Belo Horizonte: avaliação clínica e laboratorial de infecções experimentais. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2001.) was used for monocytic ehrlichiosis tests.
The first reaction mixture contained 1.2 µl of dNTPs (2.5 mM), 0.15 µl of Taq polymerase (Phoneutria, Belo Horizonte, MG, Brazil) (0.05 U), 1.5 µl of reaction buffer IB (Phoneutria, Belo Horizonte, MG, Brazil), 0.6 µl of a solution containing the mixed primers (10 µM) and 10.05 µl of sterile ultra-pure water. A 1.5 µl aliquot of the DNA template was added to the reaction mixture to achieve a final volume of 15 µl.
The second reaction mixture contained 2.0 µl of dNTPs (2.5 mM), 0.25 µl of Taq polymerase, 2.5 µl of reaction buffer IB, 1.0 µl of a solution containing the mixed primers (10 µM) and 16.75 µl of sterile ultra-pure water. An aliquot (2.5 µl) of amplicon that was obtained after the first reaction was added to the reaction mixture to achieve a final volume of 25 µl.
Amplification was performed with an Eppendorf Mastercycler (Eppendorf, São Paulo, SP, Brazil) thermocycler. The amplification program used in all nPCR assays was as follows: 94 °C for 5 min (initial denaturation step); 30 cycles of 92 °C for 1 min (denaturation), 54 °C for 1 min (annealing) and 72 °C for 2 min (extension), followed by 72 °C for 8 min (final extension step). After the amplification, the reaction mixtures were maintained at 12 °C. The PCR amplicons were separated by electrophoresis on 1% agarose gel (40 min, 100 V), which was stained with GelRed (Biotium, Hayward, CA, USA) and visualized under ultraviolet light.
The positive products of the second-round PCRs were purified using a QIAquick PCR Purification Kit (Qiagen Biotecnologia Brazil, São Paulo, Brazil), according to the manufacturer's instructions. The purified amplicons were sequenced by using ABI3130 with POP7 polymer and BigDye v3.1 (Myleus Biotechnology, Belo Horizonte, MG, Brazil) with the second reaction primers. The sequences were aligned, edited and analyzed at the URL http://asparagin.cenargen.embrapa.br/phph/and using the MEGA 6.0 software (TAMURA et al., 2013Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30 (12): 2725-2729.). The identity of each sequence was confirmed through a comparison with the sequences available at GenBank using the BLAST software (ALTSCHUL et al., 1990Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215: 403-410.). Phylogenetic analyses were conducted using the MEGA 6.0 software, and the phylogenetic position of the pathogens was inferred using the neighbor-joining method. The combination of phylogenetic clusters was assessed by bootstrap tests with 1000 replicates, and a Kimura 2-parameter model was used to test the stability of the trees.
Results
No visible macroscopic lesions were observed during the necropsy examinations of nine brown brocket deer, which led to the conclusion that the animal deaths were most likely caused by stress from animal handling. Imprint slides from deer MGI19 could not be analyzed because of the low quality of the smears; thus, slides from eight of the nine deer were analyzed. Protozoa in the form of Piroplasmida small trophozoites (<2 µm) were found in the erythrocytes of 50% (4/8) of the analyzed animals, and A. marginale corpuscles were observed in the erythrocytes of 25% (2/8) of the deer. Intraerythrocytic Piroplasmida trophozoites were detected in three spleen samples, three liver samples and one brain sample. A co-infection with A. marginale corpuscles and piroplasmid trophozoites was observed in two animals using spleen and liver slides (Table 2). The liver and spleen were the most parasitized organs, and no infected erythrocytes were observed in lymph nodes.
Hemoparasite detection in organs and whole blood from nine free-living brown brocket deer in the state of Minas Gerais, Brazil, as determined by imprint slides and nested PCR.
A molecular examination of whole blood that was published in Silveira et al. (2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66., 2012)Silveira JAG. Ocorrência de hemoparasitos e ectoparasitos em veado-catingueiro (Mazama gouazoubira Fischer, 1814), veado-campeiro (Ozotocerus bezoarticus Linnaeus, 1758) e cervo-do-pantanal (Blastocerus dichotomus Illiger, 1815): utilização de métodos parasitológicos e moleculares. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2012. revealed that 55.5% (5/9) of the samples were infected with hemoparasites; five were infected with A. marginale, four with T. cervi and one with Anaplasma sp. (which is closely related to A. platys/A. phagocytophilum). Co-infection was present in 44.4% (4/9) of the samples; three had T. cerviand A. marginale, and one had T. cervi, Anaplasma sp. and A. marginale (Table 2).
The nucleotide sequences amplified from parasites in the whole blood have been deposited in GenBank under the following accession numbers: Anaplasma marginale, JN022561.1, JN022563.1, JN022564.1 and JN022568.1; Theileria sp., HM466920.1; and T. cervi, HM466923.1 and HM466925.1 (SILVEIRA et al., 2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66., 2012Silveira JAG. Ocorrência de hemoparasitos e ectoparasitos em veado-catingueiro (Mazama gouazoubira Fischer, 1814), veado-campeiro (Ozotocerus bezoarticus Linnaeus, 1758) e cervo-do-pantanal (Blastocerus dichotomus Illiger, 1815): utilização de métodos parasitológicos e moleculares. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2012.). The organ sample sequences have been deposited under the following accession numbers: Anaplasma phagocytophilum, KF790914-KF790918 (msp4 gene) and KF790919- KF790920 (16S rRNA gene); Theileria sp. (18S rRNA gene), KF790921 - KF790922 (Table 2).
According to the nPCR assays of the organ samples, none of the animals were positive for monocytic Anaplasmataceae, 55.5% (6/9) of the samples were positive for 18S rRNA Theileria species (sharing 96-99% of their identity with Theileria strains deposited in GenBank with accession numbers GU946217.1 and AY735137.1), 55.5% (5/9) were positive for msp4 from A. phagocytophilum(98-100% shared identity, accession numbers HQ661162.1, AY829455.1, DQ104442.1 and KF111754.1), 22.2% (2/9) were positive for the 16S rRNA gene from granulocytic Anaplasmataceae agents (99% shared identity with A. phagocytophilum, accession numbers DQ104442.1, KF481943.1 and KF481930.1) and 33.3% (3/9) were positive for msp4 from A. marginale (Figures 1-3). Samples from the A. marginale-positive organs were not successfully sequenced because of their low amplified DNA concentration. The brain and liver were the most parasitized organs based on the results of nPCR assays at 3/5 (60%) and 5/9 (55.5%), respectively.
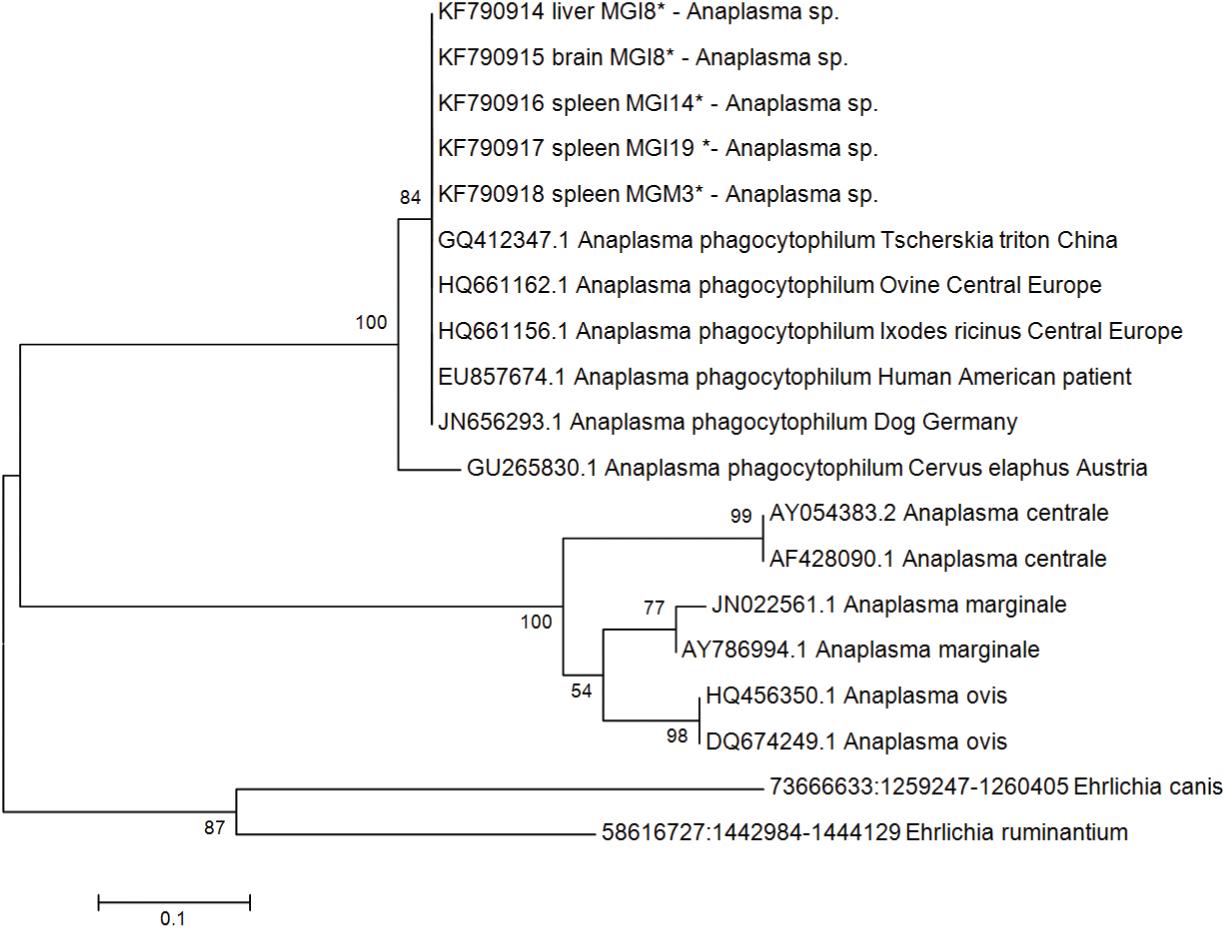
The phylogenetic tree of Anaplasma phagocytophilumas isolated from M. gouazoubira MGI8 organs and based on a partial sequence of the 16S rRNA gene. The GenBank accession numbers are shown. The tree was constructed using the neighbor-joining method, and the numbers above the internal nodes indicate the percentages of 1,000 bootstrap replicates that supported the branch. Outgroup: Wolbachia spp. *samples from this study.
The phylogenetic tree of Anaplasma phagocytophilumas isolated from M. gouazoubira organ samples based on a partial sequence of the msp4 gene. GenBank accession numbers are shown. The tree was constructed using the neighbor-joining method, and the numbers above the internal nodes indicate the percentages of 1,000 bootstrap replicates that supported the branch. Other Anaplasmataceae were used as outgroups. *samples from this study.
The phylogenetic tree of Theileria as isolated from M. gouazoubira MGI8 organs and based on the partial sequence of the 18S rRNA gene. The GenBank accession numbers are shown. The tree was constructed using the neighbor-joining method, and the numbers above the internal nodes indicate the percentages of 1,000 bootstrap replicates that supported the branch. Toxoplasma gondii, P. micans and S. muiris were used as outgroups. *samples from this study.
A phylogenetic analysis of the partial 16S rRNA and the msp4 gene showed that the phylogenetic topology generally placed samples from this study in close proximity with other A. phagocytophilum isolates, including isolates from humans and wild ruminants, reinforcing that these isolates are from A. phagocytophilum (Figures 1and 2). The 18S rRNA from the Piroplasmida phylogenetic tree showed a cluster containing T. cerviisolates from other deer, including one sequence (gb HM466930.1, Silveira et al. (2012)) from M. gouazoubira in Minas Gerais, Brazil (Figure 3).
No hemoparasites were detected in organ samples from MGI12, although the infection was detected in whole blood. However, MGI13 was not positive for any parasites in the whole blood, but it was positive for Babesia/Theileria species in the lymph node sample. Half of the lymph nodes tested (3/6) were positive for hemoparasites, and the liver samples from five animals were positive. Co-infection was most common in whole blood (4/9) compared with organ samples; however, one animal (MGI14) presented co-infection only in the organ samples (Table 2). None of the young animals were positive for hemoparasites according to the imprint slides analysis, but nPCR assays revealed that co-infection occurred in two of these animals (2/3).
Discussion
Free-ranging wild animals may be considered reservoirs of an increasing number of pathogens with zoonotic potential. When some parasites infect both susceptible wild and domestic species that share the same habitats, both present enzootic importance and must be monitored (KUIKEN et al., 2005Kuiken T, Leighton FA, Fouchier RAM, LeDuc JW, Peiris JSM, Schudel A, et al. Pathogen Surveillance in Animals. Science 2005; 309(5741): 1680-1681.). Furthermore, some hemoparasites are a serious problem for domestic ruminants and humans, and both can be in close contact with wild animals, particularly deer, in South America (DUARTE et al., 2001Duarte JMB, Merino ML, González S, Nunes ALV, Garcia JM, Szabó MPJ, et al. Order Artiodactyla, Family Cervidae (Deer). In: Fowler ME, Cubas ZS. Biology, Medicine, and Surgery of South American Wild Animals. Iowa State University Press, Iowa; 2001. p.402-422.).
According to Warns-Petit et al. (2010)Warns-Petit E, Morignat E, Artois M, Calavas D. Unsupervised clustering of wildlife necropsy data for syndromic surveillance. BMC Vet Res 2010; 6: 56., post-mortem findings are the primary data collected for general disease surveillance. No macroscopic lesions were observed during the necropsy examinations of the nine brown brocket deer that were evaluated in the present study. Death caused by stressful handling can be acute, and post-mortem macroscopic alterations may not be evident, as found in this study. However, deer are often capable of harboring some hemoparasite infections in a latent asymptomatic stage for a long time (BARKER et al., 1973Barker RW, Hoch AL, Buckner RG, Hair JA. Hematological changes in White-tailed-deer fawns, Odocoileus virginianus, infested with Theileria-infected lone star ticks. J Parasitol 1973; 59(6): 1091-1098.; KUTTLER; ZAUG, 1988; DAWSON et al., 1994Dawson JE, Childs JE, Biggie KL, Moore C, Stallknecht D, Shaddock J, et al. White-tailed-deer as a potential reservoir of Ehrlichia spp. J Wildl Dis 1994; 30(2): 162-168.; CANTU et al., 2007Cantu AC, Ortega SAJ, Mosqueda J, García-Vázquez Z, Henke SE, George JE. Immunologic and molecular identification of Babesia bovis and Babesia bigemina in Free-Ranging White-Tailed Deer in Northern Mexico. J Wildl Dis 2007; 43(3): 504-507.; DAVIDSON et al., 2001Davidson WR, Lockhart JM, Stallknecht DE, Howerth EW, Dawson JE, Rechav Y. Persistent Ehrlichia chaffeensis infection in white-tailed-deer. J Wildl Dis 2001; 37(3): 538-546.).
The blood smear is not reliable for distinguishing between some hemoparasites, such as Babesia and Theileriaspecies, especially when both species occur together in endemic areas (SILVEIRA et al., 2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66.). Although the diagnosis of a hemoparasite infection is normally achieved through the examination of blood smears, this method exhibits poor sensitivity when there is a low level of parasitemia.
Nested PCR assays have been shown to provide more consistent data for mapping the distribution of hemoparasites that affect the wild fauna of Brazil. Whole blood analysis is a useful way to detect hemoparasites by using molecular techniques, and it is more amenable to DNA extraction compared with using organ samples. However, organ samples were found to be a suitable alternative to post-mortem detection. The present study reinforces the importance and utility of molecular techniques for post-mortemdiagnostics and organ samples were found to be a suitable alternative to detection in necropsy. Erythrocytes in lymph node imprint slides have not shown infected, but half of the lymph node samples tested by nPCR were positive for hemoparasites.
No hemoparasites were detected in the whole blood of MGI13 and MGM3 deer; however, lymph nodes from MGI13 and the liver and brain of MGM3 were positive for Theileria species. Furthermore, none of the young were positive for hemoparasites in their imprint slides, but nPCR assays of blood or organ samples revealed that co-infection occurred in these animals. These facts highlight the importance of examining the lymphoid organs, liver and brain by using nPCR assays.
In relation to hemoparasites finding in this study, deer are important natural reservoir hosts of Anaplasmataceae, and they are capable of harboring latent A. marginale and A. phagocytophilum infections (CHRISTENSEN et al., 1958Christensen JF, Osebold JW, Rosen MN. Infection and antibody response in deer experimentally infected with Anaplasma marginale from bovine carriers. J Am Vet Med Assoc 1958; 32: 289-292.). Natural infection with A. phagocytophilum has been reported in wild deer, and these animals are considered reservoirs in Europe, Asia and North America (revised by Stuen et al. (2013)Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol 2013; 3: 31.). Pathogenic and apathogenic genetic variants have been found in these animals (MASSUNG et al., 2005Massung RF, Courtney JW, Hiratzka SL, Pitzer VE, Smith G, Dryden RL. Anaplasma phagocytophilum in white-tailed deer. Emerg Infect Dis 2005; 11(10): 1604-1606.; SILAGHI et al., 2011Silaghi C, Hamel D, Thiel C, Pfister K, Passos LMF, Rehbein S. Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis 2011; 11(4): 355-362.; OVERZIER et al., 2013Overzier E, Pfister K, Herb I, Mahling M, Böck G Jr, Silaghi C. Detection of tick-borne pathogens in roe deer (Capreolus capreolus), questing ticks (Ixodes ricinus) and in ticks infesting roe deer in southern Germany. Ticks Tick Borne Dis 2013; 4(4): 320-328.).
In Brazil, A. phagocytophilum has been molecularly detected in dogs, carnivorous birds, Rhipicephalus sanguineus and Amblyomma cajennense ticks (SANTOS et al., 2006Santos HA, Pires MS, Vilela JAR, Santos TM, Faccini JLH, Baldani CD, et al. Detection of Anaplasma phagocytophilum in Brazilian dogs by real-time polymerase chain reaction. J Vet Diagn Invest 2006; 23(4): 770-774., 2013Santos HA, Thomé SM, Baldani CD, Silva CB, Peixoto MP, Pires MS, et al. Molecular epidemiology of the emerging zoonosis agent Anaplasma phagocytophilum (Foggie, 1949) in dogs and ixodid ticks in Brazil. Parasit Vectors 2013; 6: 348.; MACHADO et al., 2012Machado RZ, André MA, Werther K, de Sousa E, Gavioli FA, Alves Junior JRF. Migratory and Carnivorous Birds in Brazil: Reservoirs for Anaplasma and Ehrlichia Species? Vector Borne Zoonotic Dis 2012; 12(8): 705-708.). Amblyomma cajennense was found in MGI14 and MGI19 brown brocket deer, which were positive for A. phagocytophilum, by employing msp4 gene amplification (SILVEIRA, 2012Silveira JAG. Ocorrência de hemoparasitos e ectoparasitos em veado-catingueiro (Mazama gouazoubira Fischer, 1814), veado-campeiro (Ozotocerus bezoarticus Linnaeus, 1758) e cervo-do-pantanal (Blastocerus dichotomus Illiger, 1815): utilização de métodos parasitológicos e moleculares. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2012.). However, future studies must investigate which vectors and hosts are responsible for maintaining A. phagocytophilum in nature.
According to a BLASTn analysis of the 16S rRNA gene, the MGI8 samples shared a high percentage of their identity (100%) with an A. phagocytophilum isolate from a person from Poland (gb KF111754.1) and an isolate from white-tailed deer in the USA (gb DQ104442.1). The msp4 gene analysis showed a high shared identity (98-100%) with European samples isolated from ovine, horses and Ixodes ricinus ticks. A phylogenetic tree analysis showed that the Anaplasma agent found in the present study is closely related to A. phagocytophilum at one site that is specific to this species, namely the msp4 gene, and in another site in hypervariable regions that can provide a species-specific signature, namely the 16S rRNA gene, reinforcing that these isolates are A. phagocytophilum (Figures 1 and 2). To the best of our knowledge, this is the first molecular evidence of closely related A. phagocytophilum species in Brazilian deer, and the zoonotic role of this agent is still not understood.
Liver and brain samples from the MGI8 animal were positive for A. phagocytophilum according to a PCR of 16S rRNA and msp4 targets, reinforcing the identity of the isolate. Additionally, this agent was detected in the spleens of other cervids (MGM3, MGI14 and MGI19). Previous reports have shown that various tissues and cells are susceptible to infection by A. phagocytophilum (KLEIN et al., 1997Klein MB, Miller JS, Nelson CM, Goodman JL. Primary bone marrow progenitors of both granulocytic and monocytic lineages are susceptible to infection with the agent of human granulocytic ehrlichiosis. J Infect Dis 1997; 176(5): 1405-1409.; MUNDERLOH et al., 2004Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, et al. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet Microbiol 2004; 101(1): 53-64.). The pathological change described during A. phagocytophilum infection in deer was an enlarged spleen with subcapsular bleeding in European roe deer (Capreolus capreolus) and reindeer (Rangifer tarandus) (STUEN, 2003Stuen S. Anaplasma phagocytophilum (Formerly Ehrlichia phagocytophila) Infection in Sheep and Wild Ruminants in Norway. A study on clinical manifestation, distribution and persistence [Thesis]. Oslo: School of Veterinary Science; 2003.).
The prevalence of A. marginale infection in Brazilian cervids has been reported at different values, specifically 6.3% and 48% in O. bezoarticus from the Pantanal region according to Picoloto et al. (2010)Picoloto G, Ferreira de Lima R, Olegario LAO, Carvalho CME, Lacerda ACR, Tomas WM, et al. Real time polymerase chain reaction to diagnose Anaplasma marginale in cattle and deer (Ozotoceros bezoarticus leucogaster) of the Brazilian Pantanal. Rev Bras Parasitol Vet 2010; 19(3): 186-188. and Silveira et al. (2013)Silveira JAG, Rabelo EML, Lacerda ACL, Borges PAL, Tomás WM, Pellegrin AO, et al. Molecular detection and identification of hemoparasites in pampas deer (Ozotoceros bezoarticus Linnaeus, 1758) from the Pantanal Brazil. Ticks Tick Borne Dis 2013; 4(4): 341-345., respectively, and 79.3% in M. gouzoubira from Minas Gerais state according to Silveira et al. (2012). However, the consequences of infection for deer health are not clearly understood. Nevertheless, anaplasmosis is known to be responsible for great economic losses among domestic ruminants in tropical and subtropical areas because of the intravascular anemia evident in infected animals (DUARTE et al., 2001Duarte JMB, Merino ML, González S, Nunes ALV, Garcia JM, Szabó MPJ, et al. Order Artiodactyla, Family Cervidae (Deer). In: Fowler ME, Cubas ZS. Biology, Medicine, and Surgery of South American Wild Animals. Iowa State University Press, Iowa; 2001. p.402-422.).
Theileria spp. infections in Brazilian cervids were first described by our research group. The prevalence of infections in M. gouzoubira and B. dichotomus from Minas Gerais was 47.6% (SILVEIRA et al., 2012), and it was 20% in O. bezoarticus from Pantanal (SILVEIRA et al., 2013Silveira JAG, Rabelo EML, Lacerda ACL, Borges PAL, Tomás WM, Pellegrin AO, et al. Molecular detection and identification of hemoparasites in pampas deer (Ozotoceros bezoarticus Linnaeus, 1758) from the Pantanal Brazil. Ticks Tick Borne Dis 2013; 4(4): 341-345.). Theileria cervi infections in MGI8 and MGI19 were confirmed by nucleotide sequencing (SILVEIRA et al., 2011Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66.), and the 18S rRNA in the Piroplasmida phylogenetic tree of the present study showed a cluster containing T. cervi isolates from other deer, including one sequence (gb HM466930.1, Silveira et al. (2012)) from M. gouazoubira in Minas Gerais, Brazil (Figure 3).
The pathogenesis of T. cervi infection is poorly understood, but in immunosuppressed and splenectomized deer, it can result in severe microcytic anemia (KREIER et al., 1962Kreier JP, Ristic M, Watrach AM. Theileria sp. in a deer in the United States. Am J Vet Res 1962; 23: 657-662.; FOWLER, 1993Fowler ME. Zoo and Wild Animal Medicine Current Therapy, 3rd ed. WB Saunders, Philadelphia; 1993.; YABSLEY et al., 2005Yabsley MJ, Quick TC, Little E. Theileriosis in a white-tailed-deer (Odocoileus virginianus) fawn. J Wildl Dis 2005; 41(4): 806-809.). A high percentage of parasitized red cells in the capillaries of the brain can be found in some infections with intraerythrocytic piroplasms (RISTIC, 1988Ristic M. Babesiosis of domestic animals and man. Boca Raton, CRC Press, Florida; 1988.). Although the consequence may be cerebral ischemia, the magnitude of this parasitism in deer is unknown. The present study detected hemoprotozoa in the brain samples of these deer; however, whether brain parasitism can cause nervous symptoms, especially in immunosuppressed animals, remains unclear.
Conclusion
The results of the present study showed that post-mortemdiagnostics using imprint slides and molecular assays were effective for detecting hemoparasites in deer organs.
The authors are grateful to the CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), the IBAMA (Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis), veterinarian Daniel Vilela and Professor Nelson Rodrigo da Silva Martins and to Veterinary Clinics Cães E Amigos and Leonardo Maciel. We thank American Journal Experts for improving the English in this manuscript, and we are grateful for all of the contributions that allowed us to execute the present study.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215: 403-410.
- Barker RW, Hoch AL, Buckner RG, Hair JA. Hematological changes in White-tailed-deer fawns, Odocoileus virginianus, infested with Theileria-infected lone star ticks. J Parasitol 1973; 59(6): 1091-1098.
- Bastos CV, Passos LMF, Facury-Filho EJ, Rabelo EM, de la Fuente J, Ribeiro MFB. Protection in the absence of exclusion between two Brazilian isolates of Anaplasma marginale in experimentally infected calves. Vet J 2010; 186(3): 374-378.
- Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, et al. High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol 2007; 45(6): 1771-1776.
- Cantu AC, Ortega SAJ, Mosqueda J, García-Vázquez Z, Henke SE, George JE. Immunologic and molecular identification of Babesia bovis and Babesia bigemina in Free-Ranging White-Tailed Deer in Northern Mexico. J Wildl Dis 2007; 43(3): 504-507.
- Christensen JF, Osebold JW, Rosen MN. Infection and antibody response in deer experimentally infected with Anaplasma marginale from bovine carriers. J Am Vet Med Assoc 1958; 32: 289-292.
- Costa VMM, Ribeiro MFB, Duarte ALL, Mangueira JM, Pessoa AFA, Azevedo SS, et al. Seroprevalence and risk factors for cattle anaplasmosis, babesiosis, and trypanosomiasis in a Brazilian semiarid region. Rev Bras Parasitol Vet 2013; 22(2): 207-213.
- Davidson WR, Lockhart JM, Stallknecht DE, Howerth EW, Dawson JE, Rechav Y. Persistent Ehrlichia chaffeensis infection in white-tailed-deer. J Wildl Dis 2001; 37(3): 538-546.
- Dawson JE, Childs JE, Biggie KL, Moore C, Stallknecht D, Shaddock J, et al. White-tailed-deer as a potential reservoir of Ehrlichia spp. J Wildl Dis 1994; 30(2): 162-168.
- de la Fuente J, Massung RF, Wong SJ, Chu FK, Lutz H, Meli M, et al. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J Clin Microbiol 2005; 43(3): 1309-1317.
- de la Fuente J, Ruiz-Fons F, Naranjo V, Torina A, Rodríguez O, Gortázar C. Evidence of Anaplasma infections in European roe deer (Capreolus capreolus) from southern Spain. Res Vet Sci 2008; 84(4): 382-386.
- Duarte JMB. Biologia e conservação de cervídeos Sul-americanos: Blastocerus, Ozotoceros e Mazama, FUNEP, Jaboticabal; 1997.
- Duarte JMB, Merino ML, González S, Nunes ALV, Garcia JM, Szabó MPJ, et al. Order Artiodactyla, Family Cervidae (Deer). In: Fowler ME, Cubas ZS. Biology, Medicine, and Surgery of South American Wild Animals. Iowa State University Press, Iowa; 2001. p.402-422.
- Duarte JMB. Artiodactyla – Cervidae (Veado - catingueiro, Veado-campeiro, Cervo - do - pantanal). In: Cubas ZS, Silva JCR, Catão-Dias JL. Tratado de animais selvagens - Medicina Veterinária. Roca, São Paulo; 2006. p. 641-664.
- Fowler ME. Zoo and Wild Animal Medicine Current Therapy, 3rd ed. WB Saunders, Philadelphia; 1993.
- Kawahara M, Tajima T, Torii H, Yabutani M, Ishii J, Harasawa M, et al. Ehrlichia chaffeensis infection of Sika deer, Japan. Emerg Infect Dis 2009; 15(12): 1991-1993.
- Klein MB, Miller JS, Nelson CM, Goodman JL. Primary bone marrow progenitors of both granulocytic and monocytic lineages are susceptible to infection with the agent of human granulocytic ehrlichiosis. J Infect Dis 1997; 176(5): 1405-1409.
- Kocan KM, de la Fuente J, Guglielmone AA, Meléndez RD.Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev 2003; 16(4): 698-712
- Kreier JP, Ristic M, Watrach AM. Theileria sp. in a deer in the United States. Am J Vet Res 1962; 23: 657-662.
- Kuiken T, Leighton FA, Fouchier RAM, LeDuc JW, Peiris JSM, Schudel A, et al. Pathogen Surveillance in Animals. Science 2005; 309(5741): 1680-1681.
- Kuttler KL, Zaugg JL. Characteristics of an attenuated Anaplasma marginale of deer origin as an anaplasmosis vaccine. Trop Anim Health Prod 1988; 20(2): 85-91.
- Machado RZ, Duarte JMB, Dagnone AS, Szabó MPJ. Detection of Ehrlichia chaffeensis in Brazilian marsh deer (Blastocerus dichotomus). Vet Parasitol 2006; 139(1-3): 262-266.
- Machado RZ, André MA, Werther K, de Sousa E, Gavioli FA, Alves Junior JRF. Migratory and Carnivorous Birds in Brazil: Reservoirs for Anaplasma and Ehrlichia Species? Vector Borne Zoonotic Dis 2012; 12(8): 705-708.
- Madigan JE, Pusterla N. Ehrlichial diseases. Vet Clin North Am Equine Pract 2000; 16(3): 487-499.
- Massung RF, Slater K, Owens JH, Nicholson WL, Mather TN, Solberg VB, et al. Nested PCR assay for the detection of granulocytic ehrlichiae. J Clin Microbiol 1998; 36(4): 1090-1095.
- Massung RF, Courtney JW, Hiratzka SL, Pitzer VE, Smith G, Dryden RL. Anaplasma phagocytophilum in white-tailed deer. Emerg Infect Dis 2005; 11(10): 1604-1606.
- Moreira SM. Estudo retrospectivo (1998–2001) da erliquiose canina em Belo Horizonte: avaliação clínica e laboratorial de infecções experimentais. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2001.
- Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, et al. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet Microbiol 2004; 101(1): 53-64.
- Overzier E, Pfister K, Herb I, Mahling M, Böck G Jr, Silaghi C. Detection of tick-borne pathogens in roe deer (Capreolus capreolus), questing ticks (Ixodes ricinus) and in ticks infesting roe deer in southern Germany. Ticks Tick Borne Dis 2013; 4(4): 320-328.
- Picoloto G, Ferreira de Lima R, Olegario LAO, Carvalho CME, Lacerda ACR, Tomas WM, et al. Real time polymerase chain reaction to diagnose Anaplasma marginale in cattle and deer (Ozotoceros bezoarticus leucogaster) of the Brazilian Pantanal. Rev Bras Parasitol Vet 2010; 19(3): 186-188.
- Ristic M. Babesiosis of domestic animals and man. Boca Raton, CRC Press, Florida; 1988.
- Sacchi ABV, Duarte JMB, Andre MR, Machado RZ. Prevalence and molecular characterization of Anaplasmataceae agents in free-ranging Brazilian marsh deer (Blastocerus dichotomus). Comp Immunol Microbiol Infect Dis 2012; 35(4): 325-334.
- Santos HA, Pires MS, Vilela JAR, Santos TM, Faccini JLH, Baldani CD, et al. Detection of Anaplasma phagocytophilum in Brazilian dogs by real-time polymerase chain reaction. J Vet Diagn Invest 2006; 23(4): 770-774.
- Santos HA, Thomé SM, Baldani CD, Silva CB, Peixoto MP, Pires MS, et al. Molecular epidemiology of the emerging zoonosis agent Anaplasma phagocytophilum (Foggie, 1949) in dogs and ixodid ticks in Brazil. Parasit Vectors 2013; 6: 348.
- Silaghi C, Hamel D, Thiel C, Pfister K, Passos LMF, Rehbein S. Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis 2011; 11(4): 355-362.
- Silveira JAG, Rabelo EML, Ribeiro MFB. Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Vet Parasitol 2011; 177(1-2): 61-66.
- Silveira, JAG, Rabelo EML, Ribeiro MFB. Molecular Detection of Tick-Borne Pathogens of the Family Anaplasmataceae in Brazilian Brown Brocket Deer (Mazama gouazoubira, Fischer, 1814) and Marsh Deer (Blastocerus dichotomus, Illiger, 1815). Transbound Emerg Dis 2012; 59(4): 353-360.
- Silveira JAG. Ocorrência de hemoparasitos e ectoparasitos em veado-catingueiro (Mazama gouazoubira Fischer, 1814), veado-campeiro (Ozotocerus bezoarticus Linnaeus, 1758) e cervo-do-pantanal (Blastocerus dichotomus Illiger, 1815): utilização de métodos parasitológicos e moleculares. [Tese]. Belo Horizonte: Universidade Federal de Minas Gerais; 2012.
- Silveira JAG, Rabelo EML, Lacerda ACL, Borges PAL, Tomás WM, Pellegrin AO, et al. Molecular detection and identification of hemoparasites in pampas deer (Ozotoceros bezoarticus Linnaeus, 1758) from the Pantanal Brazil. Ticks Tick Borne Dis 2013; 4(4): 341-345.
- Stuen S. Anaplasma phagocytophilum (Formerly Ehrlichia phagocytophila) Infection in Sheep and Wild Ruminants in Norway. A study on clinical manifestation, distribution and persistence [Thesis]. Oslo: School of Veterinary Science; 2003.
- Stuen S. Anaplasma phagocytophilum – the most widespread tick-borne infection in animals in Europe. Vet Res Commun 2007; 31(S1): 79-84.
- Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol 2013; 3: 31.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30 (12): 2725-2729.
- Warns-Petit E, Morignat E, Artois M, Calavas D. Unsupervised clustering of wildlife necropsy data for syndromic surveillance. BMC Vet Res 2010; 6: 56.
- Yabsley MJ, Quick TC, Little E. Theileriosis in a white-tailed-deer (Odocoileus virginianus) fawn. J Wildl Dis 2005; 41(4): 806-809.
- Zahler M, Rinder H, Schein E, Gothe R. Detection of a new pathogenic Babesia microti-like species in dogs. Vet Parasitol 2000; 89(3): 241-248.
Publication Dates
-
Publication in this collection
Apr-Jun 2014
History
-
Received
31 Jan 2014 -
Accepted
14 Mar 2014