Abstract
Phylogenies within Toxoplasmatinae have been widely investigated with different molecular markers. Here, we studied molecular phylogenies of the Toxoplasmatinae subfamily based on apicoplast and mitochondrial genes. Partial sequences of apicoplast genes coding for caseinolytic protease (clpC) and beta subunit of RNA polymerase (rpoB), and mitochondrial gene coding for cytochrome B (cytB) were analyzed. Laboratory-adapted strains of the closely related parasites Sarcocystis falcatula and Sarcocystis neurona were investigated, along with Neospora caninum, Neospora hughesi, Toxoplasma gondii (strains RH, CTG and PTG), Besnoitia akodoni, Hammondia hammondiand two genetically divergent lineages of Hammondia heydorni. The molecular analysis based on organellar genes did not clearly differentiate between N. caninum and N. hughesi, but the two lineages of H. heydorni were confirmed. Slight differences between the strains of S. falcatula and S. neurona were encountered in all markers. In conclusion, congruent phylogenies were inferred from the three different genes and they might be used for screening undescribed sarcocystid parasites in order to ascertain their phylogenetic relationships with organisms of the family Sarcocystidae. The evolutionary studies based on organelar genes confirm that the genusHammondia is paraphyletic. The primers used for amplification of clpC and rpoB were able to amplify genetic sequences of organisms of the genus Sarcocystisand organisms of the subfamily Toxoplasmatinae as well.
Keywords:
Sarcocystidae; molecular characterization; phylogeny; Toxoplasmatinae; apicoplast gene; mitochondrial gene
Resumo
A filogenia da subfamília Toxoplasmatinae tem sido amplamente investigada com diversos marcadores moleculares. Neste estudo, a filogenia molecular da subfamília Toxoplasmatinae foi analisada através de genes de apicoplasto e mitocondriais. Foram analisadas sequências parciais de genes de apicoplasto codificadores da protease caseinolítica (clpC), e da subunidade beta da RNA polimerase (rpoB) e de gene mitocondrial codificador de citocromo B (cytB). Foram investigadas cepas adaptadas em laboratório de Sarcocystis neurona eSarcocystis falcatula, parasitos estreitamente relacionados, além de Neospora caninum, Neospora hughesi, Toxoplasma gondii (cepas RH, CTG e PTG),Besnoitia akodoni, Hammondia hammondi e duas linhagens geneticamente divergentes de Hammondia heydorni. A análise molecular, baseada em genes de organelas, não diferenciou claramenteN. caninum de N. hughesi, porém foi possível confirmar as duas linhagens de H. heydorni. Foram encontradas pequenas diferenças entre as cepas adaptadas em laboratório deS. falcatula e S. neurona em todos os marcadores moleculares avaliados. Concluindo, filogenias congruentes foram reconstruídas com os três diferentes genes que podem ser úteis em triagem de parasitos sarcocistídeos não identificados, para identificar sua relação com organismos da família Sarcocystidae. Os estudos evolutivos com genes organelares confirmam que o gênero Hammondia é parafilético. Osprimers utilizados para amplificação declpC e rpoB foram capazes de amplificar sequências genéticas de organismos do gênero Sarcocystis e da subfamília Toxoplasmatinae.
Palavras-chave:
Sarcocystidae; caracterização molecular; filogenia; Toxoplasmatinae; gene apicoplasto; gene mitocondrial
Introduction
Mitochondrial genes are useful for phylogenetic analysis at different taxonomic levels of apicomplexan parasites (LIN et al., 2011Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, Xie WQ, et al. Characterization of the complete mitochondrial genomes of five species from domestic chickens. EimeriaGene 2011; 480(1-2): 28-33. http://dx.doi.org/10.1016/j.gene.2011.03.004. PMid:21402132.
http://dx.doi.org/10.1016/j.gene.2011.03...
; HE et al., 2014He L, Zhang Y, Zhang QL, Zhang WJ, Feng HH, Khan MK, et al. Mitochondrial genome of , apicomplexan parasite of water buffalo (, Linnaeus, 1758) endemic in China. Babesia orientalisBubalus babalisParasit Vectors 2014; 7(82): 1-8. http://dx.doi.org/10.1186/1756-3305-7-82. PMid:24580772.
http://dx.doi.org/10.1186/1756-3305-7-82...
). Cytochrome oxidase II and cytochrome B (cytB) coding sequences have been used for this purpose (GJERDE, 2013aGjerde B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of and (Apicomplexa: Sarcocystidae). Toxoplasma gondii, Neospora caninum, Hammondia heydorniHammondia triffittaeParasitol Res 2013a; 112(4): 1493-1511. http://dx.doi.org/10.1007/s00436-013-3296-4. PMid:23358734.
http://dx.doi.org/10.1007/s00436-013-329...
,bGjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 2013b; 43(7): 579-591. http://dx.doi.org/10.1016/j.ijpara.2013.02.004. PMid:23542092.
http://dx.doi.org/10.1016/j.ijpara.2013....
). Similar to mitochondria, apicoplasts are organelles of maternal inheritance (FERGUSON et al., 2005Ferguson DJ, Henriquez FL, Kirisits MJ, Muench SP, Prigge ST, Rice DW, et al. Maternal inheritance and stage-specific variation of the apicoplast in during development in the intermediate and definitive host. Toxoplasma gondiiEukaryot Cell 2005; 4(4): 814-826. http://dx.doi.org/10.1128/EC.4.4.814-826.2005. PMid:15821140.
http://dx.doi.org/10.1128/EC.4.4.814-826...
) that contain conserved genes that are commonly used to reconstruct evolutionary histories (FEAGIN, 1994Feagin JE. The extrachromosomal DNAs of apicomplexan parasites. Annu Rev Microbiol 1994; 48(1): 81-104. http://dx.doi.org/10.1146/annurev.mi.48.100194.000501. PMid:7826027.
http://dx.doi.org/10.1146/annurev.mi.48....
; KOHLER et al., 1997Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJM, et al. A plastid of probable green algal origin in apicomplexan parasites. Science 1997; 275(5305): 1485-1489. http://dx.doi.org/10.1126/science.275.5305.1485. PMid:9045615.
http://dx.doi.org/10.1126/science.275.53...
). Genes encoded in the apicoplast genome are useful markers for resolving the phylogeny ofPlasmodium species, because these genes have greater phylogenetic signal than the mitochondrial genome for Plasmodium phylogeny (reviewed in ARISUE & HASHIMOTO, 2015Arisue N, Hashimoto T. Phylogeny and evolution of apicoplasts and apicomplexan parasites. Parasitol Int 2015; 64(3): 254-259. http://dx.doi.org/10.1016/j.parint.2014.10.005. PMid:25451217.
http://dx.doi.org/10.1016/j.parint.2014....
). Among other genes within the plastid genome, two genes are highlighted. The gene coding for caseinolytic protease (clpC) (a member of the chaperone family) is a conserved apicoplast gene that is widely applied in phylogenetic reconstructions of apicomplexan organisms (RATHORE et al., 2001Rathore D, Wahl AM, Sullivan M, McCutchan TF. A phylogenetic comparison of gene trees constructed from plastid, mitochondrial and genomic DNA of species. PlasmodiumMol Biochem Parasitol 2001; 114(1): 89-94. http://dx.doi.org/10.1016/S0166-6851(01)00241-9. PMid:11356517.
http://dx.doi.org/10.1016/S0166-6851(01)...
; MARTINSEN et al., 2008Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol 2008; 47(1): 261-273. http://dx.doi.org/10.1016/j.ympev.2007.11.012. PMid:18248741.
http://dx.doi.org/10.1016/j.ympev.2007.1...
; LAU et al., 2009Lau AOT, McElwain TF, Brayton KA, Knowles DP, Roalson EH. Babesia bovis: a comprehensive phylogenetic analysis of plastid-encoded genes supports green algal origin of apicoplasts. Exp Parasitol 2009; 123(3): 236-243. http://dx.doi.org/10.1016/j.exppara.2009.07.007. PMid:19646439.
http://dx.doi.org/10.1016/j.exppara.2009...
). The plastid-encoded beta subunit of RNA polymerase (rpoB), which is responsible for processing polymerase activity, has also been used in phylogenetic reconstructions among organisms of the Sarcocystidae family (DUBEY et al., 2003aDubey JP, Lindsay DS, Rosenthal BM, Thomas NJ. Sarcocysts of an unidentified species of Sarcocystis in the sea otter (). Enhydra lutrisJ Parasitol 2003a; 89(2): 397-399. http://dx.doi.org/10.1645/0022-3395(2003)089[0397:SOAUSO]2.0.CO;2. PMid:12760665.
http://dx.doi.org/10.1645/0022-3395(2003...
; WENDTE et al., 2010Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, Conrad PA, et al. Limited genetic diversity among strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Sarcocystis neuronaVet Parasitol 2010; 169(1-2): 37-44. http://dx.doi.org/10.1016/j.vetpar.2009.12.020. PMid:20071081.
http://dx.doi.org/10.1016/j.vetpar.2009....
).
Here, we studied molecular phylogenies of the Toxoplasmatinae subfamily in order to target mitochondrial and apicoplast genetic sequences. The known genus forming the Toxoplasmatinae subfamily are Besnoitia,Toxoplasma, Neospora, andHammondia, closely related coccidians with similarly sized oocysts (DUBEY, 1993Dubey JP. Toxoplasma, Neospora, Sarcocystis, and other tissue cyst-forming coccidia of humans and animals. In: Kreier JP. Parasitic Protozoa. 2nd ed. San Diego: Academic Press; 1993. p. 1-158, vol. 6.). The aim of this paper was to demonstrate the suitability of these molecular markers for studying the genetic variability and phylogenetic relationships among members of the Toxoplasmatinae subfamily. Phylogenies within Toxoplasmatinae have been greatly investigated with other molecular markers (MUGRIDGE et al., 1999Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. Phylogenetic analyses based on full-length large subunit ribosomal RNA gene sequence comparison reveals that is more closely related to Neospora caninumHammondia heydorni than to Toxoplasma gondii.Int J Parasitol 1999; 29(10): 1545-1556. http://dx.doi.org/10.1016/S0020-7519(99)00150-2. PMid:10608441.
http://dx.doi.org/10.1016/S0020-7519(99)...
; ELLIS et al, 1999Ellis JT, Morrison DA, Liddell S, Jenkins MC, Mohammed OB, Ryce C, et al. The genus is paraphyletic. HammondiaParasitology 1999; 118(4): 357-362. http://dx.doi.org/10.1017/S0031182098003801. PMid:10340325.
http://dx.doi.org/10.1017/S0031182098003...
;MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
; SOARES et al., 2011Soares RM, Lopes EG, Keid LB, Sercundes MK, Martins J, Richtzenhain LJ. Identification of Hammondia heydorni oocysts by a heminested-PCR (hnPCR-AP10) based on the RAPD fragmente AP10. H. heydorniVet Parasitol 2011; 175(1-2): 168-172. http://dx.doi.org/10.1016/j.vetpar.2010.09.022. PMid:21030154.
http://dx.doi.org/10.1016/j.vetpar.2010....
).
Materials and Methods
Samples
The DNA of tachyzoites and merozoites maintained in laboratory models of the following apicomplexan parasites were used: Toxoplasma gondiistrains RH, CTG and PTG (archetypes I, II and III respectively) maintained in mice, Neospora caninum strain NC-1 maintained in VERO,Sarcocystis neurona strain 138 and Sarcocystis falcatula SF1 strain kept in CV-1 cells. Sarcocystis neurona strain 138 was obtained from the intestines of an opossum (Didelphis virginiana) that had been fed with skeletal muscles from a raccoon (Procyon lotor) that in turn had been fed with sporocysts that had been isolated in Ohio, United States (LINDSAY et al., 2004Lindsay DS, Mitchell SM, Vianna MC, Dubey JP. (protozoa: Apicomplexa) description of oocysts, sporocysts, sporozoites, excystation, and early development. Sarcocystis neuronaJ Parasitol 2004; 90(3): 461-465. http://dx.doi.org/10.1645/GE-230R. PMid:15272465.
http://dx.doi.org/10.1645/GE-230R...
). Sarcocystis falcatula SF1 was obtained from tissues of budgerigars (Melopsittacus undulatus) that had been orally inoculated with sporocysts from the intestines of an opossum (D. virginiana) that had been isolated in California, United States (MARSH et al., 1997Marsh AE, Barr BC, Tell L, Koski M, Greiner E, Dame J, et al. In vitro cultivation and experimental inoculation of Sarcocystis falcatula and Sarcocystis neurona merozoites into budgerigars (). Melopsittacus undulatusJ Parasitol 1997; 83(6): 1189-1192. http://dx.doi.org/10.2307/3284386. PMid:9406803.
http://dx.doi.org/10.2307/3284386...
). Dr. David Lindsay kindly provided both the S. neurona and the S. falcatula strain.
DNA from Besnoitia akodoni was also used. This is a parasite isolated from a rodent (Akodon montensis) that is native to Brazil and was obtained as previously described (DUBEY et al., 2003bDubey JP, Sreekumar C, Rosenthal BM, Lindsay DS, Grisard EC, Vitor RW. Biological and molecular characterization of n.sp. (Protozoa: Apicomplexa) from the rodent . Besnoitia akodoniAkodon montensis in BrazilParassitologia 2003b; 45(2): 61-70. PMid:15266998.). DNA from the Neospora hughesi Oregon isolate (DUBEY et al., 2001Dubey JP, Liddell S, Mattson D, Speer CA, Howe DK, Jenkins MC. Characterization of the Oregon isolate of Neospora hughesi from a horse. J Parasitol 2001; 87(2): 345-353. http://dx.doi.org/10.1645/0022-3395(2001)087[0345:COTOIO]2.0.CO;2. PMid:11318565.
http://dx.doi.org/10.1645/0022-3395(2001...
), Hammondia hammondi (isolate 300) and two genetically divergent lineages of Hammondia heydorni (isolates 376 and BR) were obtained as described elsewhere (MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
).
PCR and primers
The polymerase chain reaction was performed in accordance with the manufacturer’s recommendations for platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), using the primers listed in Table 1. The annealing temperature of each primer pair is given in Table 1.
Primers for clpC were designed from a consensus sequence ofclpC from EST database sequences of N. caninum and S. neurona and RNAm sequences ofT. gondii. Primers for cytochrome B coding sequences (cytB) targeting Sarcocystis spp. were designed from genomic sequences of S. neurona and S. falcatula (VALADAS et al., 2016Valadas SY, da Silva JI, Galucci E, Keid LB, Zwarg T, de Oliveira AS, et al. Diversity of Sarcocystis spp shed by opossums in Brazil inferred with phylogenetic analysis of DNA coding ITS1, Cytochrome B, and surface antigens. Exp Parasitol 2016; Feb 18. In press. http://dx.doi.org/10.1016/j.exppara.2016.02.008.
http://dx.doi.org/10.1016/j.exppara.2016...
). A CytB primer targeting members of the Toxoplasmatinae subfamily was designed from EST database sequences of N. caninum and RNAm sequences of T. gondii. TherpoB primer targeting the Toxoplasmatinae subfamily was designed from RpoB sequences from the genomic database for S. neurona, S. falcatula, N. caninum, T. gondii, Besnoitia oryctofelisi and Besnoitia darling.
The PCR products were observed through electrophoresis on 2% agarose gel. The DNA of positive samples was extracted from the gel and purified by using GE Healthcare kits (Illustra, GFX PCR DNA and GEL Band purification kits) in accordance with the manufacturer’s instructions.
Sequencing and sequence analysis
The PCR products were sequenced using the ABI PRISM BigDye® Terminator v3.1 kit (Ready Reaction Cycle Sequencing, Applied Biosystems). The DNA was purified by means of alcohol precipitation and sequencing products were analyzed using an automated sequencer (Applied Biosystems 3500 Genetic Analyzer).
Sequences were assembled and the contig sequence was obtained by means of the Phred base-calling software and the Phrap assembly tool, which are available in the Codoncode aligner v.4.2.1 suite (Codoncode Corp. Dedham, MA, USA). The sequences were aligned by means of ClustalW, which is available in the BioEdit Sequence Alignment Editor suite (HALL, 1999Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symposium Series 1999; 41: 95-98.), based on homologous sequences available in GenBank. Nucleotide sequences were submitted to GenBank under the accession numbers GenBank KP871703, KP871704, KP871707-KP871715, KP871716, KP871717, KP871723- KP871731, KP871732, KP871733, KP871734-KP871742.
Phylogenies and DNA polymorphism were evaluated by means of MEGA 6 (TAMURA et al., 2013Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30(12): 2725-2729. http://dx.doi.org/10.1093/molbev/mst197. PMid:24132122.
http://dx.doi.org/10.1093/molbev/mst197...
) and DNAsp v.5 (LIBRADO & ROZAS, 2009Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009; 25(11): 1451-1542. http://dx.doi.org/10.1093/bioinformatics/btp187. PMid:19346325.
http://dx.doi.org/10.1093/bioinformatics...
), respectively. The phylogenetic reconstructions were inferred through maximum parsimony (MP) and maximum likelihood (ML) methods. Apicoplast gene segments were analyzed individually and concatenated. ML phylogenies were based on nucleotide substitution model calculated for each set of data. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach. The MP trees were obtained using the subtree-pruning-regrafting algorithm with search level 1 in which the initial trees were obtained by the random addition of sequences (10 replicates).
In all cases, the reliability of the inferred tree was tested by bootstrap resampling technique (FELSENSTEIN, 1985Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39(4): 783-791. http://dx.doi.org/10.2307/2408678.
http://dx.doi.org/10.2307/2408678...
) after 1000 replicates. Briefly, if there are m sequences, each with n nucleotides, a phylogenetic tree can be reconstructed using some tree building method. From each sequence, n nucleotides are randomly chosen with replacements, giving rise to m rows of n columns each. These now constitute a new set of sequences. A tree is then reconstructed with these new sequences using the same tree building method as before. Next, the topology of this tree is compared to that of the original tree. Each interior branch of the original tree that is different from the bootstrap tree the sequence it partitions is given a score of 0; all other interior branches are given the value 1. This procedure of resampling the sites and the subsequent tree reconstruction is repeated several times (typically from 100 to 1000 times), and the percentage of times (the bootstrap value) each interior branch is given a value of 1 is noted.
Results and Discussion
The cytB phylogenetic reconstruction is shown in Figure 1. The two Hammondia heydorni isolate sequences were identical to each other, and the sequences of the genus Neospora were also identical to each other. Archetypes I, II and III of Toxoplasma gondii (strains RH, CTG and PTG, respectively) were identical.
Phylogenies based on cytB, clpC,rpoB. [Apico]: clpC andrpoB concatenated. [ML]: Maximum Likelihood. [MP]: Maximum Parsimony. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. For ML analysis, Tamura 3-parameter model with Gamma distribution (T92+G) was selected by MEGA 6 model selection option for cytB-based phylogeny. Tamura 3-parameter with the rate variation model allowed for some sites to be evolutionarily invariable (T92+I) was selected for clpC,rpoB, and apico -based phylogenies. The MP trees were obtained using the subtree-pruning-regrafting algorithm with search level 1 in which the initial trees were obtained by the random addition of sequences (10 replicates). There were a total of 549, 423, 434, and 857 positions in the final dataset of cytB, clpC,rpoB and Apico, respectively.
Molecular analysis using cytB consistently separates all genera within the Toxoplasmatinae subfamily and shows high similarity (~100%) within the group. The genus Neospora is formed by two known species,Neospora caninum and Neospora hughesi, which differ markedly in relation to pathogenicity and antigenicity traits (MARSH et al., 1998Marsh AE, Barr BC, Packham AE, Conrad PA. Description of a new species (Protozoa: Apicomplexa: Sarcocystidae). NeosporaJ Parasitol 1998; 84(5): 983-991. http://dx.doi.org/10.2307/3284632. PMid:9794642.
http://dx.doi.org/10.2307/3284632...
, 1999Marsh AE, Howe DK, Wang G, Barr BC, Cannon N, Conrad PA. Differentiation of Neospora hughesi from based on their immunodominant surface antigen, SAG1 and SRS2. Neospora caninumInt J Parasitol 1999; 29(10): 1575-1582. http://dx.doi.org/10.1016/S0020-7519(99)00120-4. PMid:10608444.
http://dx.doi.org/10.1016/S0020-7519(99)...
). Although Neospora caninum has a known capacity to infect cattle and cause abortion problems, Neospora hughesi is related to neurological disease in horses. Reports onN. hughesi infections are scarce (VARDELEON et al., 2001Vardeleon D, Marsh AE, Thorne JG, Loch W, Young R, Johnson PJ. Prevalence of Neospora hughesi and antibodies in horses from various geographical locations. Sarcocystis neuronaVet Parasitol 2001; 95(2-4): 273-282. http://dx.doi.org/10.1016/S0304-4017(00)00393-9. PMid:11223207.
http://dx.doi.org/10.1016/S0304-4017(00)...
; WOBESER et al., 2009Wobeser BK, Godson DL, Rejmanek D, Dowling P. Equine protozoal myeloencephalitis caused by in an adult horse in Saskatchewan. Neospora hughesiCan Vet J 2009; 50(8): 851-853. PMid:19881924.; PUSTERLA et al., 2014Pusterla N, Mackie S, Packham A, Conrad PA. Serological investigation of transplacental infection with and in broodmares. Neospora hughesiSarcocystis neuronaVet J 2014; 202(3): 649-650. http://dx.doi.org/10.1016/j.tvjl.2014.09.015. PMid:25438732.
http://dx.doi.org/10.1016/j.tvjl.2014.09...
), and little is known about this agent. The twoNeospora species are identical at cytB, and the same is valid for the two Hammondia heydorni isolates, which were previously discriminated into two distinct genetic lineages (MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
). Hammondia heydorni can be separated into two molecularly divergent lineages when compared at the alpha-tubulin, HSP 70 and ITS-1 genes (ABEL et al., 2006Abel J, Schares G, Orzeszko K, Gasser RB, Ellis JT. Hammondia isolated from dogs and foxes are genetically distinct. Parasitology 2006; 132(2): 187-192. http://dx.doi.org/10.1017/S0031182005008814. PMid:16188045.
http://dx.doi.org/10.1017/S0031182005008...
; MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
; GJERDE & DAHLGREN, 2011Gjerde B, Dahlgren SS. Hammondia triffittae n. comb. of foxes (Vulpes spp.): biological and molecular characteristics and differentiation from Hammondia heydorni of dogs. Parasitology 2011; 138(3): 303-321. http://dx.doi.org/10.1017/S0031182010001265. PMid:20854709.
http://dx.doi.org/10.1017/S0031182010001...
; SOARES et al., 2011Soares RM, Lopes EG, Keid LB, Sercundes MK, Martins J, Richtzenhain LJ. Identification of Hammondia heydorni oocysts by a heminested-PCR (hnPCR-AP10) based on the RAPD fragmente AP10. H. heydorniVet Parasitol 2011; 175(1-2): 168-172. http://dx.doi.org/10.1016/j.vetpar.2010.09.022. PMid:21030154.
http://dx.doi.org/10.1016/j.vetpar.2010....
). A third lineage of Hammondia heydorni was recently reclassified asHammondia triffitti, due to its molecular divergence from the other lineages and to its characteristic of only using foxes, and not dogs, as its definitive host. Isolates classified as H. triffitti havecytB fragments identical to those of the H. heydorni lineages used in this study (GJERDE, 2013aGjerde B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of and (Apicomplexa: Sarcocystidae). Toxoplasma gondii, Neospora caninum, Hammondia heydorniHammondia triffittaeParasitol Res 2013a; 112(4): 1493-1511. http://dx.doi.org/10.1007/s00436-013-3296-4. PMid:23358734.
http://dx.doi.org/10.1007/s00436-013-329...
). A single synonymous nucleotide substitution differentiates the sequence of SF1 from that of SN138.
ClpC phylogeny also discriminates the clades Toxoplasmatinae subfamily and Sarcocystis genus. It also resolves the phylogeny of Toxoplasmatinae but with less statistical support than the CytBphylogeny does. The phylogenic reconstruction is shown in Figure 1. Toxoplasma gondii strain RH differs from strains CTG and PTG in one synonymous substitution. A single synonymous substitution is found between N. hughesi and N. caninum sequences, whereas the two H. heydorniisolates differed in six nucleotides, among which one is a non-synonymous substitution and the rest are synonymous. Concerning the clpClocus, the topology of the phylogenetic reconstruction was similar to that obtained with cytB. ClpC was less conserved thanCytB. S. neurona 138 and S. falcatula SF1 had differences of the same magnitude as seen inT. gondii variants and the two Neosporaspecies. Meanwhile, the two H. heydorni lineages differed by magnitudes similar to those observed between T. gondii andH. hammondi, thus corroborating the divergence previously found in these two lineages of H. heydorni (MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
).
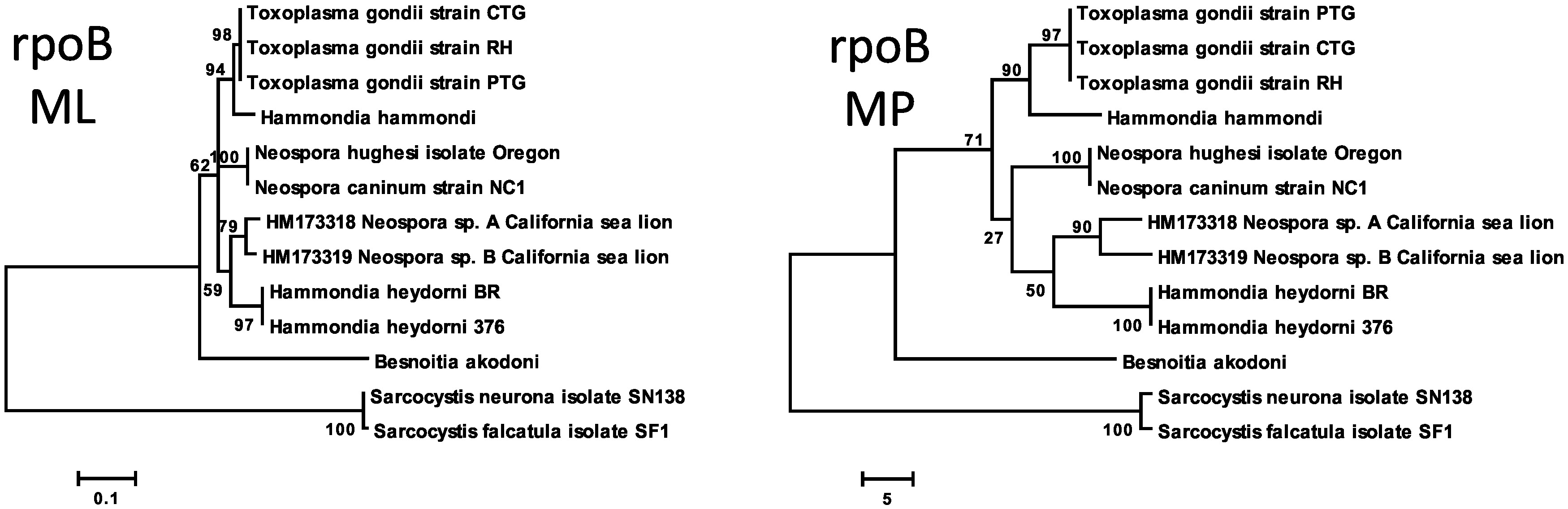
Similar to clpC analysis, rpoB phylogeny also allows reconstruction of Sarcocystidae phylogeny but with less statistical support than in the reconstruction with the CytB gene (Figure 1), in particular the node that segregates all other Toxoplasmatinae fromBesnoitia akodoni. The distance between the twoHammondia heydorni isolates is the result of a single synonymous substitution. N. hughesi and N. caninumsequences are identical at this locus. Unlike the other organelle genome sequences, the rpoB sequences presented insertions and deletions among the organisms of the Toxoplasmatinae clade. After concatenate apicoplast derived genes, the resulting phylogeny have shown stronger statistic support and corroborated each individual genealogy.
The rpoB dataset consists of oocyst sequences from a California sea lion (Zalophus californianus) that were obtained from GenBank. These were previously identified as oocysts from the genusNeospora. The phylogenetic reconstruction in Figure 2 shows that these organisms (HM173319 and HM173318) are as closely related to Neospora spp. as to Hammondia heydorni, and probably represent a new genus within Toxoplasmatinae subfamily. One synonymous difference was found between the SF1 isolate and another two S. falcatula sequences from GenBank (AY164999, AY165001). This was the same difference as between SF1 and the S. lindsayi sequence (AY164997). SF1 and SN138 differ from each other by three synonymous nucleotide substitutions. On the other hand, SN138 differs from S. neuronaAY165000 by three nucleotide differences, among which one is non-synonymous. This is a significantly greater difference than the one existing between the twoNeospora species and is also greater than between the two lineages of H. heydorni.
Phylogenies based on rpoB. [ML]: Maximum Likelihood. [MP]: Maximum Parsimony. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. For ML analysis, Tamura 3-parameter model with Gamma distribution (T92+G) was selected by MEGA 6 model selection option f. The MP tree was obtained using the subtree-pruning-regrafting algorithm with search level 1 in which the initial trees were obtained by the random addition of sequences (10 replicates). There were a total of 283 positions in the final dataset.
Regarding the Toxoplasmatinae subfamily, rpoB molecular analysis reveals great similarity within groups, since no differences of more than one nucleotide exist between sequences of the same clade. By addingNeospora-like organisms (CARLSON-BREMER et al., 2012Carlson-Bremer D, Johnson CK, Miller RH, Gulland FM, Conrad PA, Wasmuth JD, et al. Identification of two novel coccidian species shed by California sea lions (Zalophus californianus). J Parasitol 2012; 98(2): 347-354. http://dx.doi.org/10.1645/GE-2752.1. PMid:22091999.
http://dx.doi.org/10.1645/GE-2752.1...
) that are shed by aquatic mammals, the phylogenetic reconstruction through parsimony displays these sequences in clades that are divergent from both Neospora spp. and H. heydorni. These two sequences are related to each other; they form a monophyletic group, but are considerably divergent from each other, and this can be observed from the length of the branches from its common ancestry. Therefore, these two sequences likely represent distinct species, which have not been yet described in the Toxoplasmatinae subfamily.
The DNA polymorphism between sequences of Toxoplasma gondii RH,Hammondia heydorni 376, Hammondia heydorni BR,Hammondia hammondi, Neospora hughesi andNeospora caninum NC1 showed that rpoB andclpC have similar diversity values and that both genes are less conserved than cytB. The rpoB sequences are the ones with the highest diversity in non-synonymous sites (Table 2).
Nucleotide diversity of organellar gene fragments amongToxoplasma gondii (strain RH), Hammondia heydorni isolates (strains BR and 376) and Neospora caninum (strain NC1).
As expected, the phylogenies reconstructed using organellar genes of Sarcocystidae are concordant. Thereby, as in other studies (GJERDE, 2013aGjerde B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of and (Apicomplexa: Sarcocystidae). Toxoplasma gondii, Neospora caninum, Hammondia heydorniHammondia triffittaeParasitol Res 2013a; 112(4): 1493-1511. http://dx.doi.org/10.1007/s00436-013-3296-4. PMid:23358734.
http://dx.doi.org/10.1007/s00436-013-329...
, bGjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 2013b; 43(7): 579-591. http://dx.doi.org/10.1016/j.ijpara.2013.02.004. PMid:23542092.
http://dx.doi.org/10.1016/j.ijpara.2013....
), organellar genetic sequences can give a reliable estimate of the diversity among sarcocystid species. Gjerde (2013a)Gjerde B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of and (Apicomplexa: Sarcocystidae). Toxoplasma gondii, Neospora caninum, Hammondia heydorniHammondia triffittaeParasitol Res 2013a; 112(4): 1493-1511. http://dx.doi.org/10.1007/s00436-013-3296-4. PMid:23358734.
http://dx.doi.org/10.1007/s00436-013-329...
, demonstrated unequivocally that mitochondrial markers have the ability to reconstruct evolutionary history within the Sarcocystidae family. The cox2sequences of Toxoplasma gondii, Neospora caninum,Hammondia triffitti and Hammondia heydornireported in the work above had nucleotide diversity of 0.06484, whereas the diversity in synonymous and non-synonymous sites was 0.21371 and 0.01569, respectively (data not shown). RpoB and clpCsequences had similar diversity, which was both inferior to cox2and superior to cytB. However, of all the markers,rpoB sequences presented the highest diversity at non-synonymous sites, which suggests that this marker is more susceptible to positive selection pressure. On the other hand, cytB showed diversity at non-synonymous sites that was four to five times less than that of the others, and therefore was more subject to purifying selection.
In all of the evolutionary reconstructions of Toxoplasmatinae organisms, the clade holding the two Hammondia heydorni isolates has a basal position, compared with the other clades. Both H. heydorni and H. triffiti use canids as definitive hosts. The two H. heydorni isolates used in this study are molecularly different from each other (MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
) and fromH. triffiti (GJERDE & DAHLGREN, 2011Gjerde B, Dahlgren SS. Hammondia triffittae n. comb. of foxes (Vulpes spp.): biological and molecular characteristics and differentiation from Hammondia heydorni of dogs. Parasitology 2011; 138(3): 303-321. http://dx.doi.org/10.1017/S0031182010001265. PMid:20854709.
http://dx.doi.org/10.1017/S0031182010001...
). According to the results, it would be possible to assume that the relationship between canids and felids might have favored adaptation of ancestral parasites, originally from canids, to felids, i.e. the group of animals that serves as the definitive host for both H. hammondi andT. gondii. However, in all of the phylogenetic reconstruction conducted in this study and other described elsewhere, moderate statistical support, less than 75, is shown for the groups gathering Neospora spp. andT. gondii; H. heydorni and T. gondii; or H. heydorni and Neospora. The placement of Neospora spp. as sister group of T. gondii/H. hammondi was also difficult to demonstrate with Hsp70- and 18S rRNA phylogenies (ELLIS et al., 1999Ellis JT, Morrison DA, Liddell S, Jenkins MC, Mohammed OB, Ryce C, et al. The genus is paraphyletic. HammondiaParasitology 1999; 118(4): 357-362. http://dx.doi.org/10.1017/S0031182098003801. PMid:10340325.
http://dx.doi.org/10.1017/S0031182098003...
;MUGRIDGE et al., 2000Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 2000; 17(12): 1842-1853. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285. PMid:11110900.
http://dx.doi.org/10.1093/oxfordjournals...
; MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
).
Another reason for the occurrence of low statistical support for those branches is the possibility that genetic recombination might have occurred at remote times over the course of the evolution of these organisms. Genetic recombination is a plausible event in these species, due to the sexual reproduction of apicomplexan organisms.
The evolutionary history of Sarcocystidae have been extensively study using 18S rRNA and 28S rRNA (ELLIS et al., 1999Ellis JT, Morrison DA, Liddell S, Jenkins MC, Mohammed OB, Ryce C, et al. The genus is paraphyletic. HammondiaParasitology 1999; 118(4): 357-362. http://dx.doi.org/10.1017/S0031182098003801. PMid:10340325.
http://dx.doi.org/10.1017/S0031182098003...
; MUGRIDGE et al., 1999Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. Phylogenetic analyses based on full-length large subunit ribosomal RNA gene sequence comparison reveals that is more closely related to Neospora caninumHammondia heydorni than to Toxoplasma gondii.Int J Parasitol 1999; 29(10): 1545-1556. http://dx.doi.org/10.1016/S0020-7519(99)00150-2. PMid:10608441.
http://dx.doi.org/10.1016/S0020-7519(99)...
, 2000Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 2000; 17(12): 1842-1853. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285. PMid:11110900.
http://dx.doi.org/10.1093/oxfordjournals...
; MORRISON et al., 2004Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int J Parasitol 2004; 34(4): 501-514. http://dx.doi.org/10.1016/j.ijpara.2003.11.006. PMid:15013740.
http://dx.doi.org/10.1016/j.ijpara.2003....
). However, few nucleotide differences among species of theToxoplasma–Neospora–Hammondiaat these loci make difficult to resolve consistently their phylogenetic relationships. In fact, in order to be comparable to the rest of the coccidiaToxoplasma–Neospora–Hammondiawould be separate species of a single genus based only on 18S rRNA (MORRISON et al., 2004Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int J Parasitol 2004; 34(4): 501-514. http://dx.doi.org/10.1016/j.ijpara.2003.11.006. PMid:15013740.
http://dx.doi.org/10.1016/j.ijpara.2003....
). rRNA sequences cannot always be unambiguously aligned and details of the alignment are known to greatly affect the results of phylogenetic analyses involving these sequences (MUGRIDGE et al., 2000Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 2000; 17(12): 1842-1853. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285. PMid:11110900.
http://dx.doi.org/10.1093/oxfordjournals...
; MORRISON et al., 2004Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int J Parasitol 2004; 34(4): 501-514. http://dx.doi.org/10.1016/j.ijpara.2003.11.006. PMid:15013740.
http://dx.doi.org/10.1016/j.ijpara.2003....
). Thus, other universal markers are required to resolve phylogenies especially those that codes for proteins, which allow for robust alignments (MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
). Moreover, analysis of more than one gene is necessary in order to obtain a robust species signal (MUGRIDGE et al., 2000Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 2000; 17(12): 1842-1853. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285. PMid:11110900.
http://dx.doi.org/10.1093/oxfordjournals...
).
Regarding the diversity within the H. heydorni-related lineages, for these lineages to be hierarchized at species level, there needs to be certainty that they are unable to exchange genes. If two lineages do exchange genes, this event would occur during the gametogony in the intestinal epithelium of the definitive host and the infection of the host by different lineages would have to occur within a short period, because gametogony in Toxoplasmatinae does not take very long. Nevertheless, gene exchange can only be confirmed through analysis on individual oocysts, or after their clonal expansion in biological systems.
In conclusion, congruent phylogenies were inferred from the three different genes and they might be used for screening undescribed sarcocystid parasites in order to ascertain their phylogenetic relationships with organisms of the family Sarcocystidae. The evolutionary studies based on organelar genes confirm that the genus Hammondia is paraphyletic, as already stated elsewhere (ELLIS et al., 1999Ellis JT, Morrison DA, Liddell S, Jenkins MC, Mohammed OB, Ryce C, et al. The genus is paraphyletic. HammondiaParasitology 1999; 118(4): 357-362. http://dx.doi.org/10.1017/S0031182098003801. PMid:10340325.
http://dx.doi.org/10.1017/S0031182098003...
; MONTEIRO et al., 2007Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
http://dx.doi.org/10.1017/S0031182007002...
). The primers used for amplification ofclpC and rpoB were able to amplify genetic sequences of organisms of the genus Sarcocystis and organisms of the subfamily Toxoplasmatinae as well.
Ethics
The study was approved by the Ethics Committee for Use of Animals of the School of Veterinary Medicine and Animal Science of the University of Sao Paulo, Sao Paulo, Brazil, under protocol 1241/2007.
Conflict of interest statement
Acknowledgements
RMS is in receipt of grants from CNPq. SYOBV and MKS received scholarships from FAPESP (2011/10550-2 and 2007/56836-9, respectively).
References
- Abel J, Schares G, Orzeszko K, Gasser RB, Ellis JT. Hammondia isolated from dogs and foxes are genetically distinct. Parasitology 2006; 132(2): 187-192. http://dx.doi.org/10.1017/S0031182005008814. PMid:16188045.
» http://dx.doi.org/10.1017/S0031182005008814 - Arisue N, Hashimoto T. Phylogeny and evolution of apicoplasts and apicomplexan parasites. Parasitol Int 2015; 64(3): 254-259. http://dx.doi.org/10.1016/j.parint.2014.10.005. PMid:25451217.
» http://dx.doi.org/10.1016/j.parint.2014.10.005 - Carlson-Bremer D, Johnson CK, Miller RH, Gulland FM, Conrad PA, Wasmuth JD, et al. Identification of two novel coccidian species shed by California sea lions (Zalophus californianus). J Parasitol 2012; 98(2): 347-354. http://dx.doi.org/10.1645/GE-2752.1. PMid:22091999.
» http://dx.doi.org/10.1645/GE-2752.1 - Dubey JP, Liddell S, Mattson D, Speer CA, Howe DK, Jenkins MC. Characterization of the Oregon isolate of Neospora hughesi from a horse. J Parasitol 2001; 87(2): 345-353. http://dx.doi.org/10.1645/0022-3395(2001)087[0345:COTOIO]2.0.CO;2. PMid:11318565.
» http://dx.doi.org/10.1645/0022-3395(2001)087[0345:COTOIO]2.0.CO;2 - Dubey JP, Lindsay DS, Rosenthal BM, Thomas NJ. Sarcocysts of an unidentified species of Sarcocystis in the sea otter (). Enhydra lutrisJ Parasitol 2003a; 89(2): 397-399. http://dx.doi.org/10.1645/0022-3395(2003)089[0397:SOAUSO]2.0.CO;2. PMid:12760665.
» http://dx.doi.org/10.1645/0022-3395(2003)089[0397:SOAUSO]2.0.CO;2 - Dubey JP, Sreekumar C, Rosenthal BM, Lindsay DS, Grisard EC, Vitor RW. Biological and molecular characterization of n.sp. (Protozoa: Apicomplexa) from the rodent . Besnoitia akodoniAkodon montensis in BrazilParassitologia 2003b; 45(2): 61-70. PMid:15266998.
- Dubey JP. Toxoplasma, Neospora, Sarcocystis, and other tissue cyst-forming coccidia of humans and animals. In: Kreier JP. Parasitic Protozoa. 2nd ed. San Diego: Academic Press; 1993. p. 1-158, vol. 6.
- Ellis JT, Morrison DA, Liddell S, Jenkins MC, Mohammed OB, Ryce C, et al. The genus is paraphyletic. HammondiaParasitology 1999; 118(4): 357-362. http://dx.doi.org/10.1017/S0031182098003801. PMid:10340325.
» http://dx.doi.org/10.1017/S0031182098003801 - Feagin JE. The extrachromosomal DNAs of apicomplexan parasites. Annu Rev Microbiol 1994; 48(1): 81-104. http://dx.doi.org/10.1146/annurev.mi.48.100194.000501. PMid:7826027.
» http://dx.doi.org/10.1146/annurev.mi.48.100194.000501 - Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39(4): 783-791. http://dx.doi.org/10.2307/2408678.
» http://dx.doi.org/10.2307/2408678 - Ferguson DJ, Henriquez FL, Kirisits MJ, Muench SP, Prigge ST, Rice DW, et al. Maternal inheritance and stage-specific variation of the apicoplast in during development in the intermediate and definitive host. Toxoplasma gondiiEukaryot Cell 2005; 4(4): 814-826. http://dx.doi.org/10.1128/EC.4.4.814-826.2005. PMid:15821140.
» http://dx.doi.org/10.1128/EC.4.4.814-826.2005 - Gjerde B, Dahlgren SS. Hammondia triffittae n. comb. of foxes (Vulpes spp.): biological and molecular characteristics and differentiation from Hammondia heydorni of dogs. Parasitology 2011; 138(3): 303-321. http://dx.doi.org/10.1017/S0031182010001265. PMid:20854709.
» http://dx.doi.org/10.1017/S0031182010001265 - Gjerde B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of and (Apicomplexa: Sarcocystidae). Toxoplasma gondii, Neospora caninum, Hammondia heydorniHammondia triffittaeParasitol Res 2013a; 112(4): 1493-1511. http://dx.doi.org/10.1007/s00436-013-3296-4. PMid:23358734.
» http://dx.doi.org/10.1007/s00436-013-3296-4 - Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 2013b; 43(7): 579-591. http://dx.doi.org/10.1016/j.ijpara.2013.02.004. PMid:23542092.
» http://dx.doi.org/10.1016/j.ijpara.2013.02.004 - Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symposium Series 1999; 41: 95-98.
- He L, Zhang Y, Zhang QL, Zhang WJ, Feng HH, Khan MK, et al. Mitochondrial genome of , apicomplexan parasite of water buffalo (, Linnaeus, 1758) endemic in China. Babesia orientalisBubalus babalisParasit Vectors 2014; 7(82): 1-8. http://dx.doi.org/10.1186/1756-3305-7-82. PMid:24580772.
» http://dx.doi.org/10.1186/1756-3305-7-82 - Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJM, et al. A plastid of probable green algal origin in apicomplexan parasites. Science 1997; 275(5305): 1485-1489. http://dx.doi.org/10.1126/science.275.5305.1485. PMid:9045615.
» http://dx.doi.org/10.1126/science.275.5305.1485 - Lau AOT, McElwain TF, Brayton KA, Knowles DP, Roalson EH. Babesia bovis: a comprehensive phylogenetic analysis of plastid-encoded genes supports green algal origin of apicoplasts. Exp Parasitol 2009; 123(3): 236-243. http://dx.doi.org/10.1016/j.exppara.2009.07.007. PMid:19646439.
» http://dx.doi.org/10.1016/j.exppara.2009.07.007 - Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009; 25(11): 1451-1542. http://dx.doi.org/10.1093/bioinformatics/btp187. PMid:19346325.
» http://dx.doi.org/10.1093/bioinformatics/btp187 - Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, Xie WQ, et al. Characterization of the complete mitochondrial genomes of five species from domestic chickens. EimeriaGene 2011; 480(1-2): 28-33. http://dx.doi.org/10.1016/j.gene.2011.03.004. PMid:21402132.
» http://dx.doi.org/10.1016/j.gene.2011.03.004 - Lindsay DS, Mitchell SM, Vianna MC, Dubey JP. (protozoa: Apicomplexa) description of oocysts, sporocysts, sporozoites, excystation, and early development. Sarcocystis neuronaJ Parasitol 2004; 90(3): 461-465. http://dx.doi.org/10.1645/GE-230R. PMid:15272465.
» http://dx.doi.org/10.1645/GE-230R - Marsh AE, Barr BC, Packham AE, Conrad PA. Description of a new species (Protozoa: Apicomplexa: Sarcocystidae). NeosporaJ Parasitol 1998; 84(5): 983-991. http://dx.doi.org/10.2307/3284632. PMid:9794642.
» http://dx.doi.org/10.2307/3284632 - Marsh AE, Barr BC, Tell L, Koski M, Greiner E, Dame J, et al. In vitro cultivation and experimental inoculation of Sarcocystis falcatula and Sarcocystis neurona merozoites into budgerigars (). Melopsittacus undulatusJ Parasitol 1997; 83(6): 1189-1192. http://dx.doi.org/10.2307/3284386. PMid:9406803.
» http://dx.doi.org/10.2307/3284386 - Marsh AE, Howe DK, Wang G, Barr BC, Cannon N, Conrad PA. Differentiation of Neospora hughesi from based on their immunodominant surface antigen, SAG1 and SRS2. Neospora caninumInt J Parasitol 1999; 29(10): 1575-1582. http://dx.doi.org/10.1016/S0020-7519(99)00120-4. PMid:10608444.
» http://dx.doi.org/10.1016/S0020-7519(99)00120-4 - Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol 2008; 47(1): 261-273. http://dx.doi.org/10.1016/j.ympev.2007.11.012. PMid:18248741.
» http://dx.doi.org/10.1016/j.ympev.2007.11.012 - Monteiro RM, Richtzenhain LJ, Pena HF, Souza SLP, Funada MR, Gennari SM, et al. Molecular phylogenetic analysis in -like organisms based on partial Hsp70 coding sequences. HammondiaParasitology 2007; 134(9): 1195-1203. http://dx.doi.org/10.1017/S0031182007002612. PMid:17462122.
» http://dx.doi.org/10.1017/S0031182007002612 - Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int J Parasitol 2004; 34(4): 501-514. http://dx.doi.org/10.1016/j.ijpara.2003.11.006. PMid:15013740.
» http://dx.doi.org/10.1016/j.ijpara.2003.11.006 - Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. Phylogenetic analyses based on full-length large subunit ribosomal RNA gene sequence comparison reveals that is more closely related to Neospora caninumHammondia heydorni than to Toxoplasma gondii.Int J Parasitol 1999; 29(10): 1545-1556. http://dx.doi.org/10.1016/S0020-7519(99)00150-2. PMid:10608441.
» http://dx.doi.org/10.1016/S0020-7519(99)00150-2 - Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM. Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 2000; 17(12): 1842-1853. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285. PMid:11110900.
» http://dx.doi.org/10.1093/oxfordjournals.molbev.a026285 - Pusterla N, Mackie S, Packham A, Conrad PA. Serological investigation of transplacental infection with and in broodmares. Neospora hughesiSarcocystis neuronaVet J 2014; 202(3): 649-650. http://dx.doi.org/10.1016/j.tvjl.2014.09.015. PMid:25438732.
» http://dx.doi.org/10.1016/j.tvjl.2014.09.015 - Rathore D, Wahl AM, Sullivan M, McCutchan TF. A phylogenetic comparison of gene trees constructed from plastid, mitochondrial and genomic DNA of species. PlasmodiumMol Biochem Parasitol 2001; 114(1): 89-94. http://dx.doi.org/10.1016/S0166-6851(01)00241-9. PMid:11356517.
» http://dx.doi.org/10.1016/S0166-6851(01)00241-9 - Soares RM, Lopes EG, Keid LB, Sercundes MK, Martins J, Richtzenhain LJ. Identification of Hammondia heydorni oocysts by a heminested-PCR (hnPCR-AP10) based on the RAPD fragmente AP10. H. heydorniVet Parasitol 2011; 175(1-2): 168-172. http://dx.doi.org/10.1016/j.vetpar.2010.09.022. PMid:21030154.
» http://dx.doi.org/10.1016/j.vetpar.2010.09.022 - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30(12): 2725-2729. http://dx.doi.org/10.1093/molbev/mst197. PMid:24132122.
» http://dx.doi.org/10.1093/molbev/mst197 - Valadas SY, da Silva JI, Galucci E, Keid LB, Zwarg T, de Oliveira AS, et al. Diversity of Sarcocystis spp shed by opossums in Brazil inferred with phylogenetic analysis of DNA coding ITS1, Cytochrome B, and surface antigens. Exp Parasitol 2016; Feb 18. In press. http://dx.doi.org/10.1016/j.exppara.2016.02.008.
» http://dx.doi.org/10.1016/j.exppara.2016.02.008 - Vardeleon D, Marsh AE, Thorne JG, Loch W, Young R, Johnson PJ. Prevalence of Neospora hughesi and antibodies in horses from various geographical locations. Sarcocystis neuronaVet Parasitol 2001; 95(2-4): 273-282. http://dx.doi.org/10.1016/S0304-4017(00)00393-9. PMid:11223207.
» http://dx.doi.org/10.1016/S0304-4017(00)00393-9 - Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, Conrad PA, et al. Limited genetic diversity among strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Sarcocystis neuronaVet Parasitol 2010; 169(1-2): 37-44. http://dx.doi.org/10.1016/j.vetpar.2009.12.020. PMid:20071081.
» http://dx.doi.org/10.1016/j.vetpar.2009.12.020 - Wobeser BK, Godson DL, Rejmanek D, Dowling P. Equine protozoal myeloencephalitis caused by in an adult horse in Saskatchewan. Neospora hughesiCan Vet J 2009; 50(8): 851-853. PMid:19881924.
Publication Dates
-
Publication in this collection
18 Mar 2016 -
Date of issue
Jan-Mar 2016
History
-
Received
24 Nov 2015 -
Accepted
16 Feb 2016