Abstract
The swamp rats are distributed in Argentina, southern Paraguay, Uruguay and southern Brazil, with two species currently accepted: Scapteromys aquaticus Thomas, 1920 and Scapteromys tumidus Waterhouse, 1837. While S. aquaticus occurs in Argentina, Paraguay and western Uruguay, S. tumidus occurs in Brazil and Uruguay. Here we report for the first time the occurrence of S. aquaticus in gallery forest remnants in Southern Brazil. Karyologic analysis showed 2n = 32 and FNa = 40. Phylogenetic analyses, based on DNA sequences from the mitochondrial cytochrome-b gene indicate that the Brazilian and the Argentinian specimens of S. aquaticus shared one haplotype, while median joining analysis showed lack of population structure. This register, plus the karyotype data available for Brazilian population, recovered four karyomorphotypes in Brazil, corresponding to the two known species of Scapteromys and two unnamed species. This scenario indicates that more multidisciplinary studies are necessary to understand the actual diversity of Scapteromys.
New record; southern Brazil; species limits; swamp rat
SHORT COMMUNICATION
Scapteromys aquaticus (Rodentia: Sigmodontinae) in Brazil with comments on karyotype and phylogenetics relationships
Cibele R. BonvicinoI, II; Fabiano A. FernandesIII, * * Corresponding author. E-mail: fabianoaf66@yahoo.com ; Maria C. VianaI; Bernardo R. TeixeiraII; Paulo S. D'AndreaII
IDivisão de Genética, INCA. Rua André Cavalcanti 37, 20230-130 Rio de Janeiro, RJ, Brazil
IILaboratório de Biologia e Parasitologia de Mamíferos Reservatórios Silvestres, Instituto Oswaldo Cruz. Avenida Brasil 4365, 21045-900, Rio de Janeiro, RJ, Brazil
IIILaboratório de Ecoepidemiologia da Doença de Chagas, Instituto Oswaldo Cruz. Avenida Brasil 4365, 21045-900 Rio de Janeiro, RJ, Brazil
ABSTRACT
The swamp rats are distributed in Argentina, southern Paraguay, Uruguay and southern Brazil, with two species currently accepted: Scapteromys aquaticus Thomas, 1920 and Scapteromys tumidus Waterhouse, 1837. While S. aquaticus occurs in Argentina, Paraguay and western Uruguay, S. tumidus occurs in Brazil and Uruguay. Here we report for the first time the occurrence of S. aquaticus in gallery forest remnants in Southern Brazil. Karyologic analysis showed 2n = 32 and FNa = 40. Phylogenetic analyses, based on DNA sequences from the mitochondrial cytochrome-b gene indicate that the Brazilian and the Argentinian specimens of S. aquaticus shared one haplotype, while median joining analysis showed lack of population structure. This register, plus the karyotype data available for Brazilian population, recovered four karyomorphotypes in Brazil, corresponding to the two known species of Scapteromys and two unnamed species. This scenario indicates that more multidisciplinary studies are necessary to understand the actual diversity of Scapteromys.
Key words: New record; southern Brazil; species limits; swamp rat.
Species of swamp rats, Scapteromys Waterhouse, 1837 (Akodontini), are distributed in Argentina, Southern Paraguay, Uruguay and Southern Brazil (D'ELÍA & PARDIÑAS 2004) with two species: Scapteromys aquaticus Thomas, 1920 and Scapteromys tumidus Waterhouse, 1837 (MUSSER & CARLETON 2005).
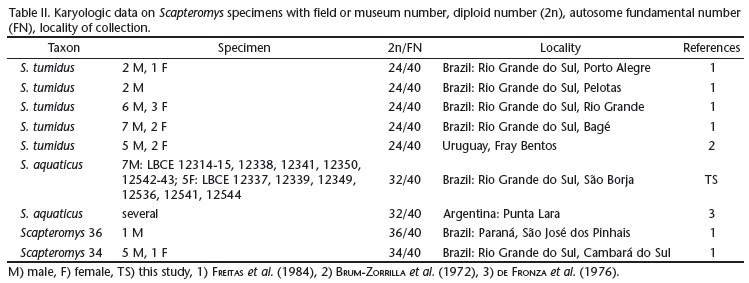
Scapteromys spp. occur in semi-aquatic habitats and areas near watercourses and swamps. There are no specific data about S. aquaticus habits but S. tumidus feeds on invertebrates (SUAREZ & BONAVENTURA 2001) and is associated with flooded microhabitats with low plant coverage (BONAVENTURA et al. 2003). Karyologic studies of this genus showed a wide variation, with four different karyotypes (FREITAS et al. 1984, D'ELÍA & PARDIÑAS 2004). Scapteromys aquaticus populations are characterized by a diploid number (2n) of 32, occurring in Argentina (BRUM-ZORRILLA et al. 1986, FRONZA et al. 1976) and southern Paraguay (D'ELÍA & PARDIÑAS 2004), while the S. tumidus population in Uruguay (BRUM-ZORRILLA et al. 1972, 1986) and southern Brazil (FREITAS et al. 1984) showed 2n = 24. Two other karyotypes have been reported in the Brazilian population, 2n = 34 in the state of Rio Grande do Sul and 2n = 36 in the state of Paraná (FREITAS et al. 1984, PEDÓ et al. 2010).
D'ELÍA & PARDIÑAS (2004) presented a systematic study of populations of Scapteromys, based on molecular and morphological evidence, and suggested two main clades: one named S. aquaticus, formed by Argentinean and Paraguayan populations together with one population from western Uruguay, and the other, S. tumidus, constituted by the remaining Uruguayan populations. Unfortunately, populations from Brazil were not analyzed.
Here, we report for the first time the occurrence of S. aquaticus with 2n = 32, extending the geographic distribution to Southern Brazil.
A zoologic and epidemiologic survey was conducted in the municipality of São Borja, Rio Grande do Sul in May 2009 (Fig. 1). Its climate is subtropical humid, with a mean annual temperature of 20°C, with minimum and maximum temperatures in December-January and July-August, respectively. São Borja is limited by the Uruguay river on the west. Altitude of study areas varied from 59-86 m above sea level. Small mammals were captured with live traps (Tomahawk® and Sherman® models) with baits containing a mixture of peanut butter, banana, oat and bacon. During this study, 14 S. aquaticus specimens were collected (7 females LBCE 12337, 12339, 12536, 12541, 12544-45, 12349, and 7 males LBCE 12314-15, 12338, 12341, 12350, 12542-43) and karyotyped for confirming identification. Specimens occurred in gallery forest remnants near Uruguay river (28°37'08"S, 56°01'12"W) and in shrubby vegetation around swamps (28°38'39"S, 55°59'54"W and 28°38'34"S, 55°59'57"W).
Chromosome preparations were obtained from short-term bone marrow cultures with 80% RPMI 1640, 20% fetal calf serum, ethidium bromide (5 mg/ml) and colchicine (10-6) for two hours at 37°C, following by hypotonic shock with KCl (0.075M) for 30 minutes, pre fixation and fixation with Carnoy (3 methyl alcohol:1 acetic acid). Estimates of fundamental number were restricted to autosome pairs.
Voucher specimens were deposited in the mammal collection of Laboratório de Biologia e Parasitologia de Mamíferos Reservatórios Silvestres (LBCE), Instituto Oswaldo Cruz, FIOCRUZ, Rio de Janeiro, Brazil (Table I). Fieldwork was carried out with authorization (License number 13373, to P.S. D'Andrea) from Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio).
DNA was isolated from livers of twelve specimens preserved in 100% ethanol with the phenol-cloroformio protocol (SAMBROOK et al. 1989). Cytochrome b DNA (cytb; ca. 801 bp) was PCR amplified with primers L14724 (IRWIN et al. 1991) and MVZ16 (SILVA & PATTON 1993), with a pre-denaturation step at 94°C for 3 min; 33 cycles of denaturation at 94°C for 20 sec, annealing at 48°C for 15 sec, extension at 72°C for 60 sec, and final extension of 72°C for 2 min. Amplicons were purified with GFXä PCR DNA and Gel Band Purification kit (GE Healthcare, Brazil) and sequenced with the same PCR primers and labeled with XL and BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing was carried out with an ABI PrismTM 3130 platform.
Phylogenetic analyses included sequence data from 28 Scapteromys deposited in GenBank: 25 S. aquaticus (AY445527 - AY445551), three S. tumidus (AY445561, AY445563, AY445565), and two other used as outgroups, one Kunsia tomentosus (Lichtenstein, 1830) (AY445526) and one Blarinomys breviceps (Winge, 1887) (AY275112).
Genetic distance estimates were carried out with complete deletion using Kimura's two parameters, with MEGA (version 5; TAMURA et al. 2011). Estimates of maximum-likelihood (ML) analysis were carried out with PAUP 4.0b10 (SWOFFORD 2003), and bootstrap analysis were based on 1,000 replicates. Bayesian analyses were carried out with Mr.Bayes (RONQUIST & HUELSENBECK 2003). NETWORK (version 4.5.1.6; available at http://www.fluxus-engineering.com) was used for reconstructing a median-joining (MJ) network (BANDELT et al. 1999) to evaluate sub-population structure and geographic distribution patterns. MJ was calculated using variable sites only.
Intraspecific analyses were carried out with DNAsp 5 (LIBRADO & ROZAS 2009) and Arlequin 3.11 (EXCOFFIER & SCHNEIDER 2005) for computing global and pairwise estimates of gene flow and for estimating haplotype and nucleotide diversity. Inferences of past events like population expansion or decline were based on estimates of neutrality tests: Tajima's D (TAJIMA 1989), Fu and Li's F and D (FU 1997). Mismatch distribution analyses was based on an assumed stepwise growth model (ROGERS & HARPENDING 1992) with DNAsp 5 (LIBRADO & ROZAS 2009).
Karyologic analysis of S. aquaticus collected in São Borja showed 2n = 32, FNa = 40 (Table II). The autosome complement is composed by five biarmed pairs, three large sized and two small sized and ten acrocentric pairs decreasing in size from median to small (Fig. 2). The X chromosome is a median sized acrocentric and the Y chromosome is a small sized biarmed chromosome.
Cytochrome b DNA, comprising 801 bp from nine specimens, showed four haplotypes, two of them shared by more than one specimen (H1 by three specimens, H4 by four specimens; Table I). These data, together with 25 S. aquaticus retrieved from GenBank, accounted for 15 haplotypes (Table I). Only one haplotype (H4), in 4 Brazilian specimens (LBCE12339, 12350, 12542, 12544), was shared by one specimen from Argentina (AY445536).
Analysis of all S. aquaticus sequences showed 21 variable sites (18 transitions and three transversions), with genetic distance estimates ranging from 0.0 to 2.7%. Genetic distance estimates between S. aquaticus haplotypes from São Borja varied from 0.0 to 0.5%, while genetic distance estimates between S. aquaticus from São Borja and other localities varied from 0.0 to 1.1%. Genetic distances estimates between Paraguayan, Uruguayan and Argentinean specimens were <0.6%.
Phylogenetic analyses confirmed the monophyly of Scapteromys (Fig. 3), grouping haplotypes from Paraguay, Argentina, Uruguay and Brazil lacking a geographic structure.
MJ network (Fig. 4) showed a star-like topology, indicating lack of geographic structure and showing the most frequent haplotype from Brazil in a central position. The majority of Argentinean and Paraguayan haplotypes (H4) were directly connected with this haplotype, while the haplotype from Uruguay was connected with one haplotype from Brazil (H3) connected to the central haplotype.
Pairwise genetic distance estimates (Fst) ranged from 0.17 (between São Borja and Argentinean haplotypes) to 0.68 (between Uruguayan and Paraguayan haplotypes). Other estimates accounted for 0.27 between São Borja and Paraguayan haplotypes, 0.57 between São Borja and Uruguayan haplotypes, 0.20 between Argentinean and Paraguayan, and 0.46 between Uruguayan and Argentinean haplotypes.
Tajima's D neutrality test and the more sensitive Fu and Li's tests resulted in negative and significant values (Tajima's D = -1.22, p < 0.05; Fu and Li's D = - 0.85, p < 0.05; Fu and Li's F = - 1.14, p < 0.05). Additional evidence of a recent history of population expansion from São Borja in Southern Brazil was provided by analysis of mismatch distribution showing a typical unimodal distribution under the exponential population expansion model, when considering only São Borja haplotypes or all haplotypes (data not show).
In the latest taxonomic review of the genus Scapteromys, the distribution of S. aquaticus included Argentina, Paraguay and the Uruguayan area of Las Cañas in Department of Río Negro (D'ELIA & PARDIÑAS 2004). The Scapteromys specimens from Brazil herein karyotyped showed 2n = 32, FNa = 40, similarly to S. aquaticus from Argentina and Paraguay (BRUM-ZORRILLA et al. 1986, FRONZA et al. 1976) and confirmed that the distribution of this species extends to the south of Rio Grande do Sul in Brazil. The only detail is the X chromosome of Brazilian specimens that was a medium size acrocentric while, in Punta Lara (Argentina), the X chromosome was found to be acrocentric and submetacentric in one female specimen, and acrocentric in other females (FRONZA et al. 1976).
Three other Scapteromys karyotypes were reported in Brazil: 2n = 24 for S. tumidus and two other karyomorphotypes, 2n = 34 and 36 in specimens without species identification (FREITAS et al. 1984). The S. aquaticus herein karyotyped were collected in the most southern region of Brazil, while the S. tumidus and Scapteromys sp. karyotyped by FREITAS et al. (1984) were collected in northern and eastern Rio Grande do Sul, Santa Catarina and Paraná states. These findings were in agreement with the proposition of FREITAS et al. (1984) that more than one species of Scapteromys occurs in Brazil, with S. aquaticus being restricted to the most southern region.
A previous review of the tribe Akodontini showed the monophyly of S. aquaticus (D'ELIA & PARDIÑAS 2004). Our findings showed that Brazilian S. aquaticus grouped with specimens from Argentina, Paraguay and Uruguay but without a geographic structure. The Brazilian (n = 4) and the Argentinean (n = 1) specimens of S. aquaticus shared a single haplotype, while haplotypes from Uruguay and Paraguay were exclusive of these populations (Fig. 4).
The star-like topology of the MJ network (Fig. 4), and the Brazilian haplotype (H4) most frequent and probably the most ancestral one (CRANDALL & TEMPLETON 1993), suggested low differentiation between localities, with the most frequent haplotypes in at least two populations (Brazilian and Argentinean) and unique haplotypes connected by one or few nucleotide changes, presumably derived from H4.
Geographic barriers, like the Uruguay river, between populations from Argentina and Brazil, usually could be associated with strong levels of genetic differentiation, but the effect of this river as main promoter of genetic divergence was not observed for S. aquaticus. The same pattern was reported between populations from Argentina and Uruguay (Fig. 1; Las Cañas - D'ELIA & PARDIÑAS 2004). This scenario demonstrated a low level of diversity in these populations suggesting a recent history of population expansion without influence of the river.
This new record plus the karyotype data available for Brazilian Scapteromys population showed four karyomorphotypes in Brazil, corresponding to the two known Scapteromys species and two unnamed species. This data indicate that the actual diversity of Scapteromys genus is underestimated and more multidisciplinary studies are necessary for understand this scenario.
ACKNOWLEDGMENTS
We are grateful to H.N. Seuánez (INCA, Brazil) for reviewing the final version of the manuscript. Clerical and technical help came from L. Monnerat and K.R. Lobo. The collaboration in fieldwork by the field team of the Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios and Laboratório de Biologia de Tripanossomatídeos (A.M. Jansen, A.L.R. Roque, and F.L. Rocha), IOC/FIOCRUZ, was most useful. Work supported by CNPq grant to CRB (302951/2007-5).
LITERATURE CITED
Submitted: 27.I.2012; Accepted: 15.X.2012.
Editorial responsibility: Walter A.P. Boeger
- BANDELT, H.J.; P. FORSTER & A. ROHL. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37-48.
- BONAVENTURA, S.M.; V. PANCOTTO; N. MADANES & V. VICARI. 2003. Microhabitat use and density of sigmodontine rodents in Spartina densiflora freshwater marshes, Argentina. Mammalia 67 (3): 367-378.
- BRUM-ZORRILLA, N.; N. LAFUENTE & P. KIBLISKY. 1972. Cytogenetic studies in the cricetid rodent Scapteromys tumidus (Rodentia-Cricetidae). Specialia 28: 1373.
- BRUM-ZORRILLA, N.; G. OLIVER; T.G. FRONZA & R. WAINBERG. 1986. Karyological studies of South American rodents (Rodentia: Cricetidae). I. Comparative chromosomic analysis in Scapteromys taxa. Caryologia 39: 131-142.
- CRANDALL, K.A. & A.R. TEMPLETON. 1993. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134 (3): 959-969.
- D' ELÍA, G. & U.F.J. PARDIÑAS. 2004. Systematics of Argentinean, Paraguayan, and Uruguayan Swamp Rats of the genus Scapteromys (Rodentia, Cricetidae, Sigmodontinae). Journal of Mammalagy 85: 897-910.
- EXCOFFIER, L.L.G. & S. SCHNEIDER. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47-50.
- FREITAS, T.R.O.; M.S. MATTEVI & L.F. OLIVEIRA. 1984. Unusual C-band patterns in three karyotypically rearranged forms of Scapteromys (Rodentia, Cricetidae) from Brazil. Cytogenetics and Cell Genetics 38: 39-44.
- FRONZA, T.G.; R.L. WAINBERG & B.E. LLORENTE. 1976. Polimorfismo del cromosoma X y significacion filogenética del cariotipo de la "Rata aquatica" Scapteromys aquaticus (Rodentia, Cricetidae) de la ribera de Punta Lara (Argentina). Mendeliana 1: 41-48.
- FU, Y. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915-925.
- HALL, T.A. 2007. Bioedit Ibis Biosciences, Carlsbad, CA.
- IRWIN, D.M.; T.D. KOCHER & A.C. WILSON. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution 32: 128-144.
- LIBRADO, P. & J. ROZAS. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451-1452.
- MUSSER, G.M. & M.D. CARLETON. 2005. Superfamily Muroidea, p. 894-1531. In: D.E. WILSON & D.M. REEDER (Eds). Mammal Species of the World. A taxonomic and geographic reference. Baltimore, The Johns Hopkins University Press, 3rd ed.
- PEDÓ, E.; T.R.O. FREITAS & S.M. HARTZ. 2010. The influence of fire and livestock grazing on the assemblage of non-flying small mammals in grassland-Araucaria Forest ecotones, southern Brazil. Zoologia 27 (4): 533-540.
- ROGERS, A.R. & H. HARPENDING. 1992. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Molecular Biology and Evolution 9 (3): 552-569.
- RONQUIST, F. & J.P. HUELSENBECK. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574.
- SAMBROOK, J.; E.F. FRITSCH & T. MANIATIS. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor, Cold Spring Harbor Laboratory Press.
- SILVA, M.N.F. & J.L. PATTON. 1993. Amazonian phylogeography: mtDNA sequences variation in arboreal echimyid rodents (Caviomorpha). Molecular and Phylogenetics Evolution 2: 243-255.
- SUÁREZ, O.V. & S.M. BONAVENTURA. 2001. Habitat use and diet in sympatric species of rodents of the low Paraná delta, Argentina. Mammalia 65 (2): 167-176.
- SWOFFORD, D.L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Others Methods). Sunderland, Sinauer Associates, version 4.0b.
- TAJIMA, F. 1989. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123: 585-595.
- TAMURA, K.; D. PETERSON; N. PETERSON; G. STECHER; M. NEI & S. KUMAR. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731-2739.
Publication Dates
-
Publication in this collection
09 May 2013 -
Date of issue
Apr 2013
History
-
Received
27 Jan 2012 -
Accepted
15 Oct 2012