Abstract
Cerradomys is a Neotropical genus of cricetid rodents with seven recognized species, Cerradomys subflavus, C. maracajuensis, C. marinhus, C. scotti, C. langguthi, C. vivoi, and C. goytaca. Species of the genus are distributed throughout the open vegetation belt across South America, from northeastern and southeastern Atlantic coast of Brazil to eastern Paraguay and Western Bolivia. Here we describe a new species of Cerradomys from the state of Tocantins in Central Brazil, based on morphological, karyological and mitochondrial DNA analyses. This species is characterized by a medium body size and long tail, dense dorsal pelage, overall dorsal color gray olive lined with yellow, color of head and dorsum continuous, ventral body color slightly yellowish, skull with deep rostral depression, mesopterygoid fossa with long and wide sphenopalatine vacuities, presence of alisphenoid strut and of complex posterolateral palatal pits, and a unique chromosomal formula (2n = 60 and FNa = 74). Phylogenetic analyses based on cytochrome b sequences, including for the first time all known Cerradomys species, indicate that the new species is more closely related to C. scotti. The new species is found in sympatry with C. marinhus, while C. marinhus, C. scotti, and C. subflavus are found in sympatry (but not in syntopy) in one locality in the state of Minas Gerais. Finally, analysis of cytochrome b sequences indicates that C. subflavus and C. goytaca are very closely related genetically and might be conspecific. Alternatively, these results can also be explained by incomplete lineage sorting due to a recent speciation event.
Biodiversity; Cerrado; karyotype; phylogeny; Sigmodontinae
SYSTEMATICS AND EVOLUTION
A new species of Cerradomys (Mammalia: Rodentia: Cricetidae) from Central Brazil, with remarks on the taxonomy of the genus
Cibele R. BonvicinoI,II; Flávia CasadoII; Marcelo WekslerIII,IV
IDivisão de Genética, Instituto Nacional de Câncer. Rua André Cavalcanti 37, 4 andar, Bairro de Fátima, Rio de Janeiro, Brazil
IILaboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios, Pavilhão Lauro Travassos, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz. Avenida Brasil 4365, Manguinhos, 21045-900 Rio de Janeiro, RJ, Brazil
IIIDepartamento de Zoologia, Universidade Federal do Estado do Rio de Janeiro. Avenida Pasteur 458, 20290-240 Rio de Janeiro, RJ, Brazil
IVCorresponding author. E-mail: marcelo.weksler@gmail.com
ABSTRACT
Cerradomys is a Neotropical genus of cricetid rodents with seven recognized species, Cerradomys subflavus, C. maracajuensis, C. marinhus, C. scotti, C. langguthi, C. vivoi, and C. goytaca. Species of the genus are distributed throughout the open vegetation belt across South America, from northeastern and southeastern Atlantic coast of Brazil to eastern Paraguay and Western Bolivia. Here we describe a new species of Cerradomys from the state of Tocantins in Central Brazil, based on morphological, karyological and mitochondrial DNA analyses. This species is characterized by a medium body size and long tail, dense dorsal pelage, overall dorsal color gray olive lined with yellow, color of head and dorsum continuous, ventral body color slightly yellowish, skull with deep rostral depression, mesopterygoid fossa with long and wide sphenopalatine vacuities, presence of alisphenoid strut and of complex posterolateral palatal pits, and a unique chromosomal formula (2n = 60 and FNa = 74). Phylogenetic analyses based on cytochrome b sequences, including for the first time all known Cerradomys species, indicate that the new species is more closely related to C. scotti. The new species is found in sympatry with C. marinhus, while C. marinhus, C. scotti, and C. subflavus are found in sympatry (but not in syntopy) in one locality in the state of Minas Gerais. Finally, analysis of cytochrome b sequences indicates that C. subflavus and C. goytaca are very closely related genetically and might be conspecific. Alternatively, these results can also be explained by incomplete lineage sorting due to a recent speciation event.
Keywords: Biodiversity; Cerrado; karyotype; phylogeny; Sigmodontinae.
The Brazilian Cerrado harbors one of the most impressive mammalian faunas of South America (Carmignotto et al. 2012, Bonvicino et al. 2012) and is considered a biodiversity hotspot (Myers et al. 2000). Besides monospecific and endemic taxa, such as Microakodontomys transitorius Hershkovitz, 1993 and the recently described Calassomys apicalis Pardiñas, Lessa, Teta, Salazar-Bravo &Câmara, 2014, some rodent genera, such as Oligoryzomys Bangs, 1900, Calomys Waterhouse, 1837, Cerradomys Weksler, Percequillo & Voss, 2006, and Thrichomys Trouessart, 1880, are relatively widely distributed and speciose in the Cerrado. These latter taxa encompass morphologically similar species that nonetheless exhibit clear karyologic and mitochondrial DNA differences (Almeida et al. 2007, Percequillo et al. 2008, Agrellos et al. 2012, Nascimento et al. 2013). In general, these species have particular distributional and ecological patterns, occurring in specific vegetation habitats within the Cerrado (e.g., Bezerra et al. 2009, Rocha et al. 2011). Given the mosaic nature of the occurrence of vegetation types in the Cerrado, the specificity of habitat use enables the sympatry of species of these genera. This sympatry is usually observed between more morphologically differentiated species, while cryptic species are allopatric (Weksler & Bonvicino 2005, Almeida et al. 2007, Percequillo et al. 2008).
Cerradomys is probably one of the most emblematic genus of this pattern. Formerly the ‘Oryzomys subflavus' group, Cerradomys species are distributed throughout an open vegetation belt, also known as the dry diagonal corridor of South America (Bonvicino 2003), and in the Brazilian Atlantic Forest (Percequillo et al. 2008), but its central distributional range coincides with the range of the Cerrado (Fig. 1). Cerradomys taxa were considered as a single species, Oryzomys subflavus (Wagner, 1842), until the end of the 20th century (e.g., Musser & Carleton 1993), but extensive morphologic (Langguth & Bonvicino 2002, Percequillo et al. 2008, Tavares et al. 2011), karyologic (Maia & Hulak 1981, Almeida & Yonenaga-Yassuda 1985, Svartman & Almeida 1992, Bonvicino et al. 1999, Andrades-Miranda et al. 2002), and molecular (Bonvicino & Moreira 2001, Percequillo et al. 2008) work revealed that seven species could be recognized for the genus: Cerradomys maracajuensis (Langguth & Bonvicino, 2002), C. marinhus (Bonvicino, 2003), C. scotti (Langguth & Bonvicino, 2002) [including Cerradomys andersoni (Brooks, Baker, Vargas, Tarifa, Aranibar & Rojas, 2004)], C. subflavus, C. langguthi Percequillo, Hingst-Zaher & Bonvicino, 2008, C. vivoi Percequillo, Hingst & Bonvicino, 2008, and C. goytaca Tavares, Pessoa & Gonçalves, 2011. Some of these species are sympatric, such as C. scotti and C. maracajuensis, and C. scotti and C. marinhus (Fig. 1).
The taxonomy of the members of this genus, however, is not yet fully explored and here we describe a new species from central Brazil, based on morphological, morphometric, and karyological evidence. Moreover, we also present a phylogenetic analysis for all Cerradomys species based on cytochrome b DNA sequence data, and discuss some taxonomic issues.
MATERIAL AND METHODS
Origin of samples and karyotypic analysis
We collected Cerradomys specimens (Appendix 1) in nine Brazilian localities in three morphoclimatic domains (sensu Ab'Saber 2003) and in the ecotone between Cerrado and Amazonia: a) Cerrado domain: Tocantins state (1) Novo Jardim (coordinates: 11°49'S and 46°38'W/collecting dates: August and November 2008, February, May-June and October 2009, February 2010); Goiás state (2) Aporé (18°58'S and 51°55'W/April and July 2003, September 2004, July-September 2005, May and July 2006, April-May 2007, July 2008, April-May 2009), (3) Luziania (16°15'S and 47°57'W/July 2006), (4) Campo Alegre de Goiás (17°38'S and 47°47'W/October 2005); Minas Gerais state (5) Uberlândia (18°55'S, 48°17'W/December 2011, March and August 2012); and Mato Grosso state (6) Campo Verde (15°33'S and 55°10'W/June 2010), (7) Campo Novo do Parecis (13°40'S and 57°53'W/July 2005). b) Caatinga Domain: Paraíba state, (8) Sousa, around rice plantation, in the gallery forest of Rio dos Peixes (06°45'S and 38°14'W/September 2011). c) Atlantic Forest Domain: Rio de Janeiro state, (9) Quissamã, in the restinga Jurubatiba, a conserved restinga vegetation (22°06'S and 41°28'W/January 2012). d) Transition between Cerrado and Amazonian domains: Mato Grosso state, (10) São José do Xingu, in semideciduous forest (10°48'S and 52°45'W/July 2005).
Chromosome preparations were obtained from short-term cell cultures following de Andrade et al. (2004). Chromosomes were ordered according to morphology and decreasing size. For new karyotypes, several metaphases were captured and analyzed in the microscope, and five metaphases were mounted for each karyotyped specimen. Here only metacentric and submetacentric chromosomes were considered as biarmed for computation autosome fundamental number.
Phylogenetic analysis
DNA was isolated from tissue samples preserved in 100% ethanol following the protocol of Sambrook et al. (1989). Parcial cytochrome b gene (ca. 801 bp) was amplified by PCR using primers L14724 (5'-CGAAGCTTGATATGAAAAACCATCGTTG-3' (Irwin et al. 1991) and MVZ16 (Smith & Patton 1983). Amplicons were purified using the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Brazil), and sequenced using the PCR primers. Sequencing was carried out with an ABI PrismTM 3730 automatic DNA sequencer.
We sequenced 34 specimens of Cerradomys: 11 of C. akroai sp. nov., 13 of C. scotti, three of C. maracajuensis, four of C. subflavus, one of C. langguthi, and two of C. goytaca (see appendix 1 for list of specimens). We also included GenBank data from two C. vivoi specimens (GenBank accession number AF181275 – museum number MN35898 and MN61666), one C. langguthi (AF181276-MN69786), two C. scotti (AF181277-MN50379, EU579482-TK61881), one C. maracajuensis (AF181278-MN44178), one C. marinhus (AF181279-MN63824) and one C. subflavus (AF181274-LV-CEG42). Hylaeamys megacephalus (Fischer, 1814) (LBCE18571; sequenced here), Euryoryzomys russatus (Wagner, 1848) (EU579486-LV-ORG67), Rhipidomys sp. (LBCE18572), and Necromys lasiurus (Lund, 1841) (LBCE8684) were used as outgroups. New sequences were deposited in GenBank with accession numbers KP122210-KP122252.
Kimura two-parameter distances were estimated between haplotypes using PAUP* 4.0b10 (Swofford 2001). For phylogenetic reconstructions, a DNA substitution model was selected using the software Modelgenerator, version 0.85 (Keane et al. 2006) and the Bayesian information criterion (BIC). The GTR model of nucleotide substitution (Rodríguez et al. 1990), corrected for site-specific rate heterogeneity using gamma distribution with four classes (Yang 1994) was used in all model-based analyses. Cladistic parsimony (MP) analysis was performed using the heuristic search algorithm implemented by PAUP*, with 1000 replicates of random taxon addition and TBR branch swapping; nodal bootstrap values (Felsenstein 1985) were calculated using 1000 pseudoreplicates. The Maximum-Likelihood (ML) trees were calculated using RaxML (Stamatakis 2006.). Bootstrap values for the likelihood analysis were calculated using 1000 pseudoreplicates. Bayesian analyses (BI) were performed using Markov chain Monte Carlo (MCMC) sampling as implemented in MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Uniform interval priors were assumed for all parameters except base composition, for which we assumed a Dirichlet prior. We performed four independent runs of 10,000,000 generations each, with two heated chains sampling for trees and parameters every 10,000 generations. The first 2,500,000 generations were discarded as burn-in, and the remaining trees were used to estimate posterior probabilities for each node. All analyses were checked for convergence by plotting the log-likelihood values against generation time for each run, using Tracer 1.6 (Rambaut & Drummond 2007), and all estimates have effective sample sizes over 200.
Morphologic analysis
We studied skins, skulls and skeletons deposited in the mammal collection of Museu Nacional (MN), Universidade Federal do Rio de Janeiro, and in the mammal collection of the Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios (LBCE), IOC, Fiocruz, Rio de Janeiro, Brazil. A list of specimens examined and collecting localities is provided in Appendix 1. All measurements are expressed in millimeters (mm), except weight that is expressed in grams (g). The following external measurements were obtained from specimen tags or from wild caught specimens during fieldwork: total length (TL) or head-and-body length (HBL), length of the tail (LT), pinnae length (Ear), length of hind foot (HF), and weight (W). When necessary, head-and-body length (HBL) was obtained by subtracting length of tail from total length.
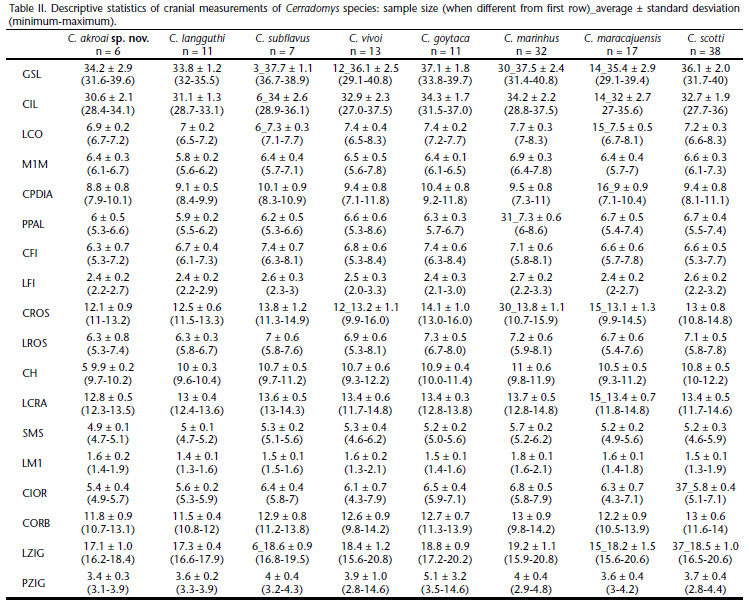
Eighteen cranial measurements (Bonvicino & Weksler 1998) were obtained with digital calipers to the nearest 0.01 mm: (GSL) greatest skull length; (CIL) condylo-incisive length, measured from the greater curvature of one upper incisor to the articular surface of the occipital condyle on the same side; (BOC) breadth of occipital condyles, measured from external border of the condyles; (BPB) breadth of palatal bridge, measured at the labial margin of maxillary bone across the third molars; (LD) length of diastema, from the crown of the first upper molar to the lesser curvature of the upper incisor on the same side; (LPB) length of palatal bridge, measured from the posterior border of the incisive foramen to the anterior border of the mesopterygoid fossa; (LIF) length of incisive foramina, greatest anterior-posterior dimension of one incisive foramina; (BIF) breadth of incisive foramina; (LR) length of rostrum, greatest dimension measured from the anterior border of the nasal bone to orbital fossa; (BR) breadth of rostrum, greatest dimension measured across the external border of the nasolacrimal capsules; (CH) cranial height, measured from dorsal surface of frontal to ventral surface of palatal bones, behind the third molar; (BB) breadth of braincase, measured across the smooth lateral surface of the braincase posterodorsal to the squamosal zygomatic processes; (LM) length of superior molars, crown length from M1 to M3; (BM1) breadth of M1, greatest crown breadth of the first maxillary molar across the paracone-protocone; (LIB) least interorbital breadth, least distance across the frontal bones; (LOF) length of orbital fossa, the greatest diameter of orbital fossa; (ZB) zygomatic breadth, greatest dimension across the squamosal root of zygomatic arches; (BZP) breadth of zygomatic plate.
Morphometric analyses of skull characters were performed for adult specimens (i.e., specimens with all teeth erupted and with at least minimal wear; Oliveira et al. 1998); males and females were grouped due to lack of sexual dimorphism in adults (Brandt & Pessoa 1994). Discriminant Analysis with estimation of canonical functions (Strauss 2010), using logarithmic-transformed data, were carried out for identifying patterns of morphometric variation within the genus, while univariate Analyses of Variance (Sokal & Rohlf 1995) were employed for comparing the new species with the sister taxon C. scotti (see phylogenetic results). Greatest skull length (GSL) was excluded from discriminant analysis due to high frequency of missing values and high correlation with condylo-incisive length (r = 0.96). Sequential Bonferroni correction (Rice 1989) was used to adjust p-values for multiple contrasts in the ANOVA. Statistical analyses were performed with STATISTICA 7.0 (StatSoft 2004).
RESULTS
Karyologic and molecular data
Karyotypic analysis of five C. akroai sp. nov. specimens showed 2n = 60 and FNa = 74 (Figs 2-11). The chromosome complement is composed by 11 pairs of biarmed chromosomes, one large sized, two median sized and 8 small sized, and 18 acrocentric pairs varying in size from large to small. The X chromosome is a large sized submetacentric and the Y chromosome a small sized acrocentric (Table I, Figs 2-11). Karyotypic analysis of five C. scotti specimens showed 2n = 58 and FNa = 70 (Table I, Figs 2-11). The nine specimens of C. marinhus showed 2n = 56 and FNa = 54, while karyotypic analysis of three C. goytaca specimens showed 2n = 54 and FNa = 62-63 (Table I, Figs 2-11).
Phylogenetic analyses of cytochrome b sequences, regardless of methodological approach (MP, ML, and BI), recovered the same general topology, with three main clades of Cerradomys (Fig. 12): I) C. akroai sp. nov. and C. scotti; II) C. maracajuensis and C. marinhus; and III) C. langguthi, plus a clade with the three remaining species of the genus (C. subflavus, C. vivoi, C. goytaca).
The clade (C. maracajuensis, C. marinhus) is found as sister group to the other two Cerradomys clades in the ML and BI, but the consensus of 8 most parsimony trees reveals a polytomy involving the three main clades (results not shown). Cerradomys akroai sp. nov. and C. scotti are found with weak or moderate support in the different phylogenetic approaches (MP bootstrap 79%, ML bootstrap 61; BI posterior probability 0.80).
The specimens identified as Cerradomys subflavus were never recovered in a monophyletic unit, as its terminals formed a polytomy with C. vivoi and C. goytaca (Fig. 12). In the parsimony analysis, C. vivoi formed a reciprocally monophyletic clade relative to the clade containing C. subflavus and C. goytaca, while in the ML and BI analyses the terminals of C. subflavus and C. goytaca are intermixed.
Genetic distance estimates (p) among C. akroai sp. nov. haplotypes varied from 0 to 1.8%, while interspecific variation between C. akroai sp. nov. and any other Cerradomys species was greater than 5.8%. Genetic distance estimates between C. scotti and C. akroai sp. nov. varied from 5.1% to 7.6% (Fig. 10).
Morphologic data
Cerradomys species were differentiated by canonical discriminant analyses (Wilks' Lambda = 0.01134, F = 5.3862, p < 0.0001, df = 119, 648), with 89% of total correct classification among species. The biplot of species scores for the first two canonical functions (which accounted for 83% of total eigenvalues' contributions) revealed that species formed three groups in morphometric space (Fig. 13), which correspond to the three major clades recovered in the phylogenetic analysis: I) C. subflavus, C. langguthi, C. goytaca and C. vivoi; II) C. scotti and C. akroai sp. nov.; and III) C. marinhus and C. maracajuensis.
The scores' range of the last species overlapped with the other groups, and C. maracajuensis was the least correctly classified species in discriminant functions, with only 63% of correct cases; C. vivoi also had a low classification score, with 62% of correct cases. Three species (C. scotti, C. goytaca, and C. akroai sp. nov.) had all specimens correctly classified, while predicted classification of remaining species varied from 78% (C. subflavus; 1 specimen misclassified as C. langguthi and 1 as C. vivoi) to 93% (C. marinhus; 2 specimens misclassified as C. maracajuensis).
In the ANOVA, C. akroai sp. nov. differed from C. scotti in 5 measurements when the Bonferroni correction was employed: CH (F = 16.0, p = 0.0003), CORB (F = 13.3, p = 0.0008), LZIG (F = 12.9, p = 0.0009), PPAL (F = 10.6, p = 0.002) and LROS (F = 9.8, p = 0.003). Five other variables showed differences using fixed p < 0.05: CIOR (F = 7.9, p = 0.007), CROS (F = 7.4, p = 0.01), CIL (F = 6.1, p = 0.02), LCO (F = 4.9, p = 0.03) and LCRA (F = 4.5; p = 0.04). Eight measurements did not differ significantly (p > 0.05) between C. scotti and C. akroai sp. nov.: GSL, M1M, CPDIA, CFI, LFI, SMS, LM1 and PZIG.
The new Cerradomys species exhibit the diagnostic characteristics of members of the genus (Weksler et al. 2006): interorbital region strongly convergent anteriorly, with well developed supraorbital crests (Figs 14-16); very long incisive foramina with lateral margins wider medially and antero-posterior margins sharp; stapedial foramen and posterior opening of alisphenoid canal vestigial or absent; squamosal-alisphenoid groove and sphenofrontal foramen absent; secondary anastomosis of internal carotid crossing the dorsal surface of pterygoid plate; capsular process of lower incisor developed. Dorsal pelage coarsely grizzled; tail longer than combined length of head and body, hind foot with small hypothenar pad, densely covered squamae distal to thenar pad, and conspicuous ungual tufts at bases of claws on dI-dV.
Cerradomys species exhibit morphological variation in integumental and cranial traits (see also Percequillo et al. 2008, Tavares et al. 2011). The fur color varies among species, from buffy-yellow grizzled with dark-brown to orange- or red-buff grizzled with black; in C. subflavus, C. langguthi, C. vivoi, and C. goytaca, the head pelage is distinctly colored from body (grayish-brown head and orange- to reddish-brown body); in remaining species, including C. akroai sp. nov., the dorsal pelage color of the head and the body is the same. The tail is very weakly bicolored dorsoventrally in C. marinhus and C. maracajuensis, weakly bicolored in C. subflavus, C. vivoi, and C. langguthi, and distinctly bicolored in C. scotti, C. goytaca, and C. akroai sp. nov. (in the proximal half); however, the extension of darker coloration in the ventral region of the tail is variable among specimens of C. subflavus, C. goytaca, C. vivoi, and C. langguthi, and thus overall degree of tail countershading is not a diagnostic character for these species.
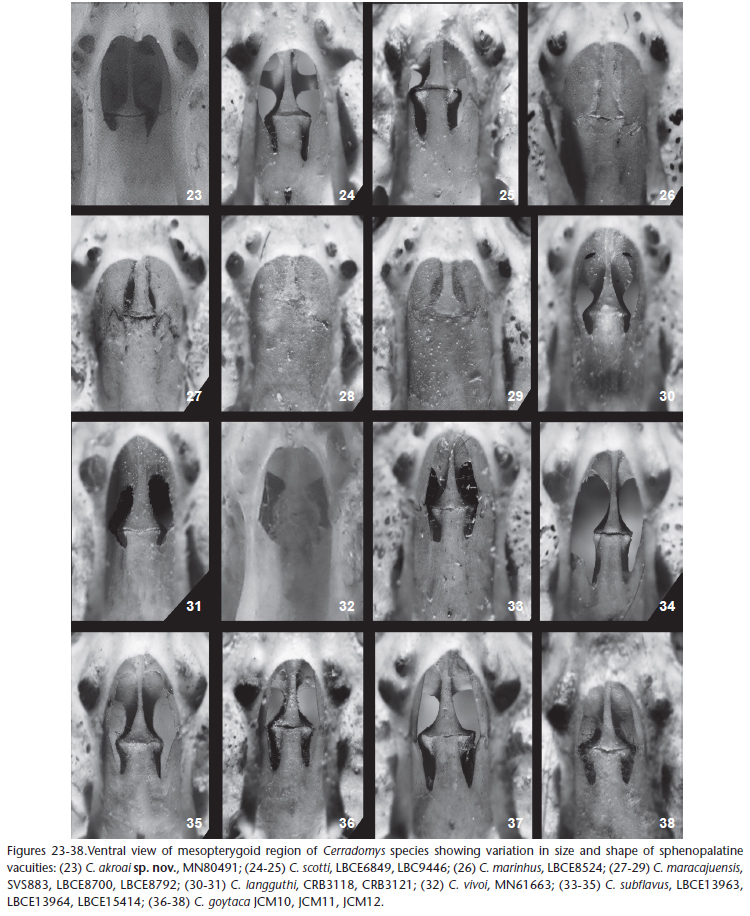
Among cranial traits, the posterior margins of incisive foramina extend to or between M1 alveoli of all species, except in some individuals of C. scotti and C. akroai sp. nov. (Figs 17-22). Complex posterolateral palatal pits are recessed at very deep and wide fossae, except in C. maracajuensis and C. marinhus; in most species, the roof of mesopterygopid fossa is perforated by sphenopalatine vacuities, usually exposing the presphenoid and basisphenoid, but C. maracajuensis and C. marinhus are characterized by very short vacuities restricted to the presphenoid or by a completely ossified roof of the mesopterygoid fossa (Figs 23-38); the alisphenoid strut is absent in all species, except C. scotti and C. akroai sp. nov. (Fig 14-16).
Given the observed differences in morphology and karyotype, and the results of the morphometric and phylogenetic analyses, we describe this form of Cerradomys as a new species.
TAXONOMY
Cerradomys akroai, sp. nov.
Holotype. MN80491, an adult female specimen collected by Flávia Casado (original field number LBCE13509) in February 6, 2010. The holotype consists of skull, partial postcranial skeleton, and skin. A bone marrow suspension cells in Carnoy's fixative (methanol: acetic acid) and a liver tissue sample preserved in ethanol are housed at Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios, Instituto Osvaldo Cruz, FIOCRUZ, under the original field number LBCE13509. Cytochrome b DNA data were deposited in GenBank (accession number KP122219).
Paratypes. Brazil, Tocantins: Novo Jardim, males MN80488 (field number LBCE13447), MN80489 (LBCE 13468), MN80490 (LBCE13483), LBCE12016, LBCE12744, LBCE13405, females MN80485 (LBCE12131), MN80486 (LBCE12756), MN80492 (LBCE13525), LBCE13390, unsexed MN80487 (LBCE13047).
Type Locality. Brazil, Tocantins: Novo Jardim municipality, in the Rio Palmeiras basin, near Porto Franco Hydroelectric Dam (ca. 11°48'S and 46°46'W). This locality is in the Cerrado domain west of the Serra Geral de Goiás.
Distribution. Known only from type locality (Fig. 1).
Diagnosis. Cerradomys akroai sp. nov. can be identified by the combination of the following characteristics: medium body size (HBL varying from 111 to 140 mm) and long tail (TL = 149 to 162 mm), dense dorsal pelage, overall dorsal color gray olive lined with yellow, head and dorsum with same color, ventral body color slightly yellowish, skull with deep rostral depression, mesopterygoid fossa with long and wide sphenopalatine vacuities with reduced basisphenoid penetration (4 presphenoid: 1 basisphenoid), alisphenoid strut present, basisphenoid foramen absent, deep palatal fossae (complex posterolateral palatal pits), and a unique chromosomal formula (2n = 60 and FNa = 74, Figs 2-11).
Description. Tail length longer than head and body (124% of head and body length); hind feet moderately narrow and long (21% of head and body length); pinnae rounded and small (Ear = 18 to 21 mm; 14% of head and body length), covered internally with short orange hairs and externally with yellow and dark hairs. Long and dense dorsum pelage gray olive lined with yellow, with many dark guard hairs, yellowish cover hairs, and gray and soft under hairs. Cover hairs long with a sub terminal yellow band; guard hairs sparse and long, with distal half entirely black or dark brown. General ventral color buffy or yellowish, slightly grizzled, and distinctively lighter than dorsal pelage, with hairs grayish-based and tipped with buffy or yellowish. Flanks bright yellow with dark guard hairs rare. Mystacial vibrissae long, reaching but not surpassing pinnae when laid back. Tail bicolored in the proximal half, covered with short and sparse brown hairs and scales on dorsal surface and unpigmented hairs and scales on ventral surface. Hind foot of medium size (24 to 32 mm), with dorsal surface white, covered with short, wholly white hairs. Ungual tufts sparse, shorter than or reaching the end of claws; ventral surface naked.
Rostrum long and broad (Figs 14-16), tapering anteriorly, with inflated capsular projection of nasolacrimal foramen, and flanked by deeply excavated zygomatic notches. Interorbital region narrow, converging anteriorly, with dorsolateral margins with sharp and well developed supraorbital crests. Braincase oblong, with prominent temporal crests. Zygomatic plate projected forward in lateral view. Incisive foramina long, with lateral margins concave and wider posteriorly; posterior margins sometimes reaching, but not extending between alveoli of upper first molars. Palate long and wide; posterolateral palatal pits numerous and complex, recessed in deep palatal fossae. Mesopterygoid fossa with anterior margin rounded or slightly acute, not reaching M3 alveoli; bony roof of mesopterygoid fossa perforated by large sphenopalatine vacuities, characterized as openings reaching the basisphenoid and wider than posterior expansion of presphenoid bone. Alisphenoid strut present. Postglenoid foramen large and nearly semicircular in shape separated from small or absent subsquamosal fenestra by a wide hamular process of squamosal. Mandible long; coronoid process large, falciform or triangular, nearly equal to condyloid process; superior notch shallow; angular process short, not surpassing the condyloid process posteriorly; inferior notch shallow; capsular process of lower incisor well developed. Cranial measurements of C. akroai and all other Cerradomys species are presented in Table II.
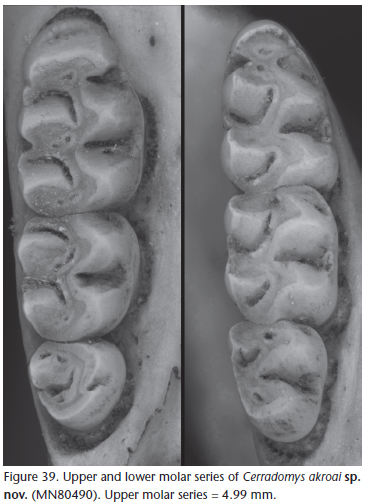
The upper incisors are opisthodont, with smoothly rounded enamel bands. The maxillary toothrows are parallel. Molars are bunodont and with labial flexi enclosed by a cingulum (Fig. 39).
The first upper molar (M1) anterocone not divided into anterolabial and anterolingual conules, anteromedian flexus not present. The anteroloph is well developed, and can be joined or not with the anterocone by labial cingulum; the protostyle is absent; mesolophs are small on M1 and M2; the mesoloph is often joined to the paracone forming a single structure. The paracone is connected by enamel bridge to the anterior moiety of the protocone. The protoflexus of M2 is poorly developed; the mesoflexus is present as single internal fossette; the paracone lacks an accessory loph. The third upper molar (M3) is reduced, and lacks a posteroloph, but has deep hypoflexus in unworn dentitions. The first lower molar (m1) anteroconid lacks an anteromedian flexid; an anterolabial cingulum is present on all lower molars; ectolophids are absent on m1 and m2; a small mesolophid is distinct on unworn m1 and m2, joined to the entoconid; a posteroflexid is well developed on m3.
Karyotype. Cerradomys akroai sp. nov. holotype shows a karyotype with 2n = 60 and FNa = 74, the highest diploid and fundamental number among Cerradomys species (Table I, Figs 2-11). See full description in results. Karyotypic data reinforces the uniqueness of C. akroai with respect to other congeneric species (Figs 2-11). This difference in the autossome complement would putatively lead to an infertile hybrid in the eventual mating of individuals between the two species (see discussion).
Comparisons. Despite the similarities shared by all congeneric species, C. akroai sp. nov. can be clearly distinguished from C. vivoi, C. langguthi, C. subflavus and C. goytaca by its external pelage coloration. In these four last species, the anterior half of dorsal head pelage is distinctively grayish to yellow-grayish and different from the remaining dorsal coloration, while in C. akroai sp. nov., C. maracajuensis, C. marinhus, and C. scotti the head coloration and dorsal body pelage coloration are similar. Cerradomys akroai sp. nov. differs from C. maracajuensis and C. marinhus by its longer, denser, and olive-brown dorsal pelage, vis-à-vis the yellow-brown dorsal pelage in the last two taxa, and by cranial features; the roof of mesopterygopid fossa is perforated by long and wide sphenopalatine vacuities, exposing the presphenoid and basisphenoid in C. akroai sp. nov. and C. scotti, while C. maracajuensis and C. marinhus are characterized by shorter vacuities restricted to the presphenoid or by a completely ossified roof of the mesopterygoid fossa. Finally, C. akroai can be distinguished from all species except C. scotti, by the presence of the alisphenoid strut and the strongly dorsoventrally bicolored tail in proximal half.
Cerradomysakroai sp. nov. is morphologically more similar to C. scotti than to any other Cerradomys species, but can be distinguished from C. scotti by its dorsal body color, which is darker in C. akroai sp. nov., and by the following cranial differences (Figs 17-22): 1) sphenopalatine vacuities in C. akroai sp. nov. with a less posterior penetration into the basisphenoid (extension of vacuities: 4 presphenoid: 1 basisphenoid) than in C. scotti (2 presphenoid: 1 basisphenoid); 2) conspicuous foramen that perforates the basisphenoid in C. scotti, but is apparently absent in C. akroai sp. nov.; 3) and posterolateral palatal pits deeper in C. scotti than in C.akroai sp. nov.
Etymology. This species is named in honor to the Akroá, an extinct Amerindian people that occupied the Novo Jardim region until the XVIII and XIX centuries (Apolinário, 2006).
DISCUSSION
Cerradomys species are characterized by a remarkable karyotypic diversity (Langguth & Bonvicino 2002, Bonvicino 2003, Percequillo et al. 2008), with several cases of intraspecific polymorphism (Table I). Cerradomys akroai sp. nov.possess the highest diploid and fundamental numbers within the genus, and is clearly diagnosed by diploid and/or fundamental autosome numbers, as well as morphology of autosomes and the sex chromosomes. The C. akroai sp. nov. karyotype, 2n = 60 and FNa = 74, is more similar to C. scotti karyotype with 2n = 58 and FNa = 70-72, but differ from it in showing one additional large sized metacentric pair. G-band coloration was not obtained for C. akroai karyotype for comparison with C. scotti; nevertheless, the difference in the autossome complement between these two taxa is related to both diploid and fundamental autosome numbers; to derive one karyotype from another involves a series of chromosomal rearrangements including inversions and fusions or fissions. Even when these chromosomal rearrangements have little effect on hybrid fitness, they might reduce gene flow through the suppression of recombination, due to mechanical pairing problems; in this form, the rearrangements extend the effects of linked isolation genes, and thus facilitate speciation (Reiseberg 2001, Kirkpatrick 2010). The effect of suppression of recombination offered by chromosomal rearrangements is more effective in species in which the number of chromosomal rearrangements is large, as is the case of C. scotti and C. akroai, where more than one chromosomal pair is involved in rearrangements.
In addition, no variation was found in diploid number among the 21 specimens of C. scotti karyotyped so far (Table I), suggesting that 2n is fixed; the FNa variation found in C. scotti (FNa = 70 or 72) is due to an pericentric inversion affecting a small chromosome pair, which is not related to the karyotype differences of the two species. Finally, C. scotti and C. akroai also differ in the morphology of the Y chromosome, a small sized acrocentric in C. akroai sp. nov., but a median sized biarmed chromosome in C. scotti. Karyological variation found in the fundamental autosome pairs of C. goytaca chromosomal complement in relation to the karyotype described by Tavares et al. (2011) could be attributed to different interpretation related to the morphology of small acrocentric pairs, and also due to a pericentric inversion affecting small chromosomes (Fig. 11). Although changes in diploid number can be easily detected in rodent karyotypes, the fundamental number is not so easily evident due to different levels of condensation of small autosomes and also to different interpretations of biarmed chromosomes. Morphology of biarmed metacentric and submetacentric chromosomes, as well telocentric chromosomes, it not problematic. However, acrocentric chromosomes have their centromere close to the short arm telomere, and the short arm has a secondary constriction, of variable length with non-coding DNA (Rieseberg 2001). In some preparations, small acrocentric chromosomes can appear as biarmed chromosomes, leading to different estimates of fundamental number.
Molecular data also support that the new karyomorph belongs to a distinct evolutionary lineage, sister to C. scotti. There are 37 fixed nucleotide differences between C. scotti and C. akroai, including one non-synonymous substitution. The average genetic distance (p) estimated for C. akroai and C. scotti is 5.7%, larger than the average distances between C. langguthi and C. subflavus (p = 4.7%), and much larger than between C. subflavus, C. goytaca, and C. vivoi (p = 1.7%; see below). Phylogenetic analyses also demonstrated the close relationship between C. maracajuensis and C. marinhus, and that a separate clade is formed by C. subflavus, C. vivoi, C. goytaca, and C. langguthi. All molecular analyses showed that C. subflavus and C. goytaca haplotypes are mixed, and that these forms are not reciprocally monophyletic, suggesting that and C. goytaca is a junior synonymous of C. subflavus. Although ML and BI analyses also showed that C. vivoi is not reciprocally monophyletic relative to C. subflavus and C. goytaca, MP analysis showed that the two groups are reciprocally monophyletic, and thus could be considered as distinct lineages. The reduced genetic distance estimate between C. goytaca and C. subflavus (average p = 0.6%) is smaller than the distances among any other species pair (average p = 2.3% between C. vivoi and C. subflavus; > 4.7% between all other species pairs); the estimate is in level with intraspecific variation of other Cerradomys species, such as C. maracujensis (p = 0.7%) and C. scotti (p = 0.6%). These analyses indicate that C. subflavus and C. goytaca might be conspecific; alternatively, the recovered phylogenetic pattern can be explained by a recent speciation event with incomplete lineage sorting. Further analyses with denser genetic and taxonomic sampling are necessary to establish a detailed relationship among these taxa.
The morphometric differentiation of Cerradomys species is coincident with the phylogenetic arrangement; the three morphometric groups recovered by the first and second canonical functions corresponded to the three clades: (C. vivoi, C. subflavus, C. goytaca, C. langguthi), (C. scotti, C. akroai), and (C. marinhus, C. maracajuensis). Cerradomys akroai is more similar to its sister species C. scotti than to any other congeneric species. They differ, however, in at least five variables, mainly in height of skull (CH), but also in orbital region (LZIG, CORB), rostrum (LROS), and palate (PPAL). The relationship between morphometric and genetic differentiation of this genus also deserves a more detailed analysis.
Cerradomysakroai, together with C.scotti, are endemic of open vegetation domain, occupying the central area of the genus geographic range (Fig. 1). Interestingly, both species are sympatric with C.marinhus in their respectively type localities. Cerradomys scotti is also sympatric with C. maracajuensis in the Brazilian states of Mato Grosso and Mato Grosso do Sul, Sapucay in Paraguay, and Santa Cruz in Bolivia (Percequillo et al. 2008). We also report for the first time the sympatry (but not syntopy) of three species – C. marinhus, C. scotti, and C. subflavus – in Uberlândia, Minas Gerais (Appendix 1).
Cerradomysakroai and C. scotti are more associated with open vegetation formation of the Cerrado domain, while C. maracajuensis, C. subflavus, C. vivoi, C. goytaca, and C. langguthi are mainly associated to forest formations that occur within the Cerrado, such as gallery forest, and in ecotones or in the Atlantic forest, while C. marinhus is more often found in the grass marshes with buriti palms called "veredas" (Eiten 1983) and flooded forests.
Our study confirms that Cerradomys is a speciose taxon with a core distribution in the open vegetation belt of Eastern South America and that could be a model for the study of the biogeography of the region. Some Cerradomys species distributional limits are shaped by the Rio São Francisco, such as C. langguthi and C. subflavus/C. vivoi, a pattern similar to that found for other open vegetation belt taxa, such as Thrichomys (Nascimento et al. 2013), Gracilinanus agilis (Burmeister, 1854) (Faria et al. 2013), and Calomys expulsus (Lund, 1841) (Nascimento et al. 2011). Other processes could have played a role on evolutionary history of the Cerradomys, such as Pleistocene climatic oscillations that lead to reduction of open vegetation areas during interglacial periods. Our understanding of the biogeographic processes that shaped the distributional limits for Cerradomys species still awaits a more extensive study with denser geographic sampling.
ACKNOWLEDGMENTS
We are grateful for Paulo S. D' Andrea, Arnaldo Maldonado and other members of the Laboratório de Biologia e Parasitologia de Mamíferos Reservatórios Silvestres; to Orílio Leoncini (in memorium), Júlio F. Vilela and Ana Flávia B. de Andrade for help in fieldwork; to João A. de Oliveira that granted us to examine specimens under their responsibility (MN) Héctor Seuánez provided insightful comments on the manuscript and also improved the language style and grammar. Maria C. Vianna and Leila S. Monnerat helped in karyologic preparations and in generating C. goytaca sequences. Alexandre R. Percequillo shared original cranial photographs. Permits for field collection were granted by SISBIO (System of Authorization and information on Biodiversity), ICMBio (Instituto Chico Mendes de Conservação da Biodiversidade) under number 11375-1. We also thank Bruce Patterson and two anonymous reviewers for helpful comments on the manuscript. This work is supported through grants from Conselho Nacional de Pesquisa e Desenvolvimento – CNPq (processes 303422/2010-6; 481286/2011-0) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro – FAPERJ (processes E-26/110.505/2012 and E-26/101.950/2012) to CRB and MW.
LITERATURE CITED
Submitted: 25.III.2014
Accepted: 31.X.2014
Appendix 1.
List of examined specimens for morphometric (M), karyologic (K) and molecular phylogenetic (P) analysis. For geographic coordinates, see Material and Methods, Percequillo et al. (2008), and Tavares et al. (2011).
Brazil, Bahia: Caetité, Minaçu (C. vivoi male MN63382M), Cocos (Fazenda Sertão do Formoso, C. scotti females MN61667MK, MN63833M, males MN61669M, MN61671M, unsexed CRB2920MK), Correntina (C. scotti females CRB2719M, CRB2731MKP), Itabuna (CEPLAC, C. vivoi holotype male MN35898MKP), Jaborandi (Fazenda Sertão do Formoso, C. marinhus females MN63823M, MN63831M, MN63833M, MN63836MK, MN63837M, MN63839MK, CRB1645M, CRB2940M, males MN63822M, MN63825M, MN63826K, MN63827M, MN63830M holotype, MN 63832MK, MN 63834MKP, MN 63835M, MN63838M, CRB1585M, CRB1673M, C. scotti male MN61672MKP, female CRB2933M, unsexed CRB2920M), Nova Viçosa (Mata do Aeroporto, C. subflavus male MN61673k).
Brazil, Goiás: Alto Paraíso (C. scotti females MN61685MK, MN61687MK, males MN61677MK, MN61682M, MN61684MK, MN61686MK), Aporé (Usina Espora (C. marinhus females LBCE6864M, LBCE8539M, LBCE8541M, LBCE12780K, males LBCE6865M, LBCE6870M, LBCE8515M, LBCE8537M, LBCE8542M, LBCE10917M, unsexed LBCE8524M, LBCE9107M, C. scotti female LBCE6849M, males LBCE9446MP, LBCE10907MKP, unsexed LBCE5848P), Campo Alegre de Goiás (C. scotti male LBCE8749MP), Cavalcante (C. scotti females MN61674MK, MN61678MK, MN61688M, males MN61675MK, MN61676MK, MN61679MK, MN61680MK, LBCE11845K, unsexed MN61681M), Corumbá de Goiás (Morro dos Cabeludos, C. scotti females MN 44177M, MN 50379P, MN 50380MK, males MN50306MK, holotype MN44176MK), Luziania (C. scotti males LBCE7545M, LBCE7546MKP), Mimoso de Goiás (C. scotti female MN67089), Serranópolis (C. maracajuensis male LBCE7475);
Brazil, Mato Grosso do Sul: Aquidauna (Fazenda Rio Negro, C. maracajuensis females LBCE4891M,LBCE5309M – C. scotti male LBCE4884MP), Dois Irmãos de Buriti (C. maracajuensis unsexed LBCE8790M), Cassilândia (PCH Planalto C. marinhus females LBCE11796K, LBCE12023K, LBCE12024K, LBCE12072K, males LBCE11476K, LBCE11783K, LBCE11955K, LBCE11960K, Corumbá (Fazenda Alegria, C. scotti males LBCE5713M, LBCE5724MP, LBCE5725MP), Maracajú (Fazenda da Mata, C. maracajuensis males MN4414M, holotype MN44178MKP, female MN4410M, C. scotti males MN4409M, AMNH 134704M), Sidrolândia (PCH Mambaí II, C. maracajuensis females LBCE8700M, males LBCE8719M, LBCE8720M), Sítio D' Abadia (C. scotti male LBCE11618MP).
Brazil, Mato Grosso: Campo Novo do Parecis (C. scotti male MN74959M), Campo Verde (C. scotti male SVS344M, C. maracajuensis females SVS847M, SVS849P, SVS883MP), Diauaruns (alto Xingú, C. maracajuensis female MN11681M, males MN11680M, MN11682M, MN11683M, MN11684M, MN11685M, unsexed MN11687M), São José do Xingú (C. maracajuensis female CRB2790MP);
Brazil, Minas Gerais: Confins (Confins Airport, C. subflavus male UFPB1926M), Juramento (Fazenda Canoas, C. vivoi females MN61661M, MN61663M, MN61666KP, males MN61662M, MN61664M, MN61665KP*), Lagoa Santa (C. subflavus females UFPB2062M, MN31386M, males UFPB1927M, UFPB1928M, unsexed LV-CEG42P), Riacho Mocambinho (C. vivoi female MN29057M, males MN29035M, MN34435M, MN43816M), Salinas (C. vivoi male MN42844M, unsexed MN42841M), São Roque de Minas (Serra da Canastra, C. subflavus LBCE15485P, LBCE15491P), Vargem do Retiro (Serra do Cipó, Ribeirão Mascates, Parque Nacional da Serra do Cipó, C. subflavus: males MN31393M, MN31394M), Uberlândia (Chacara Eldorado, C. marinhus: female LBCE17286P, males LBCE17294P, LBCE17786P, LBCE18191P; Campo Florido, C. scotti: females LBCE17287P, LBCE17370P; C. subflavus: Fazenda Viadinho, male LBCE17255P, Instituto Federal, unsexed LBCE17293P);
Brazil, Paraiba: Sapé (Corredor São João-Fazenda Pacatuba, João Pessoa, C. langguthi holotype MN69786MKP), Souza (São Gonçalo, C. langguthi male LBCE15905MP);
Brazil, Rio de Janeiro state: Quissamã (Restinga de Jurubatiba, C. goytaca LBCE-JCM09K, LBCE-JCM10MP, LBCE-JCM11MKP, LBCE-JCM12M, LBCE-JCM14MK, LBCE-JCM15MK); Parque Nacional Restingas de Jurubatiba (C. goytaca holotype male MN73177M, females MN73172M, MN73174M, males MN73180M, MN73191M, MN73183M);
Brazil, São Paulo: Itirapina (cerrado de Graúna, C. subflavus female MN43067M);
Brazil, Tocantins: Novo Jardim (Cerradomys akroai sp. nov.: females MN80485KP, MN80486KP, MN80491MKP, MN80492MKP, LBCE13390P, males MN80488MP, MN80489MP, MN80490MKP, LBCE12016P, LBCE12744P, LBCE13405P, unsexed MN80487M; C. marinhus male LBCE12997K).
Paraguay, Canindeyui: Reserva Nat. del Bosque Mbaracay (unsexed TK61881P).
- Ab' Saber, A.N. 2003. Os domínios da natureza do Brasil: potencialidades paisagísticas. São Paulo, Ateliê Editorial, 159p.
- Agrellos, R.; C.R. Bonvicino; E.S.T. Rosa; A.A.R. Marques; P.S. D'andrea & M. Weksler. 2012. The taxonomic status of the Castelo dos Sonhos Hantavirus reservoir, Oligoryzomys utiaritensis Allen 1916 (Rodentia, Cricetidae, Sigmodontinae). Zootaxa 3220: 1-28.
- Almeida, E.J.C. & Y. Yonenaga-Yassuda. 1985. Robertsonian fusion, pericentric-inversion and sex-chromosome heteromorphisms in Oryzomys subflavus (Cricetidae, Rodentia. Caryologia 38: 129-137. doi: 10.1080/00087114.1985.10797737
- Almeida, F.C.; C.R. Bonvicino & P. Cordeiro-Estrela. 2007. Phylogeny and temporal diversification of Calomys (Rodentia, Sigmodontinae): implications for the biogeography of an endemic genus of the open/dry biomes of South America. Molecular Phylogenetics and Evolution 2007: 449-466. doi: 10.1016/j.ympev.2006.07.005
- Andrade, A.F.B.; C.R. Bonvicino; D.C. Briani & S. Kasahara. 2004. Karyologic diversification and phylogenetic relationships of the genus Thalpomys (Rodentia, Sigmodontinae). Acta Theriologica 49: 181-190. doi: 10.1007/BF03192519
- Andrades-Miranda, J.; N.I. Zanchin; L.F. Oliveira; A.R. Langguth & M.S. Mattevi. 2002. (T2AG3)n telomeric sequence hybridization indicating centric fusion rearrangements in the karyotype of the rodent Oryzomys subflavus Genetica 114: 11-16. doi: 10.1023/A:1014645731798
- Apolinário, J.C. 2006. Os Akroá e outros povos indígenas nas Fronteiras do Sertão. Políticas indígena e indigenista no norte da capitania de Goiás, atual Estado do Tocantins, século XVIII. Goiânia, Ed. Kelps, 276p.
- Bezerra, A.M.R.; A.P. Carmignotto & F.H.G. Rodrigues. 2009. Small non-volant mammals of an ecotone region between the Cerrado hotspot and the Amazonian rainforest, with comments on their taxonomy and distribution. Zoological Studies 48: 861-874.
- Bonvicino, C.R. 2003. A new species of Oryzomys (Rodentia, Sigmodontinae) of the subflavus group from the Cerrado of central Brazil. Mammalian Biology 68: 78-90. doi: 10.1078/1616-5047-00066
- Bonvicino, C.R. & M.A. Moreira. 2001. Molecular phylogeny of the genus Oryzomys (Rodentia: Sigmodontinae) based on cytochrome b DNA sequences. Molecular Phylogenetics and Evolution 18: 282-292. doi: 10.1006/mpev.2000.0878
- Bonvicino, C.R. & M. Weksler. 1998. A new species of Oligoryzomys (Rodentia, Sigmodontinae) from northeastern and central Brazil. Zeitschrift Fur Saugetierkunde-International Journal of Mammalian Biology 63: 90-103.
- Bonvicino, C.R.; I.B. Otazu & P.M. Borodin. 1999. Chromosome variation in Oryzomys subflavus species group (Sigmodontinae, Rodentia) and its taxonomic implication. Cytologia 64: 327-332. doi: 10.1508/cytologia.64.327
- Bonvicino C.R.; S.M. Lindbergh; M.F. Barros & A.M.R. Bezerra. 2012. The Eastern boundary of the Brazilian Cerrado: a hotspot region. Zoological Studies 51: 1207-1218.
- Brandt, R.S. & L.M. Pessoa. 1994. Intrapopulational variability in cranial characters of Oryzomys subflavus (Wagner, 1842) (Rodentia, Cricetidae), in north-eastern Brazil. Zoologischer Anzeiger 233: 45-55.
- Carmignotto, A.P.; M. de Vivo & A. Langguth. 2012. Mammals of the Cerrado and Caatinga: distribution patterns of the tropical open biomes of Central America, p. 307-350. In: B.D. Patterson & L. Costa (Eds). Bones, clones, and biomes. Chicago, University of Chicago Press, 368p. doi: 10.7208/chicago/9780226649214.003.0014
- Eiten, G. 1983. Classificação da vegetação do Brasil. Brasília, Conselho Nacional de Desenvolvimento Científico e Tecnológico, 305p.
- Faria, M.B.; F.F. Nascimento; J.A. Oliveira & C.R. Bonvicino. 2013. Biogeographic Determinants of Genetic Diversification in the Mouse Opossum Gracilinanus agilis (Didelphimorphia: Didelphidae). Journal of Heredity 104 (5): 613-626. doi: 10.1093/jhered/est039
- Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791. doi: 10.2307/2408678
- Geise, L. & L. Pereira. 2008. Rodents (Rodentia) and marsupials (Didelphimorphia) in the municipalities of Ilhéus and Pau Brasil, state of Bahia, Brazil. Check List 4: 174-177.
- Geise, L.; R. Paresque; H. Sebastião; L.T. Shirai; D. Astúa & G. Marroig. 2010. Non-volant mammals, Parque Nacional do Catimbau, Vale do Catimbau, Buíque, state of Pernambuco, Brazil, with karyologic data. Check List 6: 180-186.
- Huelsenbeck, J.P. & F. Ronquist. 2001. MRBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755. doi: 10.1093/bioinformatics/17.8.754
- Irwin, D.M.; T.D. Kocher & A.C. Wilson. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution 32: 128-144. doi: 10.1007/BF02515385
- Keane, T.M.; C.J. Creevey; M.M. Pentony; T.J. Naughton & J.O. McLnerney. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evolutionary Biology 6: e29. doi: 10.1186/1471-2148-6-29
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol 8 (9): e1000501. doi: 10.1371/journal.pbio.1000501
- Langguth, A. & C.R. Bonvicino. 2002. The Oryzomys subflavus species group, with description of two new species (Rodentia, Muridae, Sigmodontinae). Arquivos do Museu Nacional Rio de Janeiro 60: 285-294.
- Maia, V. & A. Hulak. 1981. Robertsonian polymorphism in chromosomes of Oryzomys subflavus (Rodentia, Cricetidae). Cytogenetics and Cell Genetics 31: 33-39. doi: 10.1159/000131623
- Moreira, J.C.; E.G. Manduca; P.R. Goncalves; M.M. De Morais Jr; R.F. Pereira; G. Lessa & J.A. Dergam. 2009. Small mammals from Serra do Brigadeiro state park, Minas Gerais, southeastern Brazil: species composition and elevational distribution. Arquivos do Museu Nacional Rio de Janeiro 67: 103-118.
- Myers, N.; R.A. Mittermeier; C.G. Mittermeier; G.A.B. Fonseca & J. Kent. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853-858. doi: 10.1038/35002501
- Musser, G.G. & M.D. Carleton. 1993. Family Muridae, p. 501-756. In: D.E. Wilson & D.M. Reeder (Eds). Mammal species of the world: a taxonomic and geographic reference, 2nd edition. Washington, DC, Smithsonian Institution Press, 1207p.
- Nascimento, F.F.; L.G. Pereira; L. Geise; A.M.R. Bezerra; P.S. D'Andrea & C.R. Bonvicino. 2011. Colonization process of the Brazilian common vesper mouse, Calomys expulsus (Cricetidae, Sigmodontinae): a biogeographic hypothesis. Journal of Heredity 102 (3): 260-269. doi: 10.1093/jhered/esr012
- Nascimento, F.F.; A.L.G. Souza; A.N. Menezes; A.M. Durans; J.C. Moreira; J. Salazar-Bravo; P.S. D'Andrea & C.R. Bonvicino. 2013. The role of historical barriers in the diversification process in open vegetation formation during the Miocene/Pliocene using an ancient rodent lineage as a model. PLOS One 8 (4): e61924. doi: 10.1371/journal.pone.0061924
- Oliveira, J.A.; R.E. Strauss & S.F. dos Reis. 1998. Assessing relative age and age structure in natural populations of Bolomys lasiurus (Rodentia: Sigmodontinae) in northeastern Brazil. Journal of Mammalogy 79: 1170-1183. doi: org/10.2307/1383008
- Paresque, R.; W.P. Souza; S.L. Mendes & V. Fagundes. 2004. Composição cariotípica da fauna de roedores e marsupiais de duas áreas de Mata Atlântica do Espírito Santo, Brasil. Boletim do Museu de Biologia Mello Leitão 17: 5-33.
- Percequillo, A.R.; E. Hingst-Zaher & C.R. Bonvicino. 2008. Systematic review of genus Cerradomys Weksler, Percequillo and Voss, 2006 (Rodentia: Cricetidae: Sigmodontinae: Oryzomyini), with description of two new species from eastern Brazil. American Museum Novitates (3622): 1-46. doi: 10.1206/495.1
- Pereira, L.G. & L. Geise. 2007. Karyotype composition of some rodents and marsupials from Chapada Diamantina (Bahia, Brasil). Brazilian Journal of Biology 67: 509-518. doi: 10.1590/S1519-69842007000300016
- Rieseberg, L.H. 2001. Chromosomal rearrangements and speciation. TRENDS in Ecology & Evolution 16 (7): 351-358. doi: 10.1016/S0169-5347(01)02187-5
- Rambaut A.; A.J. Drummond. 2007 Tracer. MCMC trace analysis package, version 1.6. Available online at: http://beast.bio.ed.ac.uk/Tracer [Accessed: 11/XII/2013]
- Rice, W.R. 1989. Analyzing tables of statistical tests. Evolution 43: 223-225. doi: 10.2307/2409177
- Rocha, C.R.; R. Ribeiro; F.S.C. Takahashi & J. Marinho-Filho. 2011. Microhabitat use by rodent species in a central Brazilian cerrado. Mammalian Biology 76: 651-653. doi: 10.1016/j.mambio.2011.06.006
- Rodríguez, F.; J.L. Oliver; A. Marín & J.R. Medina. 1990. The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142: 485-501. doi: 10.1016/S0022-5193(05)80104-3
- Ronquist, F. & J.P. Huelsenbeck. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. doi: 10.1093/bioinformatics/btg180
- Sambrook, J.; E.F. Fritsch & T. Maniatis. 1989. Molecular Cloning: A Laboratory Course Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory Press. 1659 p.
- Smith, M.F. & J.L. Patton. 1983. The diversification of South American murid rodents: Evidence from mitocondrial DNA sequence data for the Akodontine tribe. Biological Journal of the Linnean Society 50: 149-177. doi: 10.1111/j.1095-8312.1993.tb00924.x
- Sokal, R.R. & F.J. Rohlf. 1995. Biometry. New York, Freeman, 3rd ed., 887p.
- Sousa, M.A.N.; A. Langguth & E.A. Gimenez. 2004. Mamíferos dos Brejos de Altitude Paraíba e Pernambuco, p. 229-254. In: K.C. Porto, J.J.P. Cabral & M. Tabareli (Eds). Brejos de altitude em Pernambuco e Paraíba História Natural, Ecologia e Conservação. Brasília, Ministério do Meio Ambiente, Série Biodiversidade 9, 324p.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688-2690. doi: 10.1093/bioinformatics/btl446
- StatSoft. 2004. STATISTICA. SataSoft, data analysis software system, version 7. Available online at: http://www.statsoft.com
- Strauss, R.E. 2010. Discriminant groups of organisms. In: A.M.T. Elewa (Ed.). Morphometrics for Nonmorphometricians. Berlin, Springer-Verlag, Lecture Notes in Earth Sciences. doi: 10.1007/978-3-540-95853-6_4
- Svartman, M. & E.J.C. Almeida. 1992. Sex-chromosomes polymorphisms in Oryzomys aff subflavus (Cricetidae, Rodentia) from central Brazil. Caryologia 45: 313-324. doi: 10.1080/00087114.1992.10797234
- Swofford D.L. 2001. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0b10 [Computer software and manual]. Sunderland, Sinauer Associates Inc.
- Tavares, W.C.; L.M. Pessoa & P.R. Goncalves. 2011. New species of Cerradomys from coastal sandy plains of southeastern Brazil (Cricetidae: Sigmodontinae). Journal of Mammalogy 92: 645-658. doi: 10.1644/10-MAMM-096.1
- Weksler, M. & C.R. Bonvicino. 2005. Taxonomy of pygmy rice rats genus Oligoryzomys Bangs, 1900 (Rodentia, Sigmodontinae) of the Brazilian Cerrado, with the description of two new species. Arquivos do Museu Nacional 63: 113-130.
- Weksler, M.; A.R. Percequillo & R.S. Voss. 2006. Ten new genera of Oryzomyine rodents (Cricetidae: Sigmodontinae). American Museum Novitates 3537: 1-29. doi: 10.1206/0003-0082(2006)3537[1:TNGOOR]2.0.CO;2
- Yang Z. 1994. Statistical properties of the maximum likelihood method of phylogenetic estimation and comparison with distance matrix methods. Systematic Biology 43: 329-342. doi: 10.1093/sysbio/43.3.329
Publication Dates
-
Publication in this collection
22 Jan 2015 -
Date of issue
Dec 2014
History
-
Accepted
31 Oct 2014 -
Received
25 Mar 2014