Abstracts
The identification of quantitative trait loci (QTL) and marker-assisted selection with a view to breeding programs have aroused great interest, including for cashew improvement. This study identified QTL for yield-related traits: nut weight, male and hermaphrodite flowers. The traits were evaluated in 71 F1 genotypes of the cross CCP 1001 x CP 96. The methods of interval mapping and multiple QTL mapping were applied to identify QTL. Eleven QTL were detected: three for nut weight, four for male flowers and four for hermaphrodite flowers. The QTL accounted for 3.79 to 12.98 % of the total phenotypic variance and had phenotypic effects of -31.81 to 34.25 %. The potential for marker-assisted selection of the QTL hf-2f and hf-3m is great and the phenotypic effects and percentage of phenotypic variation higher than of the others.
Anacardium occidentale; genetic improvement; marked-assisted selection
A identificação de locos de caráter quantitativo (QTLs) e a seleção assistida por marcadores têm despertado grande interesse em programas de melhoramento, inclusive para o cajueiro. Esta pesquisa objetivou identificar QTLs relacionados a características de componentes de produção: peso da castanha, flores masculinas e hermafroditas. Avaliaram- se 71 plantas F1 originadas do cruzamento CCP 1001 x CP 96. Para a detecção de QTLs foram utilizadas as metodologias de mapeamento de intervalo e mapeamento de QTLs múltiplos. Onze QTLs foram identificados: três para peso da castanha, quatro para flores masculinas e quatro para flores hermafroditas. Os QTLs explicaram entre 3,79 e 12,98 % da variação fenotípica total e apresentaram efeito fenotípico variando entre -31,81 e 34,25 %. Os QTLs fh-2f e fh- 3m têm grande potencial para seleção assistida por marcadores, apresentando os maiores efeitos fenotípicos e as maiores explicações da variação fenotípica.
Anacardium occidentale; melhoramento genético; seleção assistida por marcadores
NOTE
QTL detection of yield-related traits of cashew
Identificação de QTLs relacionados a caracteres de componentes de produção do cajueiro
José Jaime Vasconcelos CavalcantiI, *; Francisco Herbeth Costa dos SantosII; Fanuel Pereira da SilvaII; Cássia Renata PinheiroIII
IEmbrapa Algodão, CP 174, 58.428-095, Campina Grande, PB, Brazil. *E-mail: jaime@cnpa.embrapa.br
IIUniversidade Federal do Ceará, Departamento de Fitotecnia, CP 6035, 60.451-970, Fortaleza, CE, Brasil
IIIUniversidade de São Paulo, Departamento de Genética, CP 83, 13.418-900, Piracicaba, SP, Brasil
ABSTRACT
The identification of quantitative trait loci (QTL) and marker-assisted selection with a view to breeding programs have aroused great interest, including for cashew improvement. This study identified QTL for yield-related traits: nut weight, male and hermaphrodite flowers. The traits were evaluated in 71 F1 genotypes of the cross CCP 1001 x CP 96. The methods of interval mapping and multiple QTL mapping were applied to identify QTL. Eleven QTL were detected: three for nut weight, four for male flowers and four for hermaphrodite flowers. The QTL accounted for 3.79 to 12.98 % of the total phenotypic variance and had phenotypic effects of -31.81 to 34.25 %. The potential for marker-assisted selection of the QTL hf-2f and hf-3m is great and the phenotypic effects and percentage of phenotypic variation higher than of the others.
Key words: Anacardium occidentale, genetic improvement, marked-assisted selection.
RESUMO
A identificação de locos de caráter quantitativo (QTLs) e a seleção assistida por marcadores têm despertado grande interesse em programas de melhoramento, inclusive para o cajueiro. Esta pesquisa objetivou identificar QTLs relacionados a características de componentes de produção: peso da castanha, flores masculinas e hermafroditas. Avaliaram- se 71 plantas F1 originadas do cruzamento CCP 1001 x CP 96. Para a detecção de QTLs foram utilizadas as metodologias de mapeamento de intervalo e mapeamento de QTLs múltiplos. Onze QTLs foram identificados: três para peso da castanha, quatro para flores masculinas e quatro para flores hermafroditas. Os QTLs explicaram entre 3,79 e 12,98 % da variação fenotípica total e apresentaram efeito fenotípico variando entre -31,81 e 34,25 %. Os QTLs fh-2f e fh- 3m têm grande potencial para seleção assistida por marcadores, apresentando os maiores efeitos fenotípicos e as maiores explicações da variação fenotípica.
Palavras-chave: Anacardium occidentale, melhoramento genético, seleção assistida por marcadores.
INTRODUCTION
The cashew tree (Anacardium occidentale L.) is a species widely grown in the semi-arid regions of northeastern Brazil and is an important source of employment and income in the states of Ceara, Piaui and Rio Grande do Norte. However, one of the main difficulties in cashew orchards is the low productivity (Paula Pessoa et al. 1995, Ferreira-Silva et al. 2009). To solve this problem, the efforts of the Brazilian breeding program have been focused on the development of more productive cashew cultivars. The reduced number of genotypes in use, originated froma narrow genetic base, indicates genetic vulnerability, in view of the expansion of cashewto different agro-systems in Brazil, due to the success of the crop. In this sense, the establishment and selection of new genotypes are important to reduce this vulnerability (Cavalcanti et al. 2000, Paiva et al. 2003). This means that the genetic basis must be widened and new genotypes adapted to the climate and soil conditions of each growing environment to enhance theeconomic viability of cashew in the Brazilian agribusiness (Paiva et al. 2005).
Impressive results have been achieved by the cashewbreeding program, mainly the development of 12clones for commercial production. However, in the case of perennial plants the use of conventional improvement techniques is hampered by a number of factors, e.g.long juvenile period and time needed for assessments, high heterozygosity of genotypes, environmental effects and need of large experimental areas (Barros et al. 2009).
With the development of genetic maps and statistical approaches to the mapping of quantitative trait loci (QTL), genetic loci controlling complex traits can be identified and screened. Information of QTL mapping provides the minimum number of genetic loci that may be related to a particular trait, the location on the genetic map of these loci and estimates of their effects on the trait, which are useful information for breeders. Molecular markers associated with QTL may be useful in assisted selection to develop new cultivars (Timmerman-Vaughan et al. 2005). Based on the a priori knowledge of linkage between marker loci and yield traits, undesirable plants can be eliminated from the progeny group in early stages, i.e., when the seedlings develop the first leaves (Wang et al. 2000). This makes the selection process more efficient, reducing the number of plants and progenies to be evaluated and the time required until a clone is finally released for commercial production resulting in cost savings.
A QTL analysis must begin with the construction of a genetic linkage map with a sufficiently high density of molecular markers for a representative genome coverage (Kenis and Keulemans 2005). Genetic maps were developed for cashew by Cavalcanti and Wilkinson (2007), using a population of 85 F1 plants from the cross between the dwarf cashew clone CCP 1001 and the mother plant of common cashew CP 96. These maps represent an initial platform for the identification of QTL associated with economically important traits of this crop.
Marker-assisted selection is particularly promising for cashew improvement, since it may contribute to the solution of specific problems, being faster, cheaper and more efficient. Due to the genetic complexity of cashew yield, the detection of QTL is rather difficult, since this trait is highly polygenic and influenced by the environment. To overcome this problem, the purpose of this study was to identify QTL of yield-related traits with a simpler genetic basis i.e., controlled by fewer genes and less affected by environmental interference, e.g.: nut weight, male and hermaphrodite flowers.
MATERIALAND METHODS
The experiment was conducted in an experimental area of Embrapa Agroindústria Tropical /Embrapa Tropical agroindustry, in Pacajus, CE, Brazil, in 2006 (lat 4° 10' S, long 38° 27' W and alt 60 m asl). The climate is predominantly dry/sub-humid (C2), according to the Thornthwaite climate classification. The average rainfall is 1,100 mm per year, mostly concentrated in summer/ fall. The soil of the experimental area is Eutrophic Red Yellow Podzolic with weak A horizon and sandy/medium texture.
The cashew population consisted of 85 F1 full-sib progenies, from a controlled cross between CCP 1001 and CP 96, in September 2000, planted in March 2001. The female parent CCP 1001 is a commercial dwarf cashew clone CP 96 and the male parent genotype a giant cashew tree, both from the Cashew genebank of Embrapa Tropical Agroindustry. Of the population, 71 plants were evaluated and mapped under field conditions. To obtain the nut weight, a sample of 20 nuts per plant was weighed on a digital balance Mark 3100, and the results expressed in grams. The number of male and hermaphrodite flowers was determined based on four panicles per plant, from the mid-third of the plant, i.e., one from each cardinal point.
The QTL were mapped using Multiple QTL Mapping (MQM) analysis (Janser 1993, Janser 1994, Janser and Stam 1994), an extension of Interval Mapping (IM) analysis (Lander and Botstein 1989). The analyses considered all markers of genetic linkage maps developed by Cavalcanti and Wilkinson (2007). The maps were constructed based on the methodology of the pseudo-testcross for markers segregating at a ratio of 1:1 (Grattapaglia and Sederoff 1994) and markers with different classes of segregation (ratio 3:1, 1:2:1 and 1:1:1: 1), according to Maliepaard et al. (1997). A total of 205 genetic markers were used, with 194 AFLP and 11 SSR. Two maps were constructed separately, representing each genome of the parents (i.e. CCP 1001 and CP 96). The QTL were detected and mapped using software MapQTL 5 (Van Ooijen 2004).
The interval mapping analysis was performed to identify QTL with significant main effects. The molecular markers of these regions were used as co-factors in the subsequent analysis of multiple QTL mapping to eliminate interferences between QTL in adjacent intervals, as recommended by Van Ooijen (2004). The critical LOD score equal to 3.0 was used as basis to identify QTL candidates. The QTLs detected were described by the markers closest to the region corresponding to the QTL. The QTL location was defined by confidence intervals of 1.0 and LOD score of 2.0, corresponding to a probability of 90 and 95 %, respectively. These were located on maps using software MapChart (Voorrips 2002). The proportion of phenotypic variation explained by the trait-marker association was estimated by the formula (Ukrainetz et al. 2008):  where R2 is the coefficient of determination for the proportion of variation explained by the QTL; SSG is the sum of squares of the genotype (QTL); and SSTotal is the total sum of squares.
where R2 is the coefficient of determination for the proportion of variation explained by the QTL; SSG is the sum of squares of the genotype (QTL); and SSTotal is the total sum of squares.
The phenotypic effect of the QTL-associated marker was estimated by the equation (Broman 2001):  where PE (%): percentage of phenotypic effect;
where PE (%): percentage of phenotypic effect;  gwm: mean of the subpopulation consisting of genotypes with the marker;and
gwm: mean of the subpopulation consisting of genotypes with the marker;and  gnm: mean of the subpopulation consisting of genotypes withno marker.
gnm: mean of the subpopulation consisting of genotypes withno marker.
RESULTS AND DISCUSSION
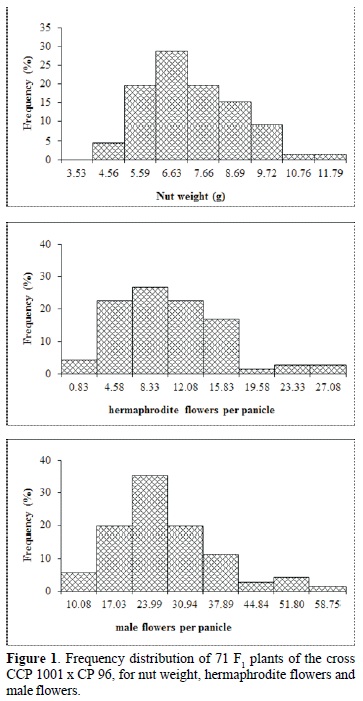
The range of variation in the plants was great for all traits (nut weight 4-11 g, male flowers 10-59, hermaphrodite flowers 1 -27), confirming the quantitative character of these traits and consequently demonstrating the viability of this population for genetic mapping studies to identify QTL associated to the traits under study (Figure 1).
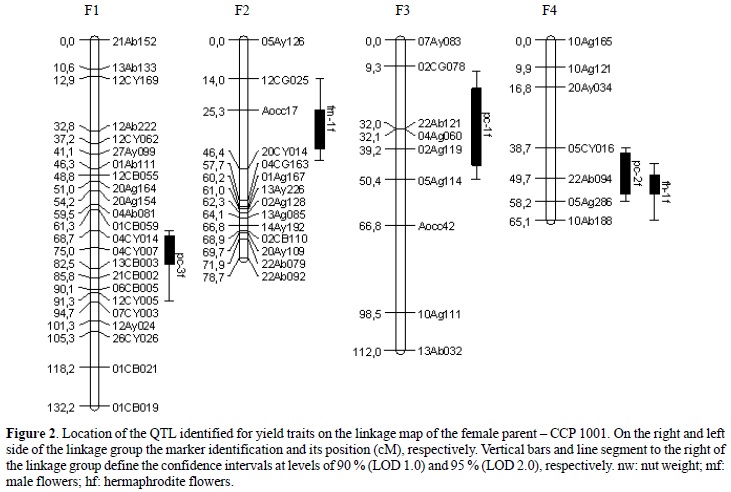
The QTL mapping analysis detected 11 loci with LOD score 3.0 (Table 1, Figures 2 and 3); three associated with QTL for nut weight (NW), four for male flowers (MF) and four for hermaphrodite flowers (HF).
The proportion of variation of a trait explained by an individual QTL is probably the most important obstacle to the implementation of marker-assisted selection. In this study, the percentages of phenotypic variation (R2) explained by the associations between the marker locus and the QTL ranged from 4.55 to 9.95 % for NW, 3.79 to 9.85 % for MF and 6, 04 to 12.98 % for HF (Table 1). Most QTL studies reveal moderate effects (R2 of 10 - 20 %) in one or few loci, while several other loci explain less than 10 % of the variation (Anderson et al. 2007), as similarly observed in this study, where only the R2 effects of the QTL hf-2f and hf-3m were moderate (12.98 and 10.11 %, respectively). Most QTL identified explained less than 10 % of the variation, i.e, small-effect QTL. Similar results have been reported in studies on QTL of yield traits in other crops (Siviero et al. 2002, Zalapa et al. 2007).
The identification of small-effect alleles by molecular markers can increase the efficiency of the selection process, improving the gains from selection by a marker-assisted selection program, using molecular data associated with phenotypic information (Melo et al. 2002). The phenotypic effect was estimated to determine the effect of indirect selection by means of a single marker for each QTL identified. The markers showed phenotypic effects ranging from 9.93 to 13.90 % for NW, -20.41 to 17.08 % for MF and -31.81 to 34.25 % for HF (Table 1). These results reflect the importance of the QTL identified in the trait expression and strongly support the existence of true QTL.
The purpose of the cashew breeding program is to increase the traitsNWand HF, and reduce MF. In this sense, for the first two,plants should be selected with the presence of markers associated with QTL with positive phenotypic effects (nw-1f, nw-2f, nw-3f, hf-1f, hf-3m and hf-4m) and absence of markers associated with QTL with negative effects (hf-2f). ForMF, plants with markers for negative QTL (mf-1f and mf-2m) and absence of markers for positive QTL (mf-3m and mf-4m) should be selected.
The low number of genotypes assessed certainly affected the estimates, particularly overestimating the QTLeffects and underestimating the number of QTL identified. Another important aspect is the effect of QTL x environment interaction, which according to Bernard (2008) may alter QTL detection and estimates when the evaluation environment is changed, although minor problems are expected for these traits, as the heritability estimates are high. Regardless of these considerations, the QTL detected in this study provide important information for the cashew breeding program.
It is therefore suggested that the QTLidentified should be validated using other populations and environments with a view to a routine use in cashew breeding program to identify desirable genotypes. This will increase the selection efficiency, as plants are selected when still in the nursery stage and the possibility of obtaining elite populations with higher yields and better product quality is greater.
CONCLUSIONS
1. Three QTL candidates were detected for nut weight, four for male flowers and four for hermaphrodite flowers, explaining 3.79 to 12.98 % of the total phenotypic variation associated with significant phenotypic effects.
2. These QTL can be used in marker-assisted selection. However, further studies are required to validate the QTL detected in this study.
ACKNOWLEDGEMENTS
The authors thank the National Council for Scientific and Technological Development (CNPq) for financial support.
Received 4 May 2010
Accepted 30 June 2011
- Anderson JA, Chao S and Liu S (2007) Molecular breeding using a major QTL for Fusarium head blight resistance in wheat. Crop Science 47: 112-119.
- Barros LM, Cavalcanti JJV, Paiva JR and Crisóstomo JR (2009) Hibridação de caju. In Borém A (ed.) Hibridação artificial de plantas Editora UFV, Viçosa, p. 214-250.
- Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Science 48: 1649-1664.
- Broman KW (2001) Review of statistical methods for QTL mapping in experimental crosses. Lab Animal 30: 44-52.
- Cavalcanti JJV and Wilkinson MJ (2007) The first genetic maps of cashew (Anacardium occidentale L.). Euphytica 157: 131-143.
- Cavalcanti JJV, Pinto CABP, Crisostómo JR and Ferreira DF (2000) Análise dialélica para avaliação de híbridos interpopulacionais de cajueiro. Pesquisa Agropecuária Brasileira 35: 1567-1575.
- Ferreira -Silva SL, Voigt EL, Viégas RA, Paiva JR and Silveira JAG (2009) Influência de porta-enxertos na resistência de mudas de cajueiro ao estresse salino. Pesquisa Agropecuária Brasileira 44: 361-367.
- Grattapaglia D and Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137: 1121-1137.
- Janser RC (1993) Interval mapping of multiple quantitative trait loci. Genetics 135: 205-211.
- Janser RC (1994) Controlling the type I and type II errors in mapping quantitative trait loci. Genetics 138: 871-881.
- Janser RC and Stam P (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447-1455.
- Kenis K and Keulemans J (2005) Genetic linkage maps of two apple cultivars (Malus x domestica Borkh.) based on AFLP and microsatellite markers. Molecular Breeding 15: 205-219.
- Lander ES and Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185-199.
- Maliepaard C, Janser J and Ooijen JWV (1997) Linkage analysis in a full-sib family of an outbreeding plant species: overview and consequences for applications. Genetical Research 70: 237-250
- Melo LC, Santos JB and Ferreira DF (2002) Mapping and stability of QTL for seed weight in common beans under different environments. Crop Breeding and Applied Biotechnology 2: 227-236.
- Paiva JR, Barros LM, Cavalcanti JJV, Lima AC, Corrêa MCM, Melo DS and Porto ZB (2005) Seleção de clones de cajueiro-anão precoce para plantio comercial no Município de Aracati, CE. Ciência Agronômica 36: 338-343.
- Paiva JR, Crisóstomo JR and Barros LM (2003) Recursos genéticos do cajueiro: coleta, conservação, caracterização e utilização. Embrapa Agroindústria Tropical, Fortaleza, 43p.
- Paula Pessoa PFA, Leite LAS and Pimentel CRM (1995) Situação atual e perspectivas da agroindústria do caju. In Araújo JPP and Silva VV (eds.) Cajucultura: modernas técnicas de produção Embrapa Agroindústria Tropical, Fortaleza, p. 23-42.
- Siviero A, Cristofani M, Boava LP and Machado MA (2002) Mapeamento de QTL associados à produção de frutos e sementes em híbridos de Citrus sunki vs. Poncirus trifoliata Revista Brasileira de Fruticultura 24: 741-743.
- Timmerman -Vaughan GM, Mills A, Whitfield C, Frew T, Butler R, Murray S, Lakeman M, Mccallum J, Russell A and Wilson D (2005) Linkage mapping of QTL for seed yield, yield components, and developmental traits in pea. Crop Science 45: 1136-1344.
- Ukrainetz NK, Ritland K and Mansfield SD (2008) Identification of quantitative trait loci for wood quality and growth across eight full-sib coastal Douglas-fir families. Tree Genetics & Genomes 4: 159-170.
- Van Ooijen JW (2004) MapQTL 5: Software for mapping of quantitative trait loci in experimental populations.
- Wageningen, Kyazma B.V. Voorrips RE (2002) MapChart. Software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93: 77-78.
- Wang D, Karle R and Iezzoni AF (2000) QTL analysis of flower and fruit traits in sour cherry. Theoretical and Applied Genetics 100: 535-544.
- Zalapa JE, Staub JE, McCreight JD, Chung SM and Cuevas H (2007) Detection of QTL for yield- related traits using recombinant inbred lines derived from exotic and elite US Western Shipping melon germplasm. Theoretical and Applied Genetics 114: 1185-1201.
Publication Dates
-
Publication in this collection
21 May 2012 -
Date of issue
Mar 2012
History
-
Received
04 May 2010 -
Accepted
30 June 2011