Abstracts
The genetic knowledge of the germplasm is crucial to the plant breeding in order todevelop new varieties. This study was carried out to evaluate the genetic similarity of 40 rice cultivarsdeveloped in Southern Brazil with the use of two tolls: genealogy and AFLP markers. The genealogy approach showed that 90% of the cultivars were developed from only six parents, revealing a narrow genetic basis among these cultivars.A little genetic diversity among them was confirmed by AFLP markers, since 111polymorphic markers identified 87% similarity among them. The geneticsimilarity among the cultivars released by Epagri in Santa Catarina state waslower, compared to that among the cultivars released by others public institutions, Embrapa and Irga. This study has demonstrated that rice cultivars developed in Southern Brazil have narrow genetic base, which suggests high genetic vulnerability risk.
Oryza sativa; genetic similarity; genealogy; AFLP marker; genetic vulnerability
O conhecimento da base genética das cultivares de arroz é crucial para programas de melhoramento como base para o desenvolvimento de novascultivares. Este estudo foirealizado para avaliar a similaridade genética de 40 cultivares de arroz desenvolvidas no Sul do Brasil com o uso de duas ferramentas:marcadores AFLP e genealogia. A abordagem genealógica mostrou que 90% das cultivares foram desenvolvidas a partir deapenas seis pais, revelando uma base genética estreita entre essas cultivares. Apouca diversidade genética entre eles foi confirmada por meio de marcadores AFLP, uma vezque 111 marcadores polimórficos identificaram 87% desimilaridade entre as cultivares. A similaridade genética entre as cultivares lançadas pela Epagri (Santa Catarina) foi menor, comparado às cultivares lançadas pelas instituições públicas Embrapa e Irga. Este estudo demonstrouque cultivares de arroz desenvolvidasno Sul do Brasil têm base genética estreita, o que sugererisco de alta vulnerabilidade genética.
Oryza sativa; similaridade genética; genealogia; marcadores AFLP; vulnerabilidade genética
NOTE
Genetic base of paddy rice cultivars of Southern Brazil
Base genética das cultivares de arroz irrigado do sul do Brasil
Juliana Vieira RaimondiI; Rubens MarschalekII; Rubens Onofre NodariIII, * * E-mail: rubens.nodari@ufsc.br
IFaculdade Avantis, Av. Marginal Leste, 3600, km 132, Bairro dos Estados, 88.339-125, Balneário Camboriú, SC, Brazil
IIEmpresa de Pesquisa Agropecuária e Extensão Rural deSanta Catarina (Epagri), Estação Experimental de Itajaí, Rodovia Antonio Heil, km 6, Itaipava, 88.301-970, Itajaí, SC, Brazil
IIIUniversidade Federal de Santa Catarina, C.P. 476, 88.040-900, Florianópolis, SC, Brazil
ABSTRACT
The genetic knowledge of the germplasm is crucial to the plant breeding in order to develop new varieties. This study was carried out to evaluate the genetic similarity of 40 rice cultivars developed in Southern Brazil with the use of two tolls: genealogy and AFLP markers. The genealogy approach showed that 90% of the cultivars were developed from only six parents, revealing a narrow genetic basis among these cultivars.A little genetic diversity among them was confirmed by AFLP markers, since 111polymorphic markers identified 87% similarity among them. The genetic similarity among the cultivars released by Epagri in Santa Catarina state was lower, compared to that among the cultivars released by others publicinstitutions, Embrapa and Irga. This study has demonstrated that rice cultivars developed in Southern Brazil have narrow genetic base, which suggests high genetic vulnerability risk.
Key words: Oryza sativa, genetic similarity, genealogy, AFLP marker, genetic vulnerability
RESUMO
O conhecimento da base genética das cultivares de arroz é crucial para programas de melhoramento como base para o desenvolvimento de novascultivares. Este estudo foirealizado para avaliar a similaridade genética de 40 cultivares de arroz desenvolvidas no Sul do Brasil com o uso de duas ferramentas:marcadores AFLP e genealogia. A abordagem genealógica mostrou que 90% das cultivares foram desenvolvidas a partir deapenas seis pais, revelando uma base genética estreita entre essas cultivares. Apouca diversidade genética entre eles foi confirmada por meio de marcadores AFLP, uma vezque 111 marcadores polimórficos identificaram 87% desimilaridade entre as cultivares. A similaridade genética entre as cultivares lançadas pela Epagri (Santa Catarina) foi menor, comparado às cultivares lançadas pelas instituições públicas Embrapa e Irga. Este estudo demonstrouque cultivares de arroz desenvolvidasno Sul do Brasil têm base genética estreita, o que sugererisco de alta vulnerabilidade genética.
Palavras-chave : Oryza sativa, similaridade genética, genealogia, marcadores AFLP, vulnerabilidade genética
INTRODUCTION
Rice (Oryzasativa L) is one of the most important human foods being a source of energyfor two thirds of the world population. According to FAO (2011) and USDA(2011), the world rice production was 674.68 million tons (mt) in the cropseason 2010/11, and the three major rice producers in 2010/11 were China Indiaand Indonesia, corresponding to 30.4%, 21%, and 8.2%, respectively. Brazil with14, 17 mt produces 2.1% of the world rice production (FAO 2011, USDA 2011), being the states of Rio Grande do Sul (RS) and Santa Catarina (SC) the biggestrice producers (61% and 8.4% of the total production), respectively (ICEPA2009).
In Southern Brazil, rice is cultivated under waterlogged conditions in SC, whereall rice area is water-seeded; the seeds are pre-germinated and broadcastedover the water-leveled fields. On the other hand, in the state of RS, 93% ofthe rice is seeded in dry land and 7% is water-seeded. The water-seededcultivated system is an important tool to control weeds, mainly red rice.Another difference between these two states is the preference for earlyvarieties in RS, while late varieties are better adapted in SC.
Previous studies have demonstrated that Brazilian rice cultivars have a narrow geneticbase (Cuevas-Perez et al. 1992, Rangel et al. 1996). This status raisesconcerns like, among others, reduction in genetic variability to improvement, approaching yield plateau, increasing vulnerability to diseases, andcontributes to genetic vulnerability. Genetic vulnerability results when anorganism lacks genes that enable it to withstand an environmental or biologicalstress or when its genes confer susceptibility to a pest, pathogen, or naturalstress (National Research Council 1992), which can leads to crop vulnerabilitythat is a measure of the susceptibility of a crop species to catastrophicdecline in its production (National Research Council 1992).
The knowledge about the pedigree of the rice cultivars presently under cultivationis also an important tool to help breeders to select parents for thedevelopment of new cultivars. Complementary to the genealogy approach, the useof molecular markers also provides fastness, consistency and efficiency, because it is free from environmental effects. Thus, the objective of thepresent work was to evaluate the genetic base of the rice cultivars currentlyin use in Southern Brazil, based on their genealogy and with the use of AFLPmolecular markers.
MATERIAL AND METHODS
Forty rice cultivars, recommended forcultivation in Southern Brazil, were used (Table 1) for the genealogical analysis.They were collected from the germplasm bank of Epagri, located at Itajaí, SC, Brazil. The genealogy ofthe cultivars were described through information provided by rice researchinstitutes (Epagri, Embrapa and IRGA), or quoted in the books of rice crosses(INGER 1991, CIAT 1995). Due to seed germination problems, only 36 cultivarswere analyzed by AFLP markers. Four new Epagri's advanced lines were alsoincluded in the molecular analysis.
The DNA was extracted from leaf tissues according to the protocol of Doyleand Doyle (1987). All the AFLP steps followed the protocol developed by Vos etal. (1995). The DNA (25 ng) was digested with Eco RI and MseI (10 U µL-1each) in buffer solution and 10 % water. Four AFLP primer combinations were tested(E40 x M62, E13x M60, E40 x M59 e E40 x M48) with markers distributed over the 12 ricechromosomes. To resolve and visualize PCR products, it was used6% polyacrylamide gel in vertical electrophoresis apparatus. Afterelectrophoresis, polyacrylamide gels underwent the fixation process, wereprocessed with silver nitrate (Blum et al. 1987), dried at room temperature, photographed and scanned. AFLP bands were organizedin the form of a binary matrix and analyzed (PAUP (Phylogenetic Analysis Using Parsimony)(Felsenstein 1985) to generate the distance matrix. The distancematrix obtained was used to construct the dissimilarity dendrogram (Kumaret al. 2008) using the distance method with the NeighbourJoining clustering algorithm (Saitou and Nei1987). The cophenetic correlation was estimated by using NTSYS(Rohlf 2000).
RESULTS AND DISCUSSION
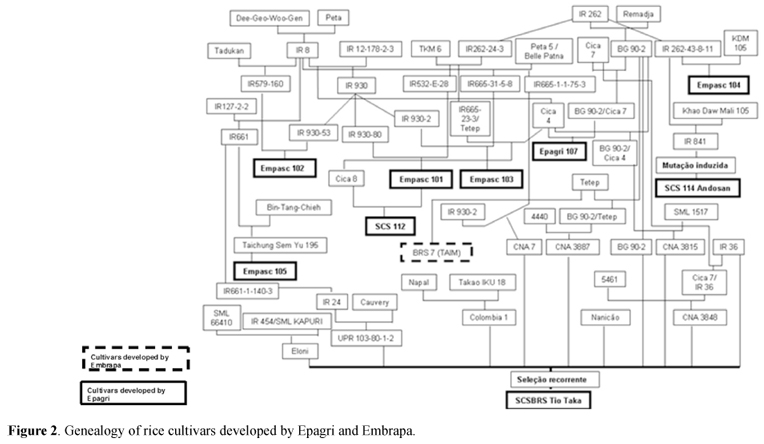
The analysis of the genealogy of rice cultivars cultivated in Southern Brazil demonstrated that 82.5% of the cultivars were originated from fiveancestors involved in two crosses (I = IR262/Remadja, II = Dee-Geo-Woo-Gen/Peta 5) and one line IR 1561. New cultivars developed by IRGA derived mainly (84.2%) from the cross II, involving only two ancestral parents (Figure 1). Cultivars developed by Epagrioriginated from cross I (12.5%), cross II (18.75%), cross III (31.25%), anddouble crosses between I and II (25%) (Figures 2 and 3). Embrapa cultivars were also derived from cross I (25%) and cross II(25%) (Figures 1 and 2). The cultivarArrank, developed by Syngenta, originated from a selection of BR Irga 410cultivar, whom also derives from the cross II.
IR8 wasthe first modern rice cultivar released in the 60's, during the greenrevolution period. IR8 was obtained from the cross between Peta and DeeGeo Woo Gen. Peta derived from crosses between Cina and Lati Sail, while DeeGeo Woo Gen derived directly from natural mutation, often utilized as source ofsemi-dwarfness characteristic to the rice plants. Following IR8, several companies throughout the world have looked for new varieties using thisgenotype as a source of genes for high yield, better nutrient absorptioncapacity and semi-dwarf plant type. For that reason, most of the rice cultivars released by Epagri, IRGA, Embrapa and Syngenta toSouthern Brazil has alleles of IR8 or theirs descends in their geneticbackground.
Th enumber of parents utilized in the development of the cultivars varied accordingto the research institution. Epagri has involved 96 parents in the developmentof 16 cultivars; while IRGA, despite having released 19 cultivars, used 38 parents. Embrapa involved 27 genitors to develop five cultivars studied in thepresent work (Figures 1 to 3).
The genealogy analysis revealed also an expressive degree of kinship among ricecommercial varieties. Specifically, there is a kinship between Embrapa andIrga cultivars (e.g. BRS Pelotas and BR Irga 410, BRS Firmeza and BR Irga 411), or Epagri cultivars (e.g. BRS-7 Taim and SCS BRS Tio Taka). The breedingprogram of Epagri released two sib cultivars, out of 16 ones: Epagri 108 andEpagri 109. In addition, the cultivar SCS 115 CL was derived from a cross between Epagri 109 andAS3510, while SCS 112 was originated from the cross Empasc 101 X Cica 8.
In the present work, it was found the narrowing of the genetic base of theimproved cultivars. The main factor was the few genetic resources used so far.In addition, the use of Pedigree as a selection method in all rice breedingprograms analyzed in this study could be contributed to develop relative siblines, half-sibs and less degree of kinship were found among the improvedreleased cultivars. The remarkable lack of diverse genetic resources used wasfound among Irga's cultivars, since 16 out of 19 ones share two parents, beingone of them IR8. In addition, other seven cultivars also derived fromIR8, which totalized 23 out of 40 cultivars thathave one shared parental.
In addition to the genealogy, the genetic similarity of these cultivarswas analyzed by 111 AFLP primers (being 91% polymorphic ones). The high genetic similarity among ricecultivars demonstrated through AFLP markers is in agreement with the narrowed geneticbase presented by the genealogical analysis. For example, the similaritybetween the cultivar BR Irga 415 and BR Irga 409 (92.7% by AFLP markers) isprobably explained by the fact that they are originated from the same cross (BRIrga 409 with Cica 9). In addition, Embrapa cultivars proved to be very similar to Irga'smaterial by AFLP markers, just as described in the genealogy studies.
The dendrogram based on those AFLP markers (Figure 4) showed the formation of three major groups(A, B and C) with 87% similarity between them. Group A includesSCS 114 Andosan, SCS 115 CL and other Epagri lines. In group B, there are alsoother Epagri cultivars and in C groups, Irga, Embrapa and Syngenta cultivars. GroupA consists of four Epagri lines, originated from crosses between divergent parents, and the cultivars SCS 115 SCS CL and SCS 114 Andosan (originated from inducedmutation). SCS 115 CL was obtained from a cross between Epagri 109 cultivar andthe access AS 3510.
Overall, the clustersanalysis based on AFLP markers also reinforced the lack of significantdiversity. However, a relatively wide divergence among cultivars was found ingroup A, compared to B and C groups, which can be due to the broad source ofgermplasm utilized in Epagri breeding program, and also to the use of inducedmutations, comparatively to the other institution breeding programs. Thus, among Epagri cultivars, the kinship isalso evident, but at minor scale.
Using the distance matrix method analysis, the greatest variability wasfound between Epagri lines and cultivars. Rice line SC 355 with cultivar Empasc100 presented 49% divergence. The divergence was 46% between the line SC 213and the cultivar Empasc 100, and between cultivars Empasc 101 and SCS 115 CL. The smallest genetic divergence was found tobe 7.3% between SCS 115 CL and SC 213, between BR Irga 409 and BR Irga 414 andBR Irga 414 and BR Irga 415. Cultivars BR Irga 409 and BR Irga 414 arehalf-sibs, according to genealogical studies.
The genetic divergence among cultivars within each institution was higheramong cultivars and lines of Epagri (0.272), followed by Embrapa (0.163) andIrga (0.145), respectively. The geneticvariability among institution cultivars was greater between Epagri and Embrapa(31%) and lower between Irga and Embrapa (17%).
The genetic similarity between Brazilian cultivars was considered highin comparison to the others. Lapitan et al. (2007) found 35% similarity amongrice cultivars marketed in the Philippines. Wang and Lu (2006) verified that thecoefficient of parentage for 100 parental lines of hybrid rice widely used inhybrid breeding and commercial production during 19762003 in China was low(0.056). The authors attributed the presence of a 49% of the all pairs ofparental lines being completely unrelated to a continual incorporation offoreign germplasm (wild rice and japonica and javanica, among others) into thegenetic base over time. Previous studies in Brazil found similar results to thepresent work, as the one done by Rangel et al. (1996). Inaddition, Malone et al. (2006) usingAFLP markers concluded that the improved Brazilian rice cultivars presenteddiversity of 57%. Based on the genealogy ofrice cultivars, Cuevas-Perez et al. (1992) found 101 ancestors as the genetic base of all improved new cultivarsdeveloped for Latin America and the Caribbean countries from 1971 to 1989. Inthat work, the authors found also that around 56% of the alleles present in thedeveloped cultivars were directly or indirectly originated from IR8. It isrelevant to point out that not always there is a 50%:50% passing alleles fromthe parents to the progenies. However, the effect of selection specific traitsusually results in the higher frequency than 50% of alleles from the parentwith best quality, which may be was the case.
Taking into account that genetic vulnerability results from the improper or inadequatedeployment of these genetic resources, in conjunction with biotic and abioticstresses (National Research Council 1993), it can be claim that the geneticbase of improved rice cultivars to Southern Brazil approaches to a situation ofgenetic vulnerability. Concerns related to the consequences of geneticvulnerability are not new neither should be considered negligible in terms offood security. Since rice is the source of calories for a great part of theincreasing Brazilian population, new rice cultivars should present higher yieldpotential. The low genetic variability among cultivars mayprevent significant increases in yield (Breseghello et al. 1999, Silva et al. 1999) if no new and divergent germplasm will be used in breedingprograms. Although highest in the past 10 years, the paddy rice productivitykept stagnant in that decade, fact that do not allow to exclude association tonarrow genetic base of the cultivars.
The main conclusion of this work is that rice cultivars developed for Southern Brazil have narrow genetic basis.The current situation characterizes a genetic vulnerability, which favorepidermises and no or small genetic gain towards selection. Thus, it is recommendedto widen the utilization of rice genetic resources in order to take advantageof the genetic variability of the Oryza genus.
ACKNOWLEDGEMENTS
The authors acknowledge EPAGRI for the financial support. The Brazilian CNPq provided scholarship to RON.
Received 22 October 2013
Accepted 09 February 2014
- Blum H, Beier H and Gross HJ (1987)Improved silver staining of plant proteins RNA and DNA in polyacrylamide gels. Electrophoresis8: 93-99.
- Breseghello F, Rangel PH and Morais OP(1999) Ganho de produtividade pelo melhoramento genético do arroz irrigado nonordeste do Brasil. Pesquisa Agropecuária Brasileira 34: 399-407.
- CIAT (1995) Registro de cruzamentos deArroz CIAT, Cali, 313p.
- Cuevas-Perez FE, Guimarães EP, Berrio LEand Gozález DI (1992) Genetic base of irrigated Rice in Latin America and theCaribbean 1971 to 1989. Crop Science 32: 1054-1059.
- Doyle JJ and Doyle JL (1987) A rapid DNAisolation procedure for small quantities of fresh leaf tissue. PhytochemicalBulletin 19: 11-15.
- FAO Food and Agriculture Organization(2011) Rice market monitor XIV(2). FAO, Rome, 37p.
- Felsenstein J (1985) Confidence limits onphylogenies: An approach using the bootstrap. Evolution 39:783-791.
- INGER - Red International para laEvaluación Genética del Arroz (1991) Cruzamentos de arroz: América LatinaCIAT, Cali, 426p.
- ICEPA - Centro de Socioeconomia ePlanejamento Agrícola (2009) Síntese anual da agricultura de Santa Catarina2009-2010 ICEPA. Florianópolis, 315p.
- Kumar S, Nei M, Dudley J and Tamura K(2008) MEGA: A biologist-centric software for evolutionary analysis of DNA andprotein sequences. Briefings in Bioinformatics 9: 299-306.
- Lapitan VC, Brar DS, Abe T and Redońa ED(2007) Assessment of genetic diversity of Philippine rice cultivars carryinggood quality traits using SSR markers. Breeding Science 57: 263-270.
- Malone G, Zimmer PD, Kopp MM, Mattos LAT, Carvalho FIF and Oliveira AC (2006) Assessment of the genetic variability amongrice cultivars revealed by amplified fragment length polymorphism AFLP. Revista Brasileira Agrociências 12: 21-25.
- National Research Council (1972) Genetic vulnerability of major crops National Academy of Sciences, WashingtonD.C., 307p.
- National Research Council (1993) Managingglobal genetic resources National Academy of Sciences, Washington, D.C., 476p.
- Rangel PHN, Guimarães EP and Neves PCF(1996) Base genética das cultivares de arroz Oryza sativa L. irrigado doBrasil. Pesquisa Agropecuária Brasileira 31: 349-347.
- Rohlf FJ (2000) NTSYS-pc: numerical taxonomy andmultivariate analysis system, version 2.1 Exeter Software, New York, 98p.
- Saitou N and Nei M (1987) Theneighbor-joining method: A new method for reconstructing phylogenetic trees. MolecularBiology and Evolution 4: 406-425.
- Silva EF, Montalván R and Ando A (1999)Genealogia dos cultivares brasileiros de arroz-de-sequeiro. Bragantia 58:281-286.
- USDA United State Department ofAgriculture (2011) Rice yearbook: Report USDA, Ithaca, 28p.
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M and Zabeau M(1995) AFLP a new technique for DNA-fingerprinting. Nucleic Acids Research 23: 4407-4414.
- Wang S and Lu Z (2006) Genetic diversityamong parental lines of Indica hybrid rice (Oryza sativa L.) in Chinabased on coefficient of parentage. Plant Breeding 125: 606-612.
Publication Dates
-
Publication in this collection
25 Nov 2014 -
Date of issue
Oct 2014
History
-
Received
22 Oct 2013 -
Accepted
09 Feb 2014