Abstract
The aim of this study was to perform chromosome counts and nuclear DNA quantification of the unconventional vegetables species: bertalha (Basella alba L.), vinagreira verde (Hibiscus sabdariffa L.), azedinha (Rumex acetosa L.), peixinho (Stachys byzantina K. Koch), and capuchinha (Tropaeolum majus L.). Metaphase chromosomes were obtained from the pre-treatment of root meristem with 8-hydroxyquinoline or colchicine and lides were prepared by the flame-drying technique and stained with 5% Giemsa. DNA quantification was performed by flow cytometry. Chromosome number and DNA content (pg) estimated for each species were: Rumex acetosa: 2n=2x=14 and 7.04 pg; Basella alba: 2n=2x=44 and 7.05 pg; Tropaeolum majus: 2n=2x=28 and 2.08 pg; Stachys byzantine: 2n=2x=30 and 1.54 pg, and Hibiscus sabdariffa: 2n=4x=72 and 5.12 pg.
Key words:
Underutilized vegetables; germplasm; plant breeding; cytogenetic analysis
INTRODUCTION
Some vegetable species have already been widely cultivated and used in human feeding; however, due to several factors, consumption of these plants has become restricted to some traditional populations. Thus, they have been denominated as traditional or unconventional (Brazil 2010BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2010) Hortaliças Não Convencionais. MAPA, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 54p. , 2013BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2013) Manual de hortaliças não convencionais. MAPA/ACS, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 99p. ). In Brazil, azedinha (Rumex acetosa L.), bertalha (Basella alba L.), capuchinha (Tropaeolum majus L.), peixinho (Stachys byzantina K. Koch), vinagreira verde (Hibiscus sabdariffa L.), among others (Brasil 2013BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2013) Manual de hortaliças não convencionais. MAPA/ACS, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 99p. , Kinupp and Lorenzi 2014Kinupp VF and Lorenzi H (2014) Plantas alimentícias não convencionais (PANC) no Brasil guia de identificação, aspectos nutricionais e receitas ilustradas:. Instituto Plantarum de Estudos da Flora, São Paulo, 768p.) are some examples of unconventional vegetables. Nevertheless, despite the government's incentive to rescue and reintroduce these species in the diet (Cardoso 1997Cardoso MO (1997) Hortaliças não-convencionais da amazônia. Embrapa, Serviço de Produção de Informação, Brasília, 152p. , Brasil 2002BRASIL - Ministério da Saúde (2002) Alimentos regionais brasileiros. Secretaria de Políticas de Saúde, Coordenação-Geral da Política de Alimentação e Nutrição, Brasília, 141p., Brasil 2010BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2010) Hortaliças Não Convencionais. MAPA, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 54p. , 2013BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2013) Manual de hortaliças não convencionais. MAPA/ACS, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 99p. ), little technical or scientific information is available in relation to the introduction of these species in Brazilian soils and to the origin of their propagative materials.
The knowledge on the taxonomy and genetic diversity of unconventional vegetable varieties grown in Brazil is also limited. For this reason, these crops have been produced, in most cases, without proper agronomic management, using mixtures of propagating materials, with no quality standards. In addition, little information on the similarities and genetic differences presented by these species is reported in the literature.
Aiming at genetic improvement, the cytogenetic characterization of species that present great genetic variability and have little evolutionary mechanisms studied is fundamental. This procedure requires more in-depth studies on the number and behavior of chromosomes and on the level of ploidy of these species and their morphogenic varieties.
For some unconventional vegetables, only the chromosome number has been studied; however, works are in general related to plants/genotypes collected in other countries. Rumex acetosa, for instance, presents complex chromosomal mechanism for the sex-determination of its plants and different levels of ploidy (Mariotti et al. 2006Mariotti B, Navajas-Pérez R, Lozano R, Parker JS, De La Herrán R, Rejón CR, Rejón MR, Garrido-Ramos M and Jamilena M (2006) Cloning and characterization of dispersed repetitive DNA derived from microdissected sex chromosomes of Rumex acetosa. Genome 49: 114-121. , Navajas-Pérez et al. 2006Navajas-Pérez R, Schwarzacher T, De La Herrán R, Rejón CR, Rejón MR and Garrido-Ramos MA (2006) The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61-71., Błocka-Wandas et al. 2007Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K and Joachimiak AJ (2007) Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction 20: 171-180. , Cuñado et al. 2007Cuñado N, Navajas-Pérez R, De La Herrán R, Rejón CR, Rejón MR, Santos JL and Garrido-Ramos MA (2007) The evolution of sex chromosomes in the genus Rumex (Polygonaceae): identification of a new species with heteromorphic sex chromosomes. Chromosome research 15: 825-833. ).
Cytogenetic studies are also incipient in the case of Basella alba. The literature reports different results for the chromosome number of this species (Hanson et al. 2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ), varying between 44, 45 and 48 chromosomes, as well as for the quantification of its DNA, ranging from 5.17 pg to 7.17 pg (Grasso and Morpurgo 1997Grasso G, Van Duren M, Lee KS and Morpurgo R (1997) DNA analysis in three populations of african spinach (Basella spp.). In FAO/IAEA (org) Improvement of basic food crops in Africa through plant breeding, including the use of induced mutations. IAEA, Viena, p. 39-43., Hanson et al. 2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ). For Tropaeolum majus, some works have indicated 28 chromosomes in varieties studied in Russia and China; however, only Nagl et al. (1976Nagl W (1976) Early embryogenesis in Tropaeolum majus L.: Evolution of DNA content and polyteny in the suspensor. Plant Science Letters 7: 1-6. ) reported the DNA quantification of this species, with 2.66 pg. In Stachys byzantine cultivated in Turkey, Baltisberger (2002Baltisberger M (2002) Cytological investigations on some Albanian plant species. Candollea 56: 245-259. ) and Martin (2011Martin E, Çetin Ö, Akcicek E and Dirmenci T (2011) New chromosome counts of genus Stachys (Lamiaceae) from Turkey. Turkish Journal of Botany 35: 671-680. ) observed 15 pairs of chromosomes; however, DNA quantification of this species has not been reported yet. For Hibiscus sabdariffa, Guerra (2009Guerra NA (2009) Estudios citogenéticos de Hibiscus sabdariffa L. (Malvaceae). Revista Científica UDO Agrícola 9: 595-598.) and Hiron et al. (2006Hiron N, Alam N, Ahmed FA, Begum R and Alam SS (2006) Differential fluorescent banding and isozyme assay of Hibiscus cannabinus L. and H. sabdariffa L. (Malvaceae). Cytologia 71: 175-180. ) observed a total of 72 chromosomes in varieties cultivated in Venezuela and in Blangadesh, respectively; however, the DNA quantification of this plant has not been reported yet.
Many of these studies report different data for the chromosome number and DNA content in the different varieties of these cultivated species. Despite the cytogenetic studies on unconventional vegetables carried out at the international level, unconventional vegetables have not been studied in details in Brazil. In relation to the DNA content, even though it tends to remain constant within the species, some exceptions may occur owing to variations in the genome of the species. In many cases, this intraspecific variation is due to chromosome variation, aneuploidy, presence of supernumerary chromosomes, loss or duplication of chromosome segments, and highly repetitive DNA (Navarini et al. 2008Navarini APG, Schifino-Wittmann MT, De Barros IBI and Almeida D (2008) Cytogenetics of Hypericum caprifoliatum Cham. and Schltdl. (Clusiaceae) populations and other species of the genus. Crop Breeding and Applied Biotechnology 8: 330-338., Olkoski and Wittmann 2011Olkoski D and Wittmann MTS (2011) Cytogenetics of Mimosa bimucronata (DC.) O. Kuntze (Mimosoideae, Leguminosae): chromosome number, polysomaty and meiosis. Crop Breeding and Applied Biotechnology 11: 27-36., Souza-Chies et al. 2014Souza-Chies TT, Burchardt P, Alves SEM, Essi L and Dos Santos EK (2014) O estudo da biodiversidade e evolução vegetal através de marcadores de DNA e citogenética: exemplos em Iridaceae e Poaceae. Ciência e Natura 36: 279-293.).
Most of the unconventional vegetable species in Brazil are of unknown origin, and they are generally propagated without quality standard control of the varieties that are traditionally grown by locals or by specific regions and cultures in the country. Knowing the quantity of nuclear DNA and the chromosome number of each species is fundamental and is considered as the initial step of a breeding program, especially for species that present great genetic variability and whose evolutionary mechanisms are little studied, such as the unconventional vegetables. Thus, cytogenetic research of unconventional vegetables grown in Brazil is necessary.
The objective of this work was to determine the chromosome number and the quantity of DNA of unconventional vegetables collected in Brazil.
MATERIAL AND METHODS
The present study analyzed the species Basella alba L. syn B. rubra, Hibiscus sabdariffa L., Rumex acetosa L., Stachys byzantina K. Koch and Tropaeolum majus L. In Brazil, these species are known as bertalha (vine spinach), vingareira verde (roselle), azedinha (sorrel), peixinho (ear lamb’s), and capuchinha (nasturtium), respectively.
All the plants come from the collection of unconventional vegetable germplasm, located in the experimental garden of the Federal University of Lavras (lat 21º 14' S, long 45º 00' W, alt 918 m asl) in Minas gerais State, Brazil. The climate of the region is classified as Cwa, according to Köppen, considered as temperate humid, with hot summer and dry winter.
Root tips of Rumex acetosa, tropaeolum majus and Stachys byzantina were subjected to mitotic block treatment with 0.002 M Colchicine for 5 h at a temperature of 25 °C. For Basella alba and Hibiscus sabdariffa, pretreatment was performed with 0.002 M 8 -hydroxyquinoline (8-H) for 24 h at 10 °C. Subsequently, root tips were fixed in Carnoy 3:1 (absolute ethanol: glacial acetic acid-1 v v-1) for 24 h at room temperature, and then stored at -20 °C until slides preparation.
To assemble the slides, roots were subjected to enzymatic digestion with pectinase and cellulase (100U 200U-1) for 35 minutes at 37 ºC. Slides were prepared using flame-drying (Dong et al. 2001Dong F, Mcgrath JM, Helgeson JP and Jiang J (2001) The genetic identity of alien chromosomes in potato breeding lines revealed by sequential GISH and FISH analyses using chromosome-specific cytogenetic DNA markers. Genome 44: 729-734.) and squash (Guerra and Souza 2002Guerra M, Souza MD (2002) Como observar cromossomos: um guia de técnicas em citogenética vegetal, animal e humana. FUNPEC, Ribeirão Preto, 131p.) techniques, and stained with 5% Giemsa for 4 minutes. The best metaphases of each species were captured on a light field microscope (Leica DMLS) equipped with microcameras (Nikon Digital Sight DS-Fi1), and the images were processed using the Corel Photo Paint software.
DNA content was determined from leaf samples of the species and from the standard reference plant (Pisum sativum L.). The analyses protocol followed the methodology of Galbraith et al. (2001Galbraith DW, Lambert GM, Macas J and Dolezel J (2001) Analysis of nuclear DNA content and ploidy in higher plants. Current Protocols In Cytometry 22: 7-6.), with the mechanical isolation of the nuclei of the foliar tissues of the plants. Subsequently, the nuclear suspension was filtered and stained with 25 μL of propidium iodide.
Analyses were performed in a FacsCaliburTM 4 Cores - BD (Becton Dickinson) flow cytometer, and the histograms were obtained using the Cell Quest and FlowJo X software (Tree Star, Ashland, OR). Nuclear DNA content (pg) of the samples was estimated based on the analysis of at least 10,000 cells by comparing with the position in relation to the G1 peak of the internal reference standard obtained for the standard Pisum sativum.
RESULTS AND DISCUSSION
DNA content ranged from 1.54 pg in Stachys byzantina to 7.05 pg in Basella alba. Chromosome number ranged from 2n=14 in Rumex acetosa to 2n=72 in Hibiscus sabdariffa (Table 1, Figures 1 and 2).
Results of the flow cytometry and nuclear DNA quantification data of the Unconventional Vegetables and of the standard (Pisum sativum L.) used in the study
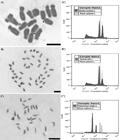
Mitotic metaphases and reflectance peak graphs of nuclear DNA of the species: A) Stachys byzantina K. Koch (2n= 30) and B) Hibiscus sabdariffa L. (2n=72). Bar: 5µm.
The chromosome number 2n=14 observed in Rumex acetosa is similar to the results presented by Ainsworth et al. (2005Ainsworth C, Rahman A, Parker J and Edwards G (2005) Intersex inflorescences of Rumex acetosa demonstrate that sex determination is unique to each flower. New phytologist 165: 711-720. ) and by Mariotti et al. (2006Mariotti B, Navajas-Pérez R, Lozano R, Parker JS, De La Herrán R, Rejón CR, Rejón MR, Garrido-Ramos M and Jamilena M (2006) Cloning and characterization of dispersed repetitive DNA derived from microdissected sex chromosomes of Rumex acetosa. Genome 49: 114-121. ), obtained for diploid and female varieties of this species, in England and Spain, respectively. According to these authors, Rumex acetosa, in its diploid varieties, presents 2n=12 + XY1Y2 and 2n=12 + XX chromosomes for male and female plants, respectively. This information indicates that the plant population evaluated in this work was composed of female plants, with 2n=12 + XX, whose sex chromosomes can be easily identified (Figure 1) by their morphology and size, according to Ainsworth et al. (2005).
Mitotic metaphase and reflectance peaks graphs in the nuclear DNA of the species A) Rumex acetosa L., (2n= 14), arrows indicate sex chromosomes, B) Basella alba L. (2n=44), and C) Tropaeolum majus L. (2n=28). Bar: 5µm.
Results obtained by flow cytometry showed that the studied variety of Rumex acetosa presented, on average, 7.04 pg of DNA. Similar result was obtained by Błocka-Wandas et al. (2007Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K and Joachimiak AJ (2007) Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction 20: 171-180. ), who reported that female plants of this species cultivated in Poland presented 7 pg of nuclear DNA. On the other hand, male plants with an additional chromosome presented, on average, 7.5 pg of nuclear DNA (Błocka-Wandas et al. 2007Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K and Joachimiak AJ (2007) Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction 20: 171-180. ).
Cytogenetic evaluations of Rumex acetosa were carried out with samples of the population of a variety cultivated locally. Therefore, these evaluations had the objective of developing a breeding program; exploring the genetic variability by crosses; producing seeds of this species; and studying a greater number of plants, in order to verify the occurrence of male plants in this population.
The Basella alba plants evaluated in this study had 44 chromosomes in all metaphases. The literature presents some divergence for the description of the chromosome number of this species. Differently from the chromosome number observed here, Hanson et al. (2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ) reported 2n=48, but they also observed plants with 2n=41 and 2n=44. Considering that the basic number of chromosomes of this genus is x=12 (Sperling and Bittrich 1993Sperling CR, Bittrich V (1993) Basellaceae. Flowering plants: dicotyledons. Springer Berlin Heidelberg: 143-146.), the species evaluated by Hanson et al. (2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ) would be tetraploid (2n=4x=48), and the Basella alba plants evaluated in this study could have been derived by descending dysploidy, with reduction from 48 to 44 chromosomes.
Several mechanisms may be involved in the variation of the number of chromosomes within a single species, especially dysploidy and polyploidy (Guerra 2008Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenetic and Genome Research 120: 339-350., Lysák and Schubert 2013Lysák MA and Schubert I (2013) Mechanisms of chromosome rearrangements. Plant Genome Diversity 2: 137-147., Souza-Chies et al. 2014Souza-Chies TT, Burchardt P, Alves SEM, Essi L and Dos Santos EK (2014) O estudo da biodiversidade e evolução vegetal através de marcadores de DNA e citogenética: exemplos em Iridaceae e Poaceae. Ciência e Natura 36: 279-293.), and variations in the levels of ploidy occur regularly during the development of all plants. Dysploidy represents an increase or decrease in the number of chromosomes without changing the quantity of DNA as a consequence of complex mechanisms, often involving structural rearrangements at subchromosomal levels, many of which are still unknown to several plant genera.
The nuclear DNA content of the studied variety Basella alba was of 7.05 pg. Hanson et al. reported the quantification of the nuclear DNA of this species in varieties from the Royal Botanic Gardens Kew, England (http://www.kew.org/), which presented a mean of 7.13 pg of cDNA. Grasso and Morpurgo (1997Grasso G, Van Duren M, Lee KS and Morpurgo R (1997) DNA analysis in three populations of african spinach (Basella spp.). In FAO/IAEA (org) Improvement of basic food crops in Africa through plant breeding, including the use of induced mutations. IAEA, Viena, p. 39-43.) also quantified nuclear DNA in three cultivated and native African varieties of Basella alba L. (syn B. rubra), ranging between 6.83, 5.73 and 6.83 pg. The variations observed for the quantification of DNA between different works can be explained by genotypic variations among varieties, as suggested by Grasso and Morpurgo (1997Grasso G, Van Duren M, Lee KS and Morpurgo R (1997) DNA analysis in three populations of african spinach (Basella spp.). In FAO/IAEA (org) Improvement of basic food crops in Africa through plant breeding, including the use of induced mutations. IAEA, Viena, p. 39-43.), or by differences in the level of ploidy, among other factors. Grasso and Morpurgo (1997Grasso G, Van Duren M, Lee KS and Morpurgo R (1997) DNA analysis in three populations of african spinach (Basella spp.). In FAO/IAEA (org) Improvement of basic food crops in Africa through plant breeding, including the use of induced mutations. IAEA, Viena, p. 39-43.) did not describe the chromosome number of the species studied; thus, it is not possible to infer if the observed variations in nuclear DNA content among the plants evaluated in different studies are related to the level of ploidy.
In the estimate obtained by Hanson et al. (2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ), this difference can be attributed to the four surplus chromosomes observed in the plants analyzed in their study, when compared with those evaluated in this study. Genome size variation has been reported for several species, and it may even occur at intraspecific level, as observed in soybean (Graham et al. 1994Graham MJ, Nickell CD and Rayburn AL (1994) Relationship between genome size and maturity group in soybean. Theoretical and Applied Genetics 88: 429-432.) and maize (Rayburn et al. 1989Rayburn AL, Auger JA, Benzinger EA and Hepburn AG (1989) Detection of intraspecific DNA content variation in Zea mays L. by flow cytometry. Journal of Experimental Botany 40: 1179-1783. ). In some cases, these variations were correlated with environmental gradients or growth conditions (Dolezel and Bartos 2005Dolezel J and Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Annals of botany 95: 99-110.).
The species Tropaeolum majus presented 28 chromosomes, represented by 2.08 pg of nuclear DNA. Probatova et al. (2006Probatova N, Rudyka E, Pavlova N, Verkhokat V and Nechaev V (2006) Chromosome numbers of plants of the primorsky territory, the amur river basin and magadan region. Botanicheskii Zhurnal 91: 491-509.) observed the same chromosome number in varieties of this species cultivated in Magadan, a region belonging to the Russian Federation, and Chen (2003Chen RY, Song WQ, Li XL, Li MX, Liang GL and Chen CB (2003) Chromosome atlas of major economic plants genome in China: Chromosome atlas of garden flowering plants in China. Tomous III, Science Press, Beijing, 646p.) had the same results in varieties cultivated in China. Considering that the basic number of chromosomes is x=14 (Raghuvanshi and Pathak 1974Raghuvanshi SS, Pathak CS (1974) Ploidy barrier in Tropaeolum Majus L. Caryologia 27: 225-235.), it can be inferred that the analyzed plant is tetraploid. No recent studies are found in relation to the quantification of nuclear DNA of this species, besides that of Nagl (1976Nagl W (1976) Early embryogenesis in Tropaeolum majus L.: Evolution of DNA content and polyteny in the suspensor. Plant Science Letters 7: 1-6. ), which presented quantification of 2.66 pg of nuclear DNA in a tetraploid variety of T. majus, cultivated in Germany. This quantification was performed by densitometric scanning, and therefore the values may not be comparable.
Stachys byzantina presented 30 chromosomes, corroborating the reports of Martin et al. (2011Martin E, Çetin Ö, Akcicek E and Dirmenci T (2011) New chromosome counts of genus Stachys (Lamiaceae) from Turkey. Turkish Journal of Botany 35: 671-680. ) and Baltisberger (2002Baltisberger M (2002) Cytological investigations on some Albanian plant species. Candollea 56: 245-259. ), who observed 15 pairs of chromosomes in varieties of this species cultivated in Turkey. These same authors reported that the basic number of chromosomes of the species is 15, indicating that the Stachys byzantina plant evaluated in his study is diploid. S. Byzantina presents wide genetic variability, among several varieties cultivated worldwide (Baltisberger 2002). This species had its nuclear DNA quantified at 1.54 pg and no reports of other studies have been found for comparison purposes.
For Hibiscus sabdariffa, 2n=72 chromosomes were observed, corroborating the description of Guerra (2009Guerra NA (2009) Estudios citogenéticos de Hibiscus sabdariffa L. (Malvaceae). Revista Científica UDO Agrícola 9: 595-598.) and Hiron et al. (2006Hiron N, Alam N, Ahmed FA, Begum R and Alam SS (2006) Differential fluorescent banding and isozyme assay of Hibiscus cannabinus L. and H. sabdariffa L. (Malvaceae). Cytologia 71: 175-180. ) for varieties of Hibiscus sabdariffa cultivated in Venezuela and Bangladesh, respectively. The plant is autotetraploid, and the chromosome complement is organized in 18 groups, with 4 chromosomes each (Hiron et at. 2006Hiron N, Alam N, Ahmed FA, Begum R and Alam SS (2006) Differential fluorescent banding and isozyme assay of Hibiscus cannabinus L. and H. sabdariffa L. (Malvaceae). Cytologia 71: 175-180. ). The quantification of nuclear DNA of H. sabdariffa was of 5.12 pg, and no other studies on quantification of the genome of this species have been reported.
In addition to the possible intraspecific variations of nuclear DNA content in plants, the methodologies for the quantification of the nuclear genome by flow cytometry are influenced by several factors, such as different methodologies to obtain and isolate cell nuclei, different equipment, software and techniques, and different chemical substances used in all the processes (Silva et al. 2014Silva RAL, Pio LAS, Pasqual M, De Oliveira ACL, Rodrigues FA and De Oliveira S (2014) An assessment of software for flow cytometry analysis in banana plants. Semina: Ciências Agrárias 35: 775-780.).
The adaptation of specific protocols for the quantification of nuclear DNA of each species requires studies to elucidate the most appropriate techniques for each case. The coefficient of variation is the most commonly used indicator to verify the quality of the analyses performed by flow cytometry, which is presented together with the statistical data of each sample evaluated in this study. According to Marie and Brown (1993Marie D and Brown SCA (1993) Cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell 78: 41-51. ), values obtained between 1 and 2% for coefficients of variation in flow cytometric analyses are considered as of high quality; thus, the evaluations performed in all species studied here were within the high quality standard, since all the results of CV were lower than 2% (Table 1).
The quantity of nuclear DNA is not always correlated with the chromosome number, and a large nuclear DNA quantity can be observed both in organisms with greater chromosome numbers and in those with smaller chromosome numbers (Hanson et al. 2005Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320. ), corroborating the results of this study. Rumex acetosa, for instance, had 7.04 pg of DNA represented by 14 chromosomes, whereas Basella alba showed similar nuclear DNA quantity (7.05 pg), represented in 44 chromosomes.
The chromosomes of Rumex acetosa are larger, when compared with those of Basella alba (Figure 1), which justifies the similarity of DNA quantity. Another example that corroborates this analysis is the comparison between chromosome number and DNA content between Tropaeolum majus (28 chromosomes and 2.08 pg nuclear DNA) and Stachys byzantina (30 chromosomes and 1.54 pg DNA). One of the possible explanations for this divergence between DNA quantity and the chromosome number is that some species have genome consisting of great quantity of highly repetitive DNA, or even duplicated regions or genes (Lysák and Schubert 2013Lysák MA and Schubert I (2013) Mechanisms of chromosome rearrangements. Plant Genome Diversity 2: 137-147.).
CONCLUSIONS
The species Rumex acetosa L., Tropaeolum majus L. and Stachys byzantina K. Koch are diploids, with 2n=14, 2n=28, and 2n=30, respectively. The chromosome number (2n=44) of Basella alba L. may have been derived by descending dysploidy from the number 2n=4x=48. Hibiscus sabdariffa L. is tetraploid with 2n=72 chromosomes. The Rumex acetosa L. plants evaluated in this study are female, diploid with 2n=14 chromosomes, represented by 2n=12 + XX. Estimates of the nuclear DNA content of the species Stachys byzantina K. Koch and Hibiscus sabdariffa L. is unpublished, being 1.54 pg and 5.12 pg, respectively.
ACKNOWLEDGEMENTS
To the Foundation for Research Support of Minas Gerais (FAPEMIG) and to CNPq/ National Council for Scientific and Technological Development (MCT) for the financial support. To the Federal University of Lavras (UFLA) for the technological equipment.
REFERENCES
- Ainsworth C, Rahman A, Parker J and Edwards G (2005) Intersex inflorescences of Rumex acetosa demonstrate that sex determination is unique to each flower. New phytologist 165: 711-720.
- Baltisberger M (2002) Cytological investigations on some Albanian plant species. Candollea 56: 245-259.
- Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K and Joachimiak AJ (2007) Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction 20: 171-180.
- BRASIL - Ministério da Saúde (2002) Alimentos regionais brasileiros. Secretaria de Políticas de Saúde, Coordenação-Geral da Política de Alimentação e Nutrição, Brasília, 141p.
- BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2010) Hortaliças Não Convencionais. MAPA, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 54p.
- BRASIL - Ministério da Agricultura, Pecuária e Abastecimento (2013) Manual de hortaliças não convencionais. MAPA/ACS, Secretaria de Desenvolvimento Agropecuário e Cooperativismo, Brasília, 99p.
- Cardoso MO (1997) Hortaliças não-convencionais da amazônia. Embrapa, Serviço de Produção de Informação, Brasília, 152p.
- Chen RY, Song WQ, Li XL, Li MX, Liang GL and Chen CB (2003) Chromosome atlas of major economic plants genome in China: Chromosome atlas of garden flowering plants in China. Tomous III, Science Press, Beijing, 646p.
- Cuñado N, Navajas-Pérez R, De La Herrán R, Rejón CR, Rejón MR, Santos JL and Garrido-Ramos MA (2007) The evolution of sex chromosomes in the genus Rumex (Polygonaceae): identification of a new species with heteromorphic sex chromosomes. Chromosome research 15: 825-833.
- Dolezel J and Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Annals of botany 95: 99-110.
- Dong F, Mcgrath JM, Helgeson JP and Jiang J (2001) The genetic identity of alien chromosomes in potato breeding lines revealed by sequential GISH and FISH analyses using chromosome-specific cytogenetic DNA markers. Genome 44: 729-734.
- Galbraith DW, Lambert GM, Macas J and Dolezel J (2001) Analysis of nuclear DNA content and ploidy in higher plants. Current Protocols In Cytometry 22: 7-6.
- Graham MJ, Nickell CD and Rayburn AL (1994) Relationship between genome size and maturity group in soybean. Theoretical and Applied Genetics 88: 429-432.
- Grasso G, Van Duren M, Lee KS and Morpurgo R (1997) DNA analysis in three populations of african spinach (Basella spp). In FAO/IAEA (org) Improvement of basic food crops in Africa through plant breeding, including the use of induced mutations. IAEA, Viena, p. 39-43.
- Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenetic and Genome Research 120: 339-350.
- Guerra M, Souza MD (2002) Como observar cromossomos: um guia de técnicas em citogenética vegetal, animal e humana. FUNPEC, Ribeirão Preto, 131p.
- Guerra NA (2009) Estudios citogenéticos de Hibiscus sabdariffa L. (Malvaceae). Revista Científica UDO Agrícola 9: 595-598.
- Hanson L, Boyd A, Johnson MA and Bennett MD (2005) First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96: 1315-1320.
- Hiron N, Alam N, Ahmed FA, Begum R and Alam SS (2006) Differential fluorescent banding and isozyme assay of Hibiscus cannabinus L. and H. sabdariffa L. (Malvaceae). Cytologia 71: 175-180.
- Kinupp VF and Lorenzi H (2014) Plantas alimentícias não convencionais (PANC) no Brasil guia de identificação, aspectos nutricionais e receitas ilustradas:. Instituto Plantarum de Estudos da Flora, São Paulo, 768p.
- Lysák MA and Schubert I (2013) Mechanisms of chromosome rearrangements. Plant Genome Diversity 2: 137-147.
- Marie D and Brown SCA (1993) Cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell 78: 41-51.
- Mariotti B, Navajas-Pérez R, Lozano R, Parker JS, De La Herrán R, Rejón CR, Rejón MR, Garrido-Ramos M and Jamilena M (2006) Cloning and characterization of dispersed repetitive DNA derived from microdissected sex chromosomes of Rumex acetosa Genome 49: 114-121.
- Martin E, Çetin Ö, Akcicek E and Dirmenci T (2011) New chromosome counts of genus Stachys (Lamiaceae) from Turkey. Turkish Journal of Botany 35: 671-680.
- Nagl W (1976) Early embryogenesis in Tropaeolum majus L.: Evolution of DNA content and polyteny in the suspensor. Plant Science Letters 7: 1-6.
- Navajas-Pérez R, Schwarzacher T, De La Herrán R, Rejón CR, Rejón MR and Garrido-Ramos MA (2006) The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61-71.
- Navarini APG, Schifino-Wittmann MT, De Barros IBI and Almeida D (2008) Cytogenetics of Hypericum caprifoliatum Cham. and Schltdl. (Clusiaceae) populations and other species of the genus. Crop Breeding and Applied Biotechnology 8: 330-338.
- Olkoski D and Wittmann MTS (2011) Cytogenetics of Mimosa bimucronata (DC.) O. Kuntze (Mimosoideae, Leguminosae): chromosome number, polysomaty and meiosis. Crop Breeding and Applied Biotechnology 11: 27-36.
- Probatova N, Rudyka E, Pavlova N, Verkhokat V and Nechaev V (2006) Chromosome numbers of plants of the primorsky territory, the amur river basin and magadan region. Botanicheskii Zhurnal 91: 491-509.
- Raghuvanshi SS, Pathak CS (1974) Ploidy barrier in Tropaeolum Majus L. Caryologia 27: 225-235.
- Rayburn AL, Auger JA, Benzinger EA and Hepburn AG (1989) Detection of intraspecific DNA content variation in Zea mays L. by flow cytometry. Journal of Experimental Botany 40: 1179-1783.
- Silva RAL, Pio LAS, Pasqual M, De Oliveira ACL, Rodrigues FA and De Oliveira S (2014) An assessment of software for flow cytometry analysis in banana plants. Semina: Ciências Agrárias 35: 775-780.
- Souza-Chies TT, Burchardt P, Alves SEM, Essi L and Dos Santos EK (2014) O estudo da biodiversidade e evolução vegetal através de marcadores de DNA e citogenética: exemplos em Iridaceae e Poaceae. Ciência e Natura 36: 279-293.
- Sperling CR, Bittrich V (1993) Basellaceae. Flowering plants: dicotyledons. Springer Berlin Heidelberg: 143-146.
Publication Dates
-
Publication in this collection
Dec 2017
History
-
Received
24 Feb 2016 -
Accepted
25 Nov 2016