Abstracts
Food contamination caused by enteric pathogens is a major cause of diarrheal disease worldwide, resulting in high morbidity and mortality and significant economic losses. Bacteria are important agents of foodborne diseases, particularly diarrheagenic Escherichia coli. The present study assessed the genetic diversity and antimicrobial resistance of E. coli isolates from pasteurized milk processed in 21 dairies in northwestern State of Parana, Brazil. The 95 E. coli isolates were subjected to antimicrobial susceptibility testing according to the recommendations of the Clinical and Laboratory Standards Institute and assessed genotypically by Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR). The highest rate of resistance was observed for cephalothin (55.78%). ERIC-PCR revealed high genetic diversity, clustering the 95 bacterial isolates into 90 different genotypic patterns. These results showed a heterogeneous population of E. coli in milk samples produced in the northwestern region of Paraná and the need for good manufacturing practices throughout the processing of pasteurized milk to reduce the risk of foodborne illnesses.
Pasteurized milk/processing; Pasteurized milk/bacterial typing; Pasteurized milk/contamination; Escherichia coli/genetic diversity; Escherichia coli/antimicrobial resistance; Food/microbiological analysis
A contaminação de alimentos por patógenos entéricos é uma das principais causas de doenças diarréicas em todo o mundo, resultando em altas taxas de morbidade e mortalidade e perdas econômicas significativas. As bactérias são importantes agentes de doenças de origem alimentar, particularmente Escherichia coli diarreiogênicas. O presente estudo teve como objetivo avaliar a diversidade genética e a resistência a antimicrobianos de E. coli isoladas de leite pasteurizado, processados em 21 laticínios na região noroeste do Paraná - Brasil. Os 95 isolados de E. coli foram submetidos a testes de suscetibilidade aos antimicrobianos de acordo com as recomendações do Clinical and Laboratory Standards Institute e avaliados genotipicamente por ERIC-PCR (Enterobacterial Repetitive Intergenic Consensus - Polymerase Chain Reaction). O principal perfil de resistência encontrado entre os isolados foi resistência à cefalotina (55,78%). ERIC-PCR revelou alta diversidade genética, agrupando os 95 isolados bacterianos em 90 diferentes perfis genotípicos. Estes resultados mostraram uma população heterogênea de E. coli em amostras de leite produzido na região noroeste do Paraná e a necessidade de boas práticas na manipulação de todo o processamento de leite pasteurizado, a fim de reduzir o risco de doenças transmitidas por alimentos.
Leite pasteurizado/processamento; Leite pasteurizado/tipagem bacteriana; Leite pasteurizado/contaminação; Escherichia coli/diversidade genética; Escherichia coli/resistência a antimicrobianos; Alimentos/análise microbiológica
INTRODUCTION
Food contamination caused by enteric pathogens is a major cause of diarrheal disease worldwide, resulting in high morbidity and mortality and significant economic losses (Centers for Disease Control and Prevention, 2013 CENTERS FOR DISEASE CONTROL AND PREVENTION. Incidence and trends of infection with pathogens transmitted commonly through food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 1996-2012. Morb. Mortal. Wkly Rep. , v.62, p.283-287, 2013.). Bacteria are important agents of foodborne diseases (Centers for Disease Control and Prevention, 2013 CENTERS FOR DISEASE CONTROL AND PREVENTION. Incidence and trends of infection with pathogens transmitted commonly through food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 1996-2012. Morb. Mortal. Wkly Rep. , v.62, p.283-287, 2013.), particularly Escherichia coli, which is widely distributed in nature. Despite being commonly found in the gastrointestinal tract of humans and other animals, it can cause a variety of diseases, some of which are life threatening in more vulnerable groups, such as the elderly, children, and immunocompromised individuals (FDA, 2012FOOD AND DRUG ADMINISTRATION. FDA. The bad bug book: foodborne pathogenic microorganisms and natural toxins. 2.ed. Washington: Center for Food Safety and Applied Nutrition, 2012. p.67-80.).
E. coli has been isolated from various foodstuffs, such as meat, vegetables, milk, and milk derivatives, which can serve as a vehicle for the transmission of diseases by food (Lee et al., 2009LEE, G.Y.; JANG, H.I.; HWANG, I.G.; RHEE, M.S. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int. J. Food Microbiol., v.134, p.196-200, 2009.; Solomakos et al., 2009SOLOMAKOS, N.; GOVARIS, A.; ANGELIDIS, A.S.; POURNARAS, S.; BURRIEL, A.R.; KRITAS, S.; PAPAGEORGIOU, D.K. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol., v.26, p.865-871, 2009.; Brooks et al., 2012BROOKS, J.C.; MARTINEZ, B.; STRATTON, J.; BIANCHINI, A.; KROKSTROMB, R.; HUTKINS, R. Survey of raw milk cheese for microbiological quality and prevalence of foodborne pathogens. Food Microbiol., v.31, p.154-158, 2012.; Maffei et al., 2013MAFFEI, D.F.; SILVEIRA, N.F.A.; CATANOZI, M.P.L.M. Microbiological quality of organic and conventional vegetables sold in Brazil. Food Control, v.29, p.226-230, 2013.).
In Brazil, some studies have reported a high prevalence of E. coli in
raw and pasteurized milk. E. coli was isolated from 41.1% of
pasteurized milk samples evaluated by Silva et
al. (2001)SILVA, Z.N.; CUNHA, A.S; LINS, M.C.; CARNEIRO, L.A.M.; ALMEIDA, A.C.F.;
QUEIROZ, M.L.P. Isolation and serological identification of enteropathogenic
Escherichia coli in pasteurized milk in Brazil. Rev. Saúde Pública, v.35, p.375-379,
2001., while in raw milk, this bacteria was found in 79,2%
(Campos et al., 2006CAMPOS, M.R.H.; KIPNIS, A.; ANDRÉ, M.C.D.P.B.; BORGES, L.J.; VIEIRA,
C.A.S.; BORGES, L.J.; SANTOS, P.P.; SERAFINI, A.B. Caracterização fenotípica pelo
antibiograma de cepas de Escherichia coli isoladas de manipuladores, de leite cru e
de queijo "Minas Frescal" em um laticínio de Goiás, Brasil. Ciên. Rural, v.36,
p.1221-1227, 2006.) and in
76% (Barreto et al., 2012BARRETO, N.S.; SANTOS, G.C.F.; CREPALDI, A.L.; SANTOS, R.A. Microbial
quality and antimicrobial susceptibility of informally traded milk in Cruz das Almas
county, Bahia. Semina: Ciências Agrárias, v.33, p.2315-2326, 2012.) of
milk samples analyzed. An important subject is the emergence of resistant foodborne
pathogens that may be transmitted to humans as food contaminants. Outbreaks of foodborne
diseases that involve resistant bacteria associated with food animal sources have been
reported (Safe Food, 2010SAFE FOOD. The problem of antimicrobial resistance in the food chain.
2010. Available at:
<http://www.safefood.eu/SafeFood/files/8a/8abb9354-4cc2-49a4-b586-bf0008eb8cf.pdf>.
Accessed on: 28 May 2013.
http://www.safefood.eu/SafeFood/files/8a...
).
Milk may get contaminated with E. coli from various sources during different stages of production and processing, which could explain the genetic diversity of these bacteria in milk. The presence of E. coli in pasteurized milk may be due to inadequate pasteurization or contamination of the product after pasteurization process. Studies of the epidemiology of foodborne microorganisms, especially the source of contamination, have been conducted using various molecular techniques, such as ribotyping, multilocus sequence typing, randomly amplified polymorphic DNA, enterobacterial repetitive intergenic consensus-polymerase chain reaction, repetitive extragenic palindromic, and pulsed-field gel electrophoresis (Foley, Lynne, Nayak, 2009FOLEY, S.L.; LYNNE, A.M.; NAYAK, R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect. Genet. Evol., v.9, p.430-440, 2009.).
Several studies have used ERIC-PCR to assess the genetic similarity of E. coli isolates from different sources. Rantsiou, Alessandria and Cocolin (2012)RANTSIOU, K.; ALESSANDRIA, V.; COCOLIN, L. Prevalence of Shiga toxin-producing Escherichia coli in food products of animal origin as determined by molecular methods. Int. J. Food Microbiology, v.154, p.37-43, 2012. assessed the genetic diversity of Shiga toxin-producing E. coli from meat and dairy products using ERIC-PCR. Wenz et al. (2006)WENZ, J.R.; BARRINGTON, G.M; GARRY, F.B.; ELLIS, R.P.; MAGNUSON, R.J. Escherichia coli isolates' serotypes, genotypes, and virulence genes and clinical coliforms mastitis severity. J. Dairy Sci., v.89, p.3408-3412, 2006. described genetic variability among E. coli isolates from dairy cattle with mastitis using ERIC-PCR. Ibenyassine et al. (2006)IBENYASSINE, K.; AITMHAND, R.; KARAMOKO, Y.; COHEN, N.; ENNAJI, M.M. Use of repetitive DNA sequences to determine the persistence of enteropathogenic Escherichia coli in vegetables and in soil grown in fields treated with irrigation water. Lett. Appl. Microbiol., v.43, p.528-533, 2006. used the same technique and evaluated the similarity of isolates from plants infected by irrigation water. Other authors have used ERIC-PCR to compare E. coli isolates from humans and animals (Leung et al., 2004LEUNG, K.T.; MACKERETH, R.; TIEN, Y.C.; TOPP, E. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig and human sources. FEMS Microbiol. Ecol., v.47, p.111-119, 2004.; Sabate et al., 2008SABATE, M.; PRATS, G.; MORENO, E.; BALLESTE, E.; BLANCH, A.R.; ANDREU, A. Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res. Microbiol., v.159, p.288-293, 2008.; Tramuta et al., 2011TRAMUTA, C.; NUCERA, D.; ROBINO, P.; SALVARANI, S.; NEBBIA, P. Virulence factors and genetic variability of uropathogenic Escherichia coli isolated from dogs and cats in Italy. J. Vet. Sci., v.12, p.49-55, 2011.; De la Fé Rodriguez et al., 2012DE LA FÉ RODRIGUEZ, P.Y.; KIIRU, J.N.; MARTIN, L.O.M.; MUÑOZ, E.C.; BUTAYE, P.; COX, E.; GODDEERIS, B.M. Characterization and clonal grouping of pathogenic Esherichia coli isolated from intestinal contents of diarrheic piglets in Villa Clara province, Cuba, according to their antibiotic resistance and ERIC-PCR profiles. Vet. Microbiol., v.154, p.425-428, 2012.). Moreover, Casarez, Pillai, Di Giovanni, (2007)CASAREZ, E.A.; PILLAI, S.D.; DI GIOVANNI, G.D. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res., v.41, p.3643-3648, 2007., Duan et al. (2009)DUAN, H.; CHAI, T.; LIU, J.; ZHANG, X.; QI, C.; GAO, J.; WANG, Y.; CAI, Y.; MIAO, Z.; YAO, M.; SCHLENKER, G. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ. Res., v.109, p.511-517, 2009., and Wan et al. (2011)WAN, L.; WANG, Z.B.; YAN, Q.G.; WANG, X.; LEI, Y.; ZUO, L.; CHENG, Y.; REN, Y.P.; GUO, W.Z. Genetic diversity of Escherichia coli isolated from commercial swines farms revealed by enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) and repetitive extragenic palindrome PCR (REP-PCR). Afr. J. Biotechnology, v.10, p.10543-10550, 2011. showed that ERIC-PCR could discriminate E. coli from water and environmental samples. Knowledge about the epidemiology of diarrheagenic E. coli in food is important for understanding its distribution and transmission, besides developing and implementing control measures in food production.
To the best of our knowledge, no studies have used ERIC-PCR to assess the diversity of E. coli in pasteurized, ready-to-consume cow milk. Thus, the aim of the present study was to evaluate the genetic diversity and antimicrobial resistance of E. coli isolates from cow milk pasteurized in the northwestern region of Paraná, Brazil, using ERIC-PCR.
MATERIAL AND METHODS
Bacterial isolates
Ninety-five E. coli isolates from pasteurized, ready-to-consume cow milk samples were analyzed. The isolates were stored in Tryptic Soy Broth (TSB; Difco, Le Pont de Claix, France) with glycerol at -20 ºC and kept at the Laboratory of Food Microbiology, Department of Clinical Analysis and Biomedicine, Maringá State University. The milk samples were obtained from 21 dairies in Paraná, Brazil, from March 2006 to November 2008 in a previous study conducted in our laboratory that evaluated the presence of coliforms in pasteurized milk (Zanella et al., 2010ZANELLA, G.N.; MIKCHA, J.M.G.; BANDO, E.; SIQUEIRA, V.L.D.; MACHINSKI, M. JR. Occurrence, antibiotic resistance of coliform bacteria and antimicrobial residues from cow's milk from Brazil. J. Food Prot., v.73, p.1684-1687, 2010.). The different dairies were identified by the abbreviations AA, APRO, CAIUÁ, CC, CIA, COC, COOP, GM, L, LAC, LOU, MOU, MP, NE, P, PN, S, SN, SO, TB, and U.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the disc diffusion technique according to the method described by the Clinical and Laboratory Standards Institute (2009). The antimicrobial agents (Oxoid, Hampshire, England) were the following: amikacin (30μg), amoxicillin-clavulanic acid (10-20 μg), ampicillin (10 μg), aztreonam (30 μg), cephalothin (30 μg), cefepime (30 μg), cefotaxime (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), gentamycin (10 μg), imipenem (10 μg), sulphamethoxazole-trimethoprim (1.25-23.75 μg), tetracycline (30 μg), and tobramycin (10 μg). The E. coli ATCC 25922 and E. coli ATCC 35218 isolates were used as controls.
ERIC-PCR
Genomic DNA of E. coli isolates was extracted from overnight bacterial growth on Nutrient Agar (Difco, Le Pont de Claix, France; Swanenburg et al., 1998SWANENBURG, M.; URLINGS, H.A.P.; KEUZENKAMP, D.A.; SNIJDERS, J.M.A. Validation of ERIC-PCR as a tool in epidemiology research of Salmonella in slaughter pigs. J. Ind. Microbiol. Biotechnol., v.21, p.141-144, 1998.). DNA quantification was performed using NanoDrop 2000.
The amplification reaction was performed by adding 100 ng of bacterial DNA to a PCR reagent mixture that contained 1 µM of the primers ERIC1R (5'-ATGTAAGCTCCTGGGGATTCAC-3') and ERIC2 (5'-AAGTAAGTGACTGGGGTGAGCG-3'; (Versalovic, Koeuth, Lupski, 1991VERSALOVIC, J.; KOEUTH, T.; LUPSKI, J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res., v.19, p.6823-6831, 1991.), PCR buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl), 1.5 mM MgCl2, 200 µM of each nucleotide (dATP, dCTP, dGTP, and dTTP), 1 U of DNA polymerase (Taq DNA polymerase; Invitrogen -Life Technologies, Brazil), and sterile deionized water to a final volume of 25 µl. The amplification was performed in a thermocycler (Gene Amp PCR System 2400, Perkin Elmer, Roche, Branchburg, NJ, USA) with denaturing at 94 ºC for 7 min, followed by 35 cycles at 94 ºC for 30 s, 52 ºC for 1min, and 72 ºC for 8 min and a final extension at 72 ºC for 16min. The PCR product underwent 1.5% agarose (Amersham Pharmacia Biotech AB, Uppsala, Sweden) gel electrophoresis at 7-10 V/cm2 for 2 h and visualized under UV light. The spectral band analysis was performed using BioNumerics software (version 4.45, Applied Maths, Sint-Martens-Latem, Belgium). The Dendrogram was constructed using the Dice coefficient, and the phylogenetic distance was determined using the Unweighted Pair Group Method with the Arithmetic Mean algorithm. Isolates with ≥95% similarity were considered closely related. The discriminatory power of ERIC-PCR was calculated based on Simpson's diversity index as indicated by Hunter and Gaston (1988)HUNTER, P.R.; GASTON, M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol., v.26, p.2465-2466, 1988..
RESULTS
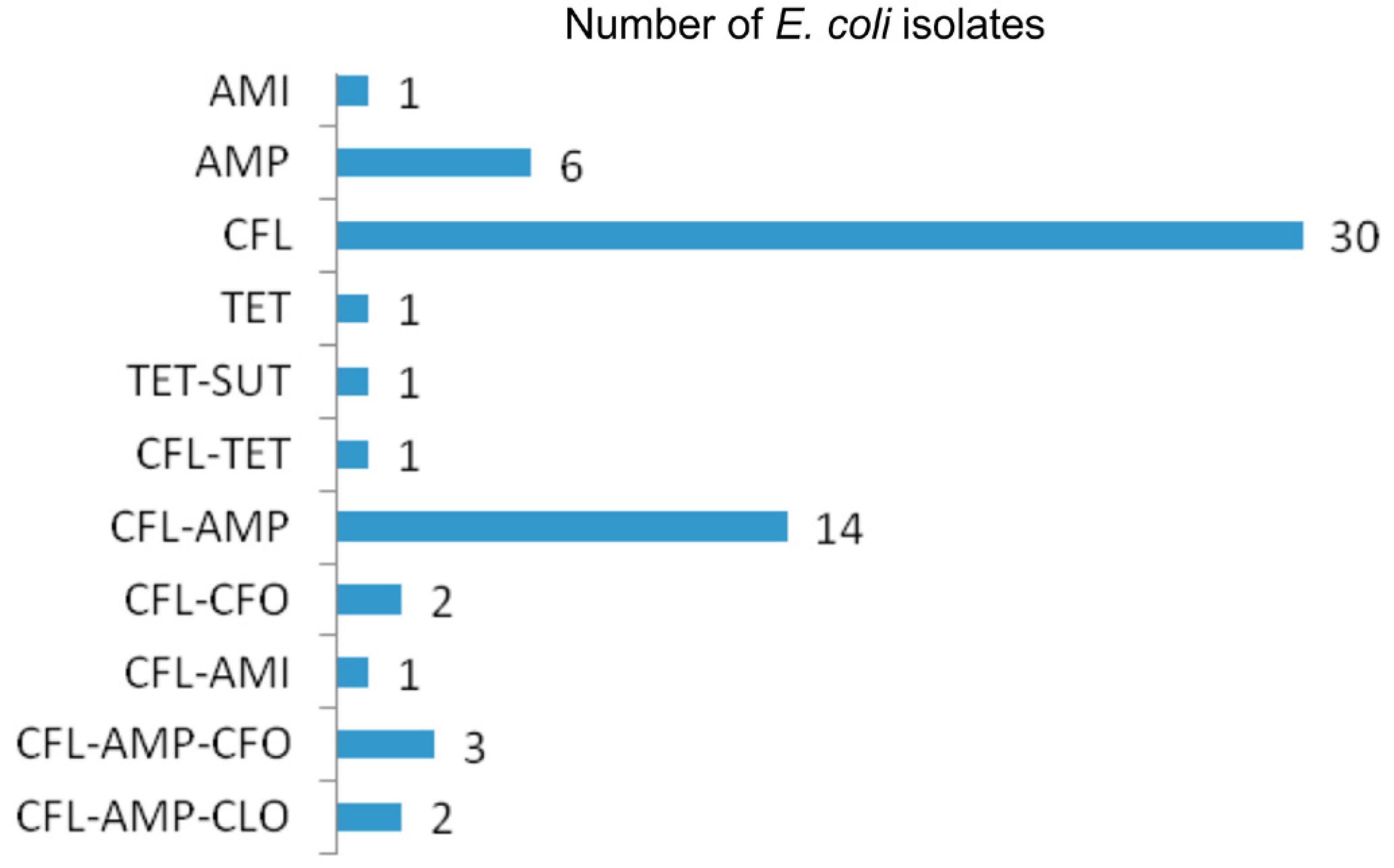
All 95 E. coli isolates were found to be susceptible to amoxicillin-clavulanic acid, aztreonam, cefepime, cefotaxime, ceftazidime, ciprofloxacin, gentamycin, imipenem, and tobramycin. Sixty-two isolates (65.26%) were resistant to at least one of the antimicrobial agents tested. Among the resistant isolates, five were simultaneously resistant to three antimicrobial agents. The highest rates of resistance were observed for cephalothin (55.78%) and ampicillin (26.31%). Resistance to cefoxitin (5.26%), tetracycline (3.15%), amikacin (2.10%), chloramphenicol (2.10%), and trimethoprim-sulphamethoxazole (1.05%) was also observed. The E. coli isolates were grouped into 11 resistance profiles. The most common resistance profiles were to cephalothin (30 isolates) and cephalothin/ampicillin (14 isolates; Figure 1).
Antimicrobial resistance profiles of E. coli isolated from pasteurized milk in State of Paraná, Brazil. AMI: amikacin, AMP: ampicillin CFL: cephalothin, TET: tetracycline, SUT: trimethoprim-sulphamethoxazole CFO: cefoxitin, CLO: chloramphenicol.
ERIC-PCR applied to 95 E. coli isolates generated patterns of four to 20 bands, with sizes that ranged from 100 to 2,000 bp. Considering the similarity level ≥95%, 90 ERIC-PCR patterns were observed. Of these, 85 (89.47%) showed unique profiles, and the remaining 10 (10.53%) were included in five clusters, comprising two isolates each (isolates 14 GM and 16 GM, 14 CC and 15 CC, 6 MP and 10 MP, 10 COC and 11 COC, and 12 L and 14 L; Figure 2).
Dendrogram that represent the genetic relationship of ERIC-PCR, milk samples and antimicrobial resistance of 95 E. coli isolated from pasteurized cow milk in State of Paraná, Brazil.
The clusters 1, 2, 4, and 5, that contained two isolates each, clustered isolates from four milk samples processed in four distinct dairies. Distinct antimicrobial resistance profiles between each of two clustered isolates were observed. Cluster 3 contained two isolates (6 MP and 10 MP) that were resistant to cephalothin and were obtained from different pasteurized milk samples in the same dairy (Figure 2). None of the clusters was obtained from milk samples processed in different dairies. The discriminatory index of ERIC-PCR was 0.999.
DISCUSSION
Pathogenic microorganisms have been isolated from milk and dairy products (D'Amico, Donnelly, 2010D'AMICO, D.J.; DONNELLY, C.W. Microbiological quality of raw milk used for small-scale artisan cheese production in Vermont: effect of farm characteristics and practices. J. Dairy Sci., v.93, n.1, p.134-147, 2010.; Costa Sobrinho et al., 2012COSTA SOBRINHO, P.S.; FARIA, C.A.M.; PINHEIRO, J.S.; ALMEIDA, H.G.; PIRES, C.V.; SANTOS, A.S. Bacteriological quality of raw milk used for production of a Brazilian farmstead raw milk cheese. Foodborne Pathog. Dis., v.9, p.138-144, 2012.). Among them, diarrheagenic E. coli is notable and can cause diseases with serious consequences, especially in children and the elderly (FDA, 2012FOOD AND DRUG ADMINISTRATION. FDA. The bad bug book: foodborne pathogenic microorganisms and natural toxins. 2.ed. Washington: Center for Food Safety and Applied Nutrition, 2012. p.67-80.). The analysis and comparison of E. coli isolates from different sources can generate important information about the origin and transmission of these bacteria (Foley, Lynne, Nayak, 2009FOLEY, S.L.; LYNNE, A.M.; NAYAK, R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect. Genet. Evol., v.9, p.430-440, 2009.).
Importantly, antimicrobial-resistant bacteria can act as reservoirs of resistance genes. Several studies of bacteria isolated from food have shown resistance to one or more antimicrobials, which is worrying because these microorganisms can be spread in the population as food contaminants (Lutgen et al., 2009LUTGEN, E.M.; MCEVOY, J.M.; SHERWOOD, J.S.; LOGUE, C.M. Antimicrobial resistance profiling and molecular subtyping of Campylobacter spp. from processed turkey. BMC Microbiol., v.9, p.203, 2009.; Solomakos et al., 2009SOLOMAKOS, N.; GOVARIS, A.; ANGELIDIS, A.S.; POURNARAS, S.; BURRIEL, A.R.; KRITAS, S.; PAPAGEORGIOU, D.K. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol., v.26, p.865-871, 2009.; Dutil et al., 2010DUTIL, L.; IRWIN, R.; FINLEY, R.; NG, L.K.; AVERY, B.; BOERLIN, P.; BOURGAULT, A.M.; COLE, L.; DAIGNAULT, D.; DESRUISSEAU, A.; DEMCZUK, W.; HOANG, L.; HORSMAN, G.B.; ISMAIL, J.; JAMIESON, F.; MAKI, A.; PACAGNELLA, A.; PILLAI, D.R. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis., v.16, p.48-54, 2010.; Rahimi, Chaleshtori, Parsaei, 2010RAHIMI, E.; CHALESHTORI, S.S.; PARSAEI, P. Prevalence and antimicrobial resistance of Escherichia coli O157 isolated from traditional cheese, ice cream and yoghurt in Iran. Afr. J. Microbiology Res., v.5, p.3706-3710, 2010.).
In the present study, 65.26% of the isolates showed resistance to at least one of the antimicrobial agents tested. Similar results were reported by Rahimi, Chaleshtori and Parsaei (2010)RAHIMI, E.; CHALESHTORI, S.S.; PARSAEI, P. Prevalence and antimicrobial resistance of Escherichia coli O157 isolated from traditional cheese, ice cream and yoghurt in Iran. Afr. J. Microbiology Res., v.5, p.3706-3710, 2010., who found that 88.9% of the isolates were resistant to one or more of the tested antimicrobials. The highest rates of resistance were observed for cephalothin (55.78%) and ampicillin (26.31%). Paneto et al. (2007)PANETO, B.R.; SHOCKEN-ITURRINO, R.P.; MACEDO, C.; SANTO, E.; MARIN, J.M. Occurrence of toxigenic Escherichia coli in raw milk cheese in Brazil. Arq. Bras. Med. Vet. Zootec., v.59, p.508-512, 2007. evaluated E. coli isolated from cheese and provided indices of resistance to ampicillin and cephalothin that were similar to those observed in the present study. Rahimi, Chaleshtori and Parsaei, (2010)RAHIMI, E.; CHALESHTORI, S.S.; PARSAEI, P. Prevalence and antimicrobial resistance of Escherichia coli O157 isolated from traditional cheese, ice cream and yoghurt in Iran. Afr. J. Microbiology Res., v.5, p.3706-3710, 2010. found that 44.4% of the E. coli isolates from dairy products showed ampicillin resistance. Solomakos et al. (2009)SOLOMAKOS, N.; GOVARIS, A.; ANGELIDIS, A.S.; POURNARAS, S.; BURRIEL, A.R.; KRITAS, S.; PAPAGEORGIOU, D.K. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol., v.26, p.865-871, 2009. found that all of the E. coli isolates from milk were resistant to ampicillin, whereas Campos et al. (2006)CAMPOS, M.R.H.; KIPNIS, A.; ANDRÉ, M.C.D.P.B.; BORGES, L.J.; VIEIRA, C.A.S.; BORGES, L.J.; SANTOS, P.P.; SERAFINI, A.B. Caracterização fenotípica pelo antibiograma de cepas de Escherichia coli isoladas de manipuladores, de leite cru e de queijo "Minas Frescal" em um laticínio de Goiás, Brasil. Ciên. Rural, v.36, p.1221-1227, 2006. found increased resistance to ampicillin and cephalothin and resistance to tetracycline in E. coli isolated from milk and cheese.
With regard to resistance profiles, the E. coli isolates were differentiated into 11 profiles. Heterogeneous profiles of antimicrobial resistance were also described by Rangel and Marin (2009)RANGEL, P.; MARIN, J.M. Analysis of Escherichia coli isolated from bovine mastitic milk. Pesq. Vet. Bras., v.29, p.363-368, 2009. and Solomakos et al. (2009)SOLOMAKOS, N.; GOVARIS, A.; ANGELIDIS, A.S.; POURNARAS, S.; BURRIEL, A.R.; KRITAS, S.; PAPAGEORGIOU, D.K. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol., v.26, p.865-871, 2009. in E. coli isolated from milk.
In the present study, ERIC-PCR showed high genetic diversity among the 95 E. coli isolates, which were clustered into 90 profiles. Among the clusters generated by ERIC-PCR, eight clustered E. coli isolates from four milk samples processed in four different dairies, suggesting a single clone contaminant per milk sample (Figure 2). Nevertheless, one cluster that comprised two E. coli isolates from different milk samples processed in the same dairy could suggest a common source of contamination. These results indicate failures in pasteurization process or contamination in post-pasteurization processing and show the need for control measures to minimize E. coli contamination. The present results are consistent with Wenz et al. (2006)WENZ, J.R.; BARRINGTON, G.M; GARRY, F.B.; ELLIS, R.P.; MAGNUSON, R.J. Escherichia coli isolates' serotypes, genotypes, and virulence genes and clinical coliforms mastitis severity. J. Dairy Sci., v.89, p.3408-3412, 2006., who studied E. coli isolates from dairy cattle with mastitis, and Rantsiou, Alessandria and Cocolin (2012)RANTSIOU, K.; ALESSANDRIA, V.; COCOLIN, L. Prevalence of Shiga toxin-producing Escherichia coli in food products of animal origin as determined by molecular methods. Int. J. Food Microbiology, v.154, p.37-43, 2012., who used ERIC-PCR to assess the genetic similarity of E. coli isolated from meat products and unpasteurized dairy products. E. coli is widely distributed in nature, and the sources of milk contamination can be diverse, explaining the high genotypic diversity observed in the present study.
ERIC-PCR is effective in typing E. coli isolates from animals (Mohapatra, Broersma, Mazumder, 2007MOHAPATRA, B.R.; BROERSMA, K.; MAZUMDER, A. Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiol. Lett., v.277, p.98-106, 2007.; Prabu et al., 2010; Wan et al., 2011; De la Fé Rodriguez et al., 2012DE LA FÉ RODRIGUEZ, P.Y.; KIIRU, J.N.; MARTIN, L.O.M.; MUÑOZ, E.C.; BUTAYE, P.; COX, E.; GODDEERIS, B.M. Characterization and clonal grouping of pathogenic Esherichia coli isolated from intestinal contents of diarrheic piglets in Villa Clara province, Cuba, according to their antibiotic resistance and ERIC-PCR profiles. Vet. Microbiol., v.154, p.425-428, 2012.) and water (Casarez, Pillai, Di Giovanni, 2007CASAREZ, E.A.; PILLAI, S.D.; DI GIOVANNI, G.D. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res., v.41, p.3643-3648, 2007.). However, some reports indicated that ERIC-PCR is not effective in typing E. coli isolated from humans, animals, and food (Giammanco et al., 2002GIAMMANCO, G.M.; PIGNATO, S.; GRIMONT, F.; GRIMONT, P.A.D.; CAPRIOLI, A.; MORABITO, S.; GIAMMANCO, G. Characterization of Shiga toxin-producing Escherichia coli O157:H7 isolated in Italy and in France. J. Clin. Microbiol., v.40, p.4619-4624, 2002.; Leung et al., 2004LEUNG, K.T.; MACKERETH, R.; TIEN, Y.C.; TOPP, E. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig and human sources. FEMS Microbiol. Ecol., v.47, p.111-119, 2004.; Costa et al., 2006COSTA, M.M.; SILVA, M.S.; SPRICIGO, D.A.; WITT, N.M.; MARCHIORO, S.B.; KOLLING, L.; VARGAS, A.P.C. Epidemiology, molecular characterization and resistance to antimicrobials of Escherichia coli isolated from South-Brazilian pig herds. Pesq. Vet. Bras., v.26, p.5-8, 2006.).
The present study showed heterogeneous profiles of antimicrobial resistance and a high genotypic diversity using ERIC-PCR in E. coli isolated from dairy milk processed in northwestern Paraná. The high genetic diversity observed in the present study can be justified by several sources of contamination of cow milk by E. coli during the entire production process.
ACKNOWLEDGEMENTS
This research received financial support from Fundação Araucária/Apoio ao Desenvolvimento Científico e Tecnológico do Paraná.
REFERENCES
- BARRETO, N.S.; SANTOS, G.C.F.; CREPALDI, A.L.; SANTOS, R.A. Microbial quality and antimicrobial susceptibility of informally traded milk in Cruz das Almas county, Bahia. Semina: Ciências Agrárias, v.33, p.2315-2326, 2012.
- BROOKS, J.C.; MARTINEZ, B.; STRATTON, J.; BIANCHINI, A.; KROKSTROMB, R.; HUTKINS, R. Survey of raw milk cheese for microbiological quality and prevalence of foodborne pathogens. Food Microbiol., v.31, p.154-158, 2012.
- CAMPOS, M.R.H.; KIPNIS, A.; ANDRÉ, M.C.D.P.B.; BORGES, L.J.; VIEIRA, C.A.S.; BORGES, L.J.; SANTOS, P.P.; SERAFINI, A.B. Caracterização fenotípica pelo antibiograma de cepas de Escherichia coli isoladas de manipuladores, de leite cru e de queijo "Minas Frescal" em um laticínio de Goiás, Brasil. Ciên. Rural, v.36, p.1221-1227, 2006.
- CASAREZ, E.A.; PILLAI, S.D.; DI GIOVANNI, G.D. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res., v.41, p.3643-3648, 2007.
- CENTERS FOR DISEASE CONTROL AND PREVENTION. Incidence and trends of infection with pathogens transmitted commonly through food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 1996-2012. Morb. Mortal. Wkly Rep. , v.62, p.283-287, 2013.
- CLINICAL AND LABORATORY STANDARDS INSTITUTE. CLSI. Performance standards for antimicrobial susceptibility testing: Wayne, 2009. 149 p. (M100-S16 Nineteenth Informational Supplement).
- COSTA, M.M.; SILVA, M.S.; SPRICIGO, D.A.; WITT, N.M.; MARCHIORO, S.B.; KOLLING, L.; VARGAS, A.P.C. Epidemiology, molecular characterization and resistance to antimicrobials of Escherichia coli isolated from South-Brazilian pig herds. Pesq. Vet. Bras., v.26, p.5-8, 2006.
- COSTA SOBRINHO, P.S.; FARIA, C.A.M.; PINHEIRO, J.S.; ALMEIDA, H.G.; PIRES, C.V.; SANTOS, A.S. Bacteriological quality of raw milk used for production of a Brazilian farmstead raw milk cheese. Foodborne Pathog. Dis., v.9, p.138-144, 2012.
- DE LA FÉ RODRIGUEZ, P.Y.; KIIRU, J.N.; MARTIN, L.O.M.; MUÑOZ, E.C.; BUTAYE, P.; COX, E.; GODDEERIS, B.M. Characterization and clonal grouping of pathogenic Esherichia coli isolated from intestinal contents of diarrheic piglets in Villa Clara province, Cuba, according to their antibiotic resistance and ERIC-PCR profiles. Vet. Microbiol., v.154, p.425-428, 2012.
- D'AMICO, D.J.; DONNELLY, C.W. Microbiological quality of raw milk used for small-scale artisan cheese production in Vermont: effect of farm characteristics and practices. J. Dairy Sci., v.93, n.1, p.134-147, 2010.
- DUAN, H.; CHAI, T.; LIU, J.; ZHANG, X.; QI, C.; GAO, J.; WANG, Y.; CAI, Y.; MIAO, Z.; YAO, M.; SCHLENKER, G. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ. Res., v.109, p.511-517, 2009.
- DUTIL, L.; IRWIN, R.; FINLEY, R.; NG, L.K.; AVERY, B.; BOERLIN, P.; BOURGAULT, A.M.; COLE, L.; DAIGNAULT, D.; DESRUISSEAU, A.; DEMCZUK, W.; HOANG, L.; HORSMAN, G.B.; ISMAIL, J.; JAMIESON, F.; MAKI, A.; PACAGNELLA, A.; PILLAI, D.R. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis., v.16, p.48-54, 2010.
- FOOD AND DRUG ADMINISTRATION. FDA. The bad bug book: foodborne pathogenic microorganisms and natural toxins. 2.ed. Washington: Center for Food Safety and Applied Nutrition, 2012. p.67-80.
- FOLEY, S.L.; LYNNE, A.M.; NAYAK, R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect. Genet. Evol., v.9, p.430-440, 2009.
- GIAMMANCO, G.M.; PIGNATO, S.; GRIMONT, F.; GRIMONT, P.A.D.; CAPRIOLI, A.; MORABITO, S.; GIAMMANCO, G. Characterization of Shiga toxin-producing Escherichia coli O157:H7 isolated in Italy and in France. J. Clin. Microbiol., v.40, p.4619-4624, 2002.
- HUNTER, P.R.; GASTON, M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol., v.26, p.2465-2466, 1988.
- IBENYASSINE, K.; AITMHAND, R.; KARAMOKO, Y.; COHEN, N.; ENNAJI, M.M. Use of repetitive DNA sequences to determine the persistence of enteropathogenic Escherichia coli in vegetables and in soil grown in fields treated with irrigation water. Lett. Appl. Microbiol., v.43, p.528-533, 2006.
- LEE, G.Y.; JANG, H.I.; HWANG, I.G.; RHEE, M.S. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int. J. Food Microbiol., v.134, p.196-200, 2009.
- LEUNG, K.T.; MACKERETH, R.; TIEN, Y.C.; TOPP, E. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig and human sources. FEMS Microbiol. Ecol., v.47, p.111-119, 2004.
- LUTGEN, E.M.; MCEVOY, J.M.; SHERWOOD, J.S.; LOGUE, C.M. Antimicrobial resistance profiling and molecular subtyping of Campylobacter spp. from processed turkey. BMC Microbiol., v.9, p.203, 2009.
- MAFFEI, D.F.; SILVEIRA, N.F.A.; CATANOZI, M.P.L.M. Microbiological quality of organic and conventional vegetables sold in Brazil. Food Control, v.29, p.226-230, 2013.
- MOHAPATRA, B.R.; BROERSMA, K.; MAZUMDER, A. Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiol. Lett., v.277, p.98-106, 2007.
- PANETO, B.R.; SHOCKEN-ITURRINO, R.P.; MACEDO, C.; SANTO, E.; MARIN, J.M. Occurrence of toxigenic Escherichia coli in raw milk cheese in Brazil. Arq. Bras. Med. Vet. Zootec., v.59, p.508-512, 2007.
- RAHIMI, E.; CHALESHTORI, S.S.; PARSAEI, P. Prevalence and antimicrobial resistance of Escherichia coli O157 isolated from traditional cheese, ice cream and yoghurt in Iran. Afr. J. Microbiology Res., v.5, p.3706-3710, 2010.
- RANGEL, P.; MARIN, J.M. Analysis of Escherichia coli isolated from bovine mastitic milk. Pesq. Vet. Bras., v.29, p.363-368, 2009.
- RANTSIOU, K.; ALESSANDRIA, V.; COCOLIN, L. Prevalence of Shiga toxin-producing Escherichia coli in food products of animal origin as determined by molecular methods. Int. J. Food Microbiology, v.154, p.37-43, 2012.
- SABATE, M.; PRATS, G.; MORENO, E.; BALLESTE, E.; BLANCH, A.R.; ANDREU, A. Virulence and antimicrobial resistance profiles among Escherichia coli strains isolated from human and animal wastewater. Res. Microbiol., v.159, p.288-293, 2008.
- SAFE FOOD. The problem of antimicrobial resistance in the food chain. 2010. Available at: <http://www.safefood.eu/SafeFood/files/8a/8abb9354-4cc2-49a4-b586-bf0008eb8cf.pdf>. Accessed on: 28 May 2013.
» http://www.safefood.eu/SafeFood/files/8a/8abb9354-4cc2-49a4-b586-bf0008eb8cf.pdf - SILVA, Z.N.; CUNHA, A.S; LINS, M.C.; CARNEIRO, L.A.M.; ALMEIDA, A.C.F.; QUEIROZ, M.L.P. Isolation and serological identification of enteropathogenic Escherichia coli in pasteurized milk in Brazil. Rev. Saúde Pública, v.35, p.375-379, 2001.
- SOLOMAKOS, N.; GOVARIS, A.; ANGELIDIS, A.S.; POURNARAS, S.; BURRIEL, A.R.; KRITAS, S.; PAPAGEORGIOU, D.K. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol., v.26, p.865-871, 2009.
- SWANENBURG, M.; URLINGS, H.A.P.; KEUZENKAMP, D.A.; SNIJDERS, J.M.A. Validation of ERIC-PCR as a tool in epidemiology research of Salmonella in slaughter pigs. J. Ind. Microbiol. Biotechnol., v.21, p.141-144, 1998.
- TRAMUTA, C.; NUCERA, D.; ROBINO, P.; SALVARANI, S.; NEBBIA, P. Virulence factors and genetic variability of uropathogenic Escherichia coli isolated from dogs and cats in Italy. J. Vet. Sci., v.12, p.49-55, 2011.
- VERSALOVIC, J.; KOEUTH, T.; LUPSKI, J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res., v.19, p.6823-6831, 1991.
- WAN, L.; WANG, Z.B.; YAN, Q.G.; WANG, X.; LEI, Y.; ZUO, L.; CHENG, Y.; REN, Y.P.; GUO, W.Z. Genetic diversity of Escherichia coli isolated from commercial swines farms revealed by enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) and repetitive extragenic palindrome PCR (REP-PCR). Afr. J. Biotechnology, v.10, p.10543-10550, 2011.
- WENZ, J.R.; BARRINGTON, G.M; GARRY, F.B.; ELLIS, R.P.; MAGNUSON, R.J. Escherichia coli isolates' serotypes, genotypes, and virulence genes and clinical coliforms mastitis severity. J. Dairy Sci., v.89, p.3408-3412, 2006.
- ZANELLA, G.N.; MIKCHA, J.M.G.; BANDO, E.; SIQUEIRA, V.L.D.; MACHINSKI, M. JR. Occurrence, antibiotic resistance of coliform bacteria and antimicrobial residues from cow's milk from Brazil. J. Food Prot., v.73, p.1684-1687, 2010.
Publication Dates
-
Publication in this collection
Apr-Jun 2014
History
-
Received
19 June 2013 -
Accepted
04 Oct 2013