Abstract
The genetic diversity of Theobroma speciosum is important because its use in breeding programs, once the species is closely related to species of great economic value such as Theobroma cacao (cocoa) and Theobroma grandiflorum (cupuaçu). Thus, the objective of this work is to characterize the intra and interpopulational genetic diversity of Theobroma speciosum in natural populations in the Brazilian Amazon. Ninety individuals of T. speciosum from four populations localized in different states of legal Amazon were selected and genotyped. The data were obtained by fluorescence microsatellite analysis and the number of alleles, number of private alleles, fixation index, observed and expected heterozygosity were analyzed. Bayesian analysis, AMOVA and PCOa were used to reveal the molecular genetic structure of the populations, using the programs Structure and GenAIEx 6.5, respectively. All populations studied present great levels of gene diversity, although, there was a greater similarity among the AUR, API and MAC populations, while RBC population presented higher heterozygosity and less inbreeding than the others, becoming a possible refuge area in the Amazon, and the most important population for T. speciosum conservation.
Key words:
Amazon; cacauhy; genetic variability
Resumo
O estudo da diversidade genética de Theobroma speciosum é importante, pois pode ser utilizado em programas de melhoramento, uma vez que a espécie está intimamente relacionada a espécies de grande valor econômico, como Theobroma cacao (cacau) e Theobroma grandiflorum (cupuaçu). Assim, o objetivo deste trabalho é caracterizar a diversidade genética intra e interpopulacional de Theobroma speciosum em populações naturais na Amazônia brasileira. Foram selecionados e genotipados 90 indivíduos de T. speciosum provenientes de quatro populações situadas em diferentes estados da Amazônia Legal. Os dados foram obtidos por meio da análise de microssatélite e a diversidade genética foi caracterizada através do número de alelos, número de alelos privados, índice de fixação e heterozigosidade observada e esperada. A análise bayesiana, a AMOVA e PCOa foram utilizadas para revelar a estrutura genética molecular das populações, através dos programas Structure e GenAIEx 6.5, respectivamente. Todas as populações estudadas apresentaram níveis de diversidade gênica, contudo, as populações AUR, API e MAC apresentaram grande similaridade, enquanto a população de Rio Branco apresentou maior heterozigosidade e menor endogamia do que as outras, se tornando um ponto de refúgio na Amazônia, e a mais importante população para a conservação de T. speciosum.
Palavras-chave:
Amazônia; cacauí; variabilidade genética
Introduction
In the Amazon region there is a large variety of environments and an enormous potential of natural resources, this potential is found in the most diverse species of the botanical families found in the region, such as, Anacardiaceae, Araceae, Arecaceae, Asteraceae, Bignoniaceae, Fabaceae, Lauraceae, Lecythidaceae, Malvaceae, Poaceae, and Rubiaceae (Steege et al. 2016Steege HT, Vaessen RW, Cárdenas-López D, Sabatier D, Antonelli A, Oliveira SM, Pitmannigel CA, Jorgensen PM & Salomão RP (2016) The discovery of the Amazonian tree flora with na updated checklist of all known tree taxa. Scientific Reports 6: 29549.).
Wild species of the genus Theobroma (Malvaceae) are endemic in the Amazon region (Dias 2001Dias LAS (2001) Melhoramento genético do cacaueiro. Funape, Viçosa. 578p.) (see Figure S1, available on supplementary material <https://doi.org/10.6084/m9.figshare.13696195.v1>) and require research for their inclusion in breeding programs, since they represent genetic resources with potential for the development of varieties more productive and resistant to pests and diseases (Almeida et al. 2009Almeida CMVC, Dias LAS & Silva AP (2009) Caracterização agronômica de acessos de cacau. Pesquisa Agropecuária Brasileira 44: 368-373.). The wild species Theobroma speciosum Willd. ex Spreng., is among the species of the genus least explored and with great potential, since it presents the fat content most similar to cocoa, making it a potential substitute (Silva & Martins 2004Silva AR & Martins MB (2004) A new anthophilic species of Drosophila Fallén belonging to the bromeliae group of species (Diptera, Drosophilidae). Revista Brasileira de Zoologia 21: 435-437.).
However, native species reminiscent of the genus Theobroma is suffering from strong genetic erosion due to anthropic action (Alves et al. 2013Alves RM, Silva CRS, Silva MSC, Silva DCS & Sebbenn AM (2013) Diversidade genética em coleções amazônicas de germoplasma de cupuaçuzeiro [Theobroma grandiflorum (Willd. ex Spreng.) Schum.]. Revista Brasileira de Fruticultura 35: 818-828.), which has led to the isolation of the populations in small fragments, reducing the number of reproductive individuals and the populational density (Young & Boyle 2000Young A & Boyle T (2000) Forest fragmentation. In: Young A, Boshier D & Boyle T (eds.) Forest conservation genetics: principles and practice. CSIRO Publishing, Collingwood. Pp. 123-132.). According to Laurance & Vasconcelos (2009)Laurance WF & Vasconcelos H (2009) Consequências ecológicas da fragmentação florestal na Amazônia. Oecologia Brasiliensis 13: 434-451., forest fragmentation causes innumerable effects because it alters population size and dynamics, community composition and dynamics, trophic interactions, and ecosystem processes. Considering that fact, measures that reduce the rate of deforestation are urgent in the fragmented Amazonian landscape.
The use of population genetics to quantify the diversity of tropical tree populations indicates some important directions that aim to minimize environmental impacts, as well as, to analyze the conservation level of populations (Frankham et al. 2002Frankham R, Ballou JD & Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge. 617p.). The knowledge of how genetic variation is partitioned between and within populations may have important implications not only in evolutionary and ecological biology but also in conservation biology (Balloux & Lugon-Moulin 2002Balloux F & Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Molecular Ecolology 11: 155-165.). Genetic diversity is an important factor for the survival of populations in different environments, and it is recognized as a fundamental component of biodiversity (Mace et al. 1996Mace GM, Smith TB, Bruford MW & Wayne RK (1996) An overview of the issues. In: Smith TB & Wayne RK (eds.) Molecular genetic approaches in conservation. Oxford University Press, New York. Pp. 3-12.). Thus, studying the genetic diversity in tree species is crucial, due to the importance they present in the structuring of ecosystems.
Recent studies using ISSR markers have helped to evaluate the genetic diversity of some species of the genus Theobroma. In their study about cultivations of Theobroma grandiflorum (Willd. ex Spreng.) Schum.) in northern Mato Grosso, Silva et al. (2016)Silva BM, Rossi AAB, Dardengo JFE, Araujo VAAC, Rossi FS, Oliveira LO & Clarindo WR (2016) Diversidade genética estimada com marcadores entre sequências simples repetidas em cultivos comerciais de Cupuaçuzeiro. Ciência Rural 46: 108-113. claim that most of the genetic diversity is contained inside the T. grandiflorum cultures. While analyzing natural populations of T. speciosum and T. subincanum Mart, Giustina et al. (2014)Giustina LD, Luz LN, Vieira FS, Rossi FS, Soares-Lopes CRA, Pereira TNS & Rossi AAB (2014) Population structure and genetic diversity in natural populations of Theobroma speciosum Willd. ex Spreng (Malvaceae). Genetics and Molecular Research 13: 3510-3519. and Rivas et al. (2013)Rivas LH, Giustina LD, Luz LN, Karsburg IV, Pereira TNS & Rossi AAB (2013) Genetic diversity in natural populations of Theobroma subincanum Mart. in the Brazilian Amazon. Genetics and Molecular Research 12: 4998-5006. found a greater genetic differentiation among the populations. Additionally, Silva et al. (2015)Silva BM, Rossi AAB, Dardengo JFE, Silva CR, Silva IV, Silva ML & Silva CJ (2015) Genetic structure of natural populations of Theobroma in the Juruena National Park, Mato Grosso state, Brazil. Genetics and Molecular Research 14: 10365-10375., used SSR (simple sequence repeat) markers to study native populations of T. speciosum and T. subincanum in the Juruena National Park - MT, and verified that the analyzed accesses present a high genetic diversity and therefore may be useful in the formation of germplasm banks. This work is the first to analyze T. speciosum populations from different states in legal Amazon and use the fluorescence technique, which is more effective once it allows greater accuracy in the detection of alleles (Alekcevetch 2013Alekcevetch JC (2013) Estudo de diversidade genética, por meio de marcadores moleculares de uma população de Coffea canephora var. Conilon. Dissertação de Mestrado. Universidade Federal de Lavras, Lavras. 115p.).
The observed differences in genetic diversity among populations of different states may be indicative of retraction and population expansion, as predicted by Haffer (1969)Haffer J (1969) Speciation in Amazonian forest birds. Science 165: 131-137. and Vanzolini & Williams (1970)Vanzolini PE & Williams EE (1970) South American anoles: the geographic differentiation and evolution of Anolis chrysolepis species group (Sáuria, Iguanidae). Arquivos de Zoologia 19: 1-298. refuge theory. Areas of endemism of butterflies, birds or plants are found in various parts of the Amazon, with overlapping areas of endemism of two or more groups at some points. These areas of endemism are possible refuge points, where less environmental variation occurred (Haffer & Prance 2002Haffer J & Prance GT (2002) Impulsos climáticos da evolução na Amazônia durante o Cenozóico: sobre a teoria dos Refúgios da diferenciação biótica. Estudos Avançados 16: 175-206.). Thus, there may have been sites with lower population fluctuations and are expected to have populations with higher heterozygosity and higher number of private alleles, as in the centers of origin of the species (Alves et al. 2007Alves RM, Sebbenn AM, Artero AS, Clement C & Figueira A (2007) High levels of genetic divergence and inbreeding in populations of cupuassu (Theobroma grandiflorum). Tree Genetics & Genomes 3: 289-298.).
The purpose of this work was to characterize the intra and interpopulational genetic diversity of T. speciosum in natural populations in the Brazilian Amazonin order to answer the following questions: (a) How is genetic diversity partitioned among and within populations? (b) How can the results obtained in this study improve the knowledge about T. speciosum and collaborate for the species conservation? The information obtained in this study can contribute, along with the other studies on the species, in the design of strategies for the conservation, improvement and management of T. speciosum, as they will provide a better understanding of the distribution of their current diversity and genetic structure.
Material and Methods
Study area and Sampling
To characterize genetic diversity, 90 T. speciosum individuals were identified, sampled, and georeferenced in four populations in the brazilian Amazon (Tab. 1; Fig. 1). The four populations presents natural individuals of T. specioum with an aggregated pattern, population density average of 57.5 ind.ha-1 and are in protected areas. Since it has this distribution pattern, we try not to sampled trees in a distance smaller than 70 m (Dardengo et al. 2016Dardengo JFE, Rossi AAB, Silva BM, Silva IV, Silva CJ & Sebbenn AM (2016) Diversity and spatial genetic structure of a natural population of Theobroma speciosum (Malvaceae) in the Brazilian Amazon. International Journal of Tropical Biology 64: 1091-1099.).
Theobroma speciosum populations sampled with their codes, geographical coordinates and number of specimes sampled. NS = number of species.
Geographical location of the four populations of Theobroma speciosum under study in brazilian Amazon.
DNA extraction and Polymerase chain reaction (PCR) amplification
Total genomic DNA was extracted using the cetyltrimethylammonium bromide method as described by Doyle & Doyle (1987)Doyle JJ & Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11-15. with the modifications: increase of 1% PVP concentration, 3% of CTAB and 2.7% of β-mercaptoetanol in buffer extraction and decrease of incubation time in 30 minutes. DNA was applied to an agarose gel (1% w/v) and stained with ethidium bromide for quantification. Bands were compared with a standard DNA (lambda phage) of known concentration. The gels were then examined using an ultraviolet transilluminator (UVB LTB-21x26) and photographed.
Twenty-three microsatellite loci (simple sequence repeats) that were characterized by Lanaud et al. (1999)Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A & Lagoda PJL (1999) Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology 8: 2141-2143. were tested in an initial PCR amplification using one T. speciosum individual. Of the 23 loci tested, 6 were selected for genetic diversity analysis. The amplification protocol followed that described by Lanaud et al. (1999)Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A & Lagoda PJL (1999) Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology 8: 2141-2143., with some modifications: one initial cycle at 94 °C for 4 min, followed by 32 cycles at 94 °C for 30 s, 46° or 51 °C (depending on the primer used) for 1 min, 72 °C for 1 min, and a final cycle at 72°C for 5 min.
Each forward primer was labeled with either 6-FAM, HEX, or NED (Applied Biosystems, São Paulo, Brazil) fluorescence (Tab. 2). The fragments were separated on a 96 capillary sequencer ABI PRISM 3130 × 1 DNA Analyzer (Applied Biosystems, Sao Paulo, Brazil), and PCR products were sized relative to a molecular size marker (ROX 500-Applied Biosystems, São Paulo, Brazil). The fragments were scored using GeneMarker v. 2.6.3 (Soft Genetics LLC).
Number of alleles (Na), expected heterozygosity (He), observed heterozygosity (Ho), fixation index (f) and the polymorphism information content (PIC) of six simple sequence repeat primers in 90 Theobroma speciosum individuals from four populations in Brazilian Amazon. * = P < 0,05
Data analysis
We used the Power Marker program (Liu & Muse 2005Liu K & Muse S (2005) Power Marker: integrated analysis environment for genetic marker data. Bioinformatics 21: 2128-2129.) to assess allelic frequency, genetic diversity, the observed and expected heterozygosity, fixation index (Weir & Cockerham 1984Weir BS & Cockerham CC (1984) Estimating F-statistics for the analysis ofpopulation structure. Evolution 38: 1358-1370.) and the polymorphism information content (PIC). The frequency of null alleles and score were estimated using the MICROCHEKER v. 2.2.3 (Oosterhout et al. 2004Oosterhou TCV, Hutchinson WF, Wills DPM & Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535-538.). Nei et al. (1973)Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, PNAS, USA 70: 3321-3323. matrix of genetic distance between T. speciosum trees was estimated using the same program. This matrix was imported by MEGA 4 (Tamura et al. 2007Tamura K, Dudley J, Nei M & Kumar S (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596-1599.) to construct a dendrogram of mean distance using the unweighted pair group method with arithmetic mean (UPGMA).
The Structure program (Pritchard et al. 2000Pritchard J, Stephens M & Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959.), which is based on Bayesian statistics, was used to indicate the number of genetic groups (K). We conducted 20 runs for each K value, with 200,000 burn-ins and 500,000 Markov chain Monte Carlo simulations. To determine the most probable value of K, we used the criteria proposed by Pritchard & Wen (2004)Pritchard JK & Wen W (2004) Documentation for structure software: Version 2.2. Available at <https://web.stanford.edu/group/pritchardlab/software/structure22/readme.pdf>. Access on 27 January 2021.
https://web.stanford.edu/group/pritchard...
and Evano et al. (2005)Evano G, Regnaut S & Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611-2620.. Principal coordinate analysis (PCA), deviations of the Hardy-Weinberg equilibrium and analysis of molecular variance (AMOVA) were performed using the GenAlEx 6.5 program (Peakall & Smouse 2006Peakall R & Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology 6: 288-295.).
Results
Genetic diversity by microsatellite loci
The six primers used in the analysis were polymorphic and amplified 86 alleles, with a mean of 14.33 alleles per locus. The highest number of alleles (21) was found at locus mTcCIR10 and mTcCIR19, and the lowest (6) at locus mTcCIR7 and mTcCIR28. All of the loci had high PIC values that varied between 0.20 and 0.88, with a mean of 0.70, besides the loci mTcCIR28 presents a low value (0.20). The mean observed heterozygosity was 0.47, and it ranged from 0.07 (mTcCIR28) to 0.69 (mTcCIR19). The mean expected heterozygosity was 0.72. The observed heterozygosity was lower than the expected heterozygosity for all locus (Tab. 2). There is no significant evidence for the presence of a null allele at the loci evaluated and all loci deviated the proportions of the Hardy-Weinberg equilibrium.
Genetic diversity by population
The total number of alleles in the studied populations varied from 53 for the API population to 32 in the RBC population (Tab. 3). The RBC population had the lowest number of alleles, but it was the one with the highest number of private alleles (5), and the observed heterozygosity was higher than expected, indicating a higher presence of heterozygotes than expected under the Hardy-Weinberg equilibrium condition.
Number of alleles (Na), number of private alleles (Apriv), expected heterozygosity (He), observed heterozygosity (Ho), fixation index (f) and the polymorphism information content (PIC) from four populations in Brazilian Amazon. * = P < 0,05
In the AUR, MAC and API populations, the heterozygosity observed was lower than expected, suggesting excess of homozygotes. This pattern becomes clearer by observing the fixation index (f). The negative and significantly different from zero value of the f-index in the RBC population suggests selection for heterozygotes. The positive and significantly different value of zero of the index f in the other populations suggests inbreeding. The content of polymorphic information was low only for the RBC population, being above 0.60 for the other populations (Tab. 3).
Genetic structure and population differentiation
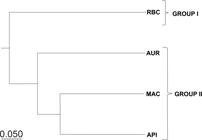
Analyzing the dendrogram (Fig. 2; see Table S1, available on supplementary material <https://doi.org/10.6084/m9.figshare.13696195.v1>), it is observed that the RBC population presented greater genetic dissimilarity in relation to the other analyzed populations, forming an exclusive group. The Bayesian analysis performed by the “Structure” program corroborates the result obtained by the UPGMA method, with the formation of two distinct groups (k = 2) (Fig. 3). The individuals from the Acre RBC population were assigned to a different group (green), the other samples from the states of Pará (AUR), Amapá (MAC) and Mato Grosso (API) were allocated in another group (red).
Dendrogram obtained by the UPGMA method, based on the genetic distances of Nei (1973)Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, PNAS, USA 70: 3321-3323., based on 4 populations of Theobroma speciosum in the Brazilian Amazon.
a. Map with the representation of the population grouping percentage in the two groups (K = 2) generated by the “Structure”. b. Delta K for visualization of the best K. c. Division of 90 Theobroma speciosum trees into two groups based on molecular data from six simple sequence repeat loci using the Structure program. Individuals are represented by vertical columns and are shaded according to their group (two genetic groups, K = 2). RBC = Rio Branco - Acre; AUR = Aurora do Pará - Pará; API = Apiacás - Mato Grosso; MAC = Macapá - Amapá.
The PCA explained 23.79% of the total variation, with 12.89% for the first component, 10.90% for the second (Fig. 4). As in the other clusters, the isolation of the RBC population was observed in relation to the other populations. AMOVA revealed that 91% of the total variance occurred within populations and 09% between populations (Tab. 4).
Principal coordinates analysis of 90 Theobroma speciosum individuals from Brazilian Amazon. RBC = Rio Branco - Acre; AUR = Aurora do Pará - Pará; API = Apiacás - Mato Grosso; MAC = Macapá - Amapá.
Analysis of molecular variance (AMOVA) of the four populations of Theobroma speciosum studied based on 06 SSR markers. d.f. = degrees of freedom; SS = sum of squares; CV = coefficient of variation; TV = total variation; and P = chances of a variance component greater than the observed values by chance. The probabilities were calculated using 1000 random permutations.
Discussion
Genetic diversity by locus and population
The high expected heterozygosity obtained in this study can be explained considering that most tropical tree species present a large number of alleles per locus and, consequently, a high expected heterozygosity (Alves et al. 2007Alves RM, Sebbenn AM, Artero AS, Clement C & Figueira A (2007) High levels of genetic divergence and inbreeding in populations of cupuassu (Theobroma grandiflorum). Tree Genetics & Genomes 3: 289-298.). Except for the RBC population, it is possible to observe higher values for the expected heterozygosity (He) in comparison with the observed heterozigosity (Ho), on the analysis by locus (Tab. 3) as well as on the analysis by population (Tab. 4).
Corroborating with these results, Zhang et al. (2012)Zhang D, Martínez WJ, Johnson ES, Somarriba E, Phillips- Mora W, Astorga C, Mischke S & Meinhardt LW (2012) Genetic diversity and spatial structure in a new distinct Theobroma cacao L. population in Bolivia. Genetic Resources and Crop Evolution 59: 239-252., in a study with populations of T. cacao, obtained higher values for He (0.56) compared to Ho (0.38). The species T. subincanum and T. speciosum (in a study with 13 microsatellite markers) presented values of 0.95 and 0.96 for He and 0.16 and 0.19 for Ho, respectively (Silva et al. 2015Silva BM, Rossi AAB, Dardengo JFE, Silva CR, Silva IV, Silva ML & Silva CJ (2015) Genetic structure of natural populations of Theobroma in the Juruena National Park, Mato Grosso state, Brazil. Genetics and Molecular Research 14: 10365-10375.). In a study with T. speciosum, Dardengo et al. (2016)Dardengo JFE, Rossi AAB, Silva BM, Silva IV, Silva CJ & Sebbenn AM (2016) Diversity and spatial genetic structure of a natural population of Theobroma speciosum (Malvaceae) in the Brazilian Amazon. International Journal of Tropical Biology 64: 1091-1099. and Varella et al. (2016)Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10. also obtained equivalent values, He = 0.88 and 0.97; Ho = 0.34 and 0.25, respectively.
Theobroma speciosum seed dispersal is generally performed by medium-sized mammals, such as monkeys, which consume the fruits and discard the seeds while they are still on the trees, contributing to the occurrence of aggregate seed shadows in the immediate vicinity of the mother plants, resulting in a spatial aggregation of individuals which share a recent common ancestral (Dardengo et al. 2017Dardengo JFE, Rossi AAB, Silva CJ & Silveira M (2017) Spatial structure of Theobroma subincanum Mart. and Theobroma speciosum Will. ex Spreng. in the Parque Nacional do Juruena, Mato Grosso state, Brazil. Revista Árvore 41: e:410101.). This aggregation could explain the low heterozygosity rates observed in most of the populations analyzed in this study. Furthermore, the study of T. speciosum by Dardengo et al. (2016)Dardengo JFE, Rossi AAB, Silva BM, Silva IV, Silva CJ & Sebbenn AM (2016) Diversity and spatial genetic structure of a natural population of Theobroma speciosum (Malvaceae) in the Brazilian Amazon. International Journal of Tropical Biology 64: 1091-1099. showed that the seeds of the species are dispersed up to approximately 70 m away from the mother tree, which can cause crossbreeding between relatives, generating biparental inbreeding, since the species presents mechanisms of self-incompatibility (Souza & Venturieri 2010Souza MS & Venturieri AG (2010) Floral Biology of Cacauhy (Theobroma speciosum - Malvaceae). Brazilian Archives of Biology and Technology 53: 861-872.).
However, more precise information regarding the effective performance of the different dispersers, the distance between the mother tree, the seed germination site and the genetic structure in the different stages of development of T. speciosum may lead to a better understanding of the causes of the excess of homozygotes in the T. speciosum populations AUR, API and MAC. Nybom (2004)Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143-1155. reviewed 106 studies of intraspecific genetic diversity in native plants based on microsatellite markers and reported an average of 9.9 alleles per locus. However, Rivas et al. (2013)Rivas LH, Giustina LD, Luz LN, Karsburg IV, Pereira TNS & Rossi AAB (2013) Genetic diversity in natural populations of Theobroma subincanum Mart. in the Brazilian Amazon. Genetics and Molecular Research 12: 4998-5006. studied native populations of Theobroma subincanum and obtained an average of 6.69 number of bands per locus.
In their study with T. speciosum in three urban forest fragments, Varella et al. (2016)Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10. obtained a mean of 9.33 alleles using 09 SSR loci. This value is similar to that described in the Nybom (2004)Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143-1155. review, but still lower than the average of alleles found in this study (14.33). This difference can be explained due to the fact that the others authors analyzed populations that were geographically close to each other and the present research studied populations that were geographically very distant from each other (Amapá, Acre, Mato Grosso and Pará state), having as a consequence a greater diversity.
The fixation index represents one of the most important parameters in population genetics, by measuring the balance between homozygotes and heterozygotes in the populations (Kageyama et al. 2003Kageyama PY, Sebbenn AM, Ribas LA, Gandara FB, Castellen M, Perecim MB & Vencovsky R (2003) Diversidade genética em espécies arbóreas tropicais de diferentes estágios sucessionais por marcadores genéticos. Scientia Florestalis 64: 93-107.), in this study the index was positive and significantly different from zero for all analyzed loci and for all analyzed populations (Tabs. 3; 4), with the exception of the RBC population, due the deviations of the Hardy-Weinberg equilibrium proportions caused by the excess of homozygotes, probably due to inbreeding, since T. speciosum is considered a self-incompatible species (Souza & Venturieri 2010Souza MS & Venturieri AG (2010) Floral Biology of Cacauhy (Theobroma speciosum - Malvaceae). Brazilian Archives of Biology and Technology 53: 861-872.).
According to Table 4, we observed that the population of Rio Branco (RBC) presents a pattern different from the others, has a lower number of alleles, but with more private alleles, with observed heterozygosity higher than expected and consequently the lowest endogamy among the populations analyzed, with negative fixation index (Carvalho et al. 2010Carvalho ACM, Freitas MLM, Moraes SMB, Moraes MLT, Stranghetti V, Alzate-Marin AL & Sebbenn AM (2010) Diversidade genética, endogamia e fluxo gênico em pequena população fragmentada de Copaifera langsdorffii. Revista Brasileira de Botânica 33: 599-606.).
Analyzing the values of Polimorphic Information Content, it was observed that the locus mTcCIR28 presented a value below from the others (PIC = 0.20), which would support an exclusion of the loco in other studies with T. speciosum species, once which according to Botstein et al. (1980)Botstein D, White RL, Skolmick H & Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisn. American Journal of Human Genetics 32: 314-331., markers with PIC values below 0.25 may be considered as minimally informative. However, the population analysis showed a high PIC value for most of the populations analyzed (except for the RBC population), thus indicating the existence of a high genetic diversity and revealing the quality of the markers used.
Diversity among populations
Geographically, MAC and AUR population are the closest to each other (Fig. 1). However, differently from that predicted by the isolation by distance model, the dendrogram (Fig. 2) revealed that the MAC and API populations are genetically more similar to each other. More studies in these areas have to be done to explain this unexpected result, probably there is a geographical barrier between the MAC and AUR populations that could substantially limit the gene flow between these populations.
The genetic differentiation of the RBC population is reflected in the structure of the populations obtained by UPGMA (Fig. 2), Bayesian (Fig. 3) and principal coordinates (Fig. 4), in which the population of Rio Branco is seen isolated from the others. Although according to Varella et al. (2016)Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10. the grouping made by “Structure” (Bayesian method) has the tendency to generate a deeper differentiation of subgroups, it is possible to observe a correspondence in the grouping of the individuals realized by the three methodologies used, as well as in the study of Varella et al. (2016)Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10. with T. speciosum, Silva et al. (2016)Silva BM, Rossi AAB, Dardengo JFE, Araujo VAAC, Rossi FS, Oliveira LO & Clarindo WR (2016) Diversidade genética estimada com marcadores entre sequências simples repetidas em cultivos comerciais de Cupuaçuzeiro. Ciência Rural 46: 108-113. with T. grandiflorum and Silva et al. (2015)Silva BM, Rossi AAB, Dardengo JFE, Silva CR, Silva IV, Silva ML & Silva CJ (2015) Genetic structure of natural populations of Theobroma in the Juruena National Park, Mato Grosso state, Brazil. Genetics and Molecular Research 14: 10365-10375. with T. subincanum and T. speciosum.
The genetic diversity partition made by AMOVA indicated that most of the genetic diversity (91%) is in the intrapopulation component, which can be explained by the fact that perennial species of cross fertilization, as well as T. speciosum, accumulate greater genetic diversity within their populations, and according to Hamrick et al. (1991)Hamrick JL, Godt MJW, Murawski DA & Loveless MD (1991) Correlations between species traits and allozyme diversity: in implications for conservation biology. In: Falk DA & Holsinger KE (eds.) Genetic and conservation of rare plants. Oxford University Press, New York. Pp. 75-86., present less differentiation between populations.
Thus, according to Nybom & Bartish (2000)Nybom H & Bartish I (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3: 93-114., the results pointed out by AMOVA corroborate those found for other tropical allogamous species. As for example, T. grandiflorum in the study by Silva et al. (2016)Silva BM, Rossi AAB, Dardengo JFE, Araujo VAAC, Rossi FS, Oliveira LO & Clarindo WR (2016) Diversidade genética estimada com marcadores entre sequências simples repetidas em cultivos comerciais de Cupuaçuzeiro. Ciência Rural 46: 108-113. where 34.91% of genetic diversity was contained between crops and Mauritia flexuosa studied by Rossi et al. (2014)Rossi FS, Rossi AAB, Dardengo JFE, Brauwers LR, Silva ML & Sebbenn AM (2014) Diversidade genética em populações naturais de Mauritia flexuosa L.f. (Arecaceae) com uso de marcadores ISSR. Scientia Forestalis 42: 631-639., which presented only 15.9% of the genetic variation among populations. However, Giustina et al. (2014)Giustina LD, Luz LN, Vieira FS, Rossi FS, Soares-Lopes CRA, Pereira TNS & Rossi AAB (2014) Population structure and genetic diversity in natural populations of Theobroma speciosum Willd. ex Spreng (Malvaceae). Genetics and Molecular Research 13: 3510-3519. and Rivas et al. (2013)Rivas LH, Giustina LD, Luz LN, Karsburg IV, Pereira TNS & Rossi AAB (2013) Genetic diversity in natural populations of Theobroma subincanum Mart. in the Brazilian Amazon. Genetics and Molecular Research 12: 4998-5006. when analyzing through ISSR locus natural populations of T. speciosum and T. subincanum, respectively, found a greater interpopulational genetic differentiation.
The population of Acre, among the analyzed populations, was the one that presented heterozygosity observed above the expected and greater number of private alleles and is located near of one of the possible refuges in Amazonia, as described by Haffer & Prance (2002)Haffer J & Prance GT (2002) Impulsos climáticos da evolução na Amazônia durante o Cenozóico: sobre a teoria dos Refúgios da diferenciação biótica. Estudos Avançados 16: 175-206.. However, there are few studies testing the theory of refuges in the Amazon, mainly with plants. The data of this work also do not allow making inferences about this theory, but the obtained results are an indicative that this also is a hypothesis to be more investigated.
Implications for conservation
All the populations studied presented high levels of gene diversity and although the RBC population presented lower alleles than the others, it was the one with the highest number of exclusive alleles (5), and its average of alleles per locus (5, 33) was superior to those found by Lanaud et al. (1999)Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A & Lagoda PJL (1999) Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology 8: 2141-2143. analyzing genotypes of T. cacao and Alves et al. (2013)Alves RM, Silva CRS, Silva MSC, Silva DCS & Sebbenn AM (2013) Diversidade genética em coleções amazônicas de germoplasma de cupuaçuzeiro [Theobroma grandiflorum (Willd. ex Spreng.) Schum.]. Revista Brasileira de Fruticultura 35: 818-828. in accessions of T. grandiflorum, which obtained averages of 4.4 alleles per locus for T. cacao and 3.21 for T. grandiflorum. Thus, considering the average number of alleles per locus and the presence of private alleles, it can be affirmed that all the populations studied have value for in situ genetic conservation of T. speciosum, as well as for the collection of germoplasm aiming its conservation ex situ and collection of seeds, or for the formation of seedlings destined to the restoration of degraded areas or for the forest improvement.
It is important to maintain and protect the genetic diversity of T. speciosum throughout the Amazonian landscapes studied, in order to avoid the fragmentation and predatory exploitation of the fruits, which can prevent their dispersion and consequently the natural establishment of the species (Varella et al. 2016Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10.). In addition, it is possible to identify the genetic diversity of the species in the next generations (Varella et al. 2016Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10.).
Ecology and genetics information on natural populations of tropical tree species are essential for understanding the genetic structure of populations and therefore, for the design of strategies for conservation, breeding and sustainable management (definition of reserve areas, adequate management of species, recuperation of degraded areas, seed collection for plantations with native species) (Kageyama et al. 2003Kageyama PY, Sebbenn AM, Ribas LA, Gandara FB, Castellen M, Perecim MB & Vencovsky R (2003) Diversidade genética em espécies arbóreas tropicais de diferentes estágios sucessionais por marcadores genéticos. Scientia Florestalis 64: 93-107.). Thus, the results obtained in this study are important for the adoption of strategies for the conservation of the Amazon Forest, generating indicators for establishment and management of genetic reserves in situ, as well as for the implantation of gene flow corridors between small reserves, once was indicated in this study, that the populations have a conexion, showed by the cluster of structure program.
Conclusions
All populations studied present levels of gene diversity, high average number of alleles per locus and presence of private alleles, so the establishment of permanent conservation units could be a valuable tool to preserve genetic diversity among the individuals of these natural populations. Although, there was a greater similarity among the AUR, API and MAC populations, while individuals from the RBC population presented higher heterozygosity and less inbreeding than the others, suggesting that their geographical position may have been little affected by environmental changes, becoming a point of refuge in the Amazon, and the most important population for T. speciosum conservation.
-
See supplementary material at <https://doi.org/10.6084/m9.figshare.13696195.v1>
Acknowledgements
The authors acknowledge Glenn Hawes, M.Ed. English, University of Georgia, for editing this manuscript, BIONORTE - MT (Projeto Conhecimento, Uso Sustentável e Bioprospecção da Biodiversidade na Amazônia Meridional - Processo: 554330/2010-5) and CAPES for the financial support.
References
- Alekcevetch JC (2013) Estudo de diversidade genética, por meio de marcadores moleculares de uma população de Coffea canephora var. Conilon Dissertação de Mestrado. Universidade Federal de Lavras, Lavras. 115p.
- Almeida CMVC, Dias LAS & Silva AP (2009) Caracterização agronômica de acessos de cacau. Pesquisa Agropecuária Brasileira 44: 368-373.
- Alves RM, Silva CRS, Silva MSC, Silva DCS & Sebbenn AM (2013) Diversidade genética em coleções amazônicas de germoplasma de cupuaçuzeiro [Theobroma grandiflorum (Willd. ex Spreng.) Schum.]. Revista Brasileira de Fruticultura 35: 818-828.
- Alves RM, Sebbenn AM, Artero AS, Clement C & Figueira A (2007) High levels of genetic divergence and inbreeding in populations of cupuassu (Theobroma grandiflorum). Tree Genetics & Genomes 3: 289-298.
- Balloux F & Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Molecular Ecolology 11: 155-165.
- Botstein D, White RL, Skolmick H & Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisn. American Journal of Human Genetics 32: 314-331.
- Carvalho ACM, Freitas MLM, Moraes SMB, Moraes MLT, Stranghetti V, Alzate-Marin AL & Sebbenn AM (2010) Diversidade genética, endogamia e fluxo gênico em pequena população fragmentada de Copaifera langsdorffii Revista Brasileira de Botânica 33: 599-606.
- Dardengo JFE, Rossi AAB, Silva BM, Silva IV, Silva CJ & Sebbenn AM (2016) Diversity and spatial genetic structure of a natural population of Theobroma speciosum (Malvaceae) in the Brazilian Amazon. International Journal of Tropical Biology 64: 1091-1099.
- Dardengo JFE, Rossi AAB, Silva CJ & Silveira M (2017) Spatial structure of Theobroma subincanum Mart. and Theobroma speciosum Will. ex Spreng. in the Parque Nacional do Juruena, Mato Grosso state, Brazil. Revista Árvore 41: e:410101.
- Dias LAS (2001) Melhoramento genético do cacaueiro. Funape, Viçosa. 578p.
- Doyle JJ & Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11-15.
- Frankham R, Ballou JD & Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge. 617p.
- Evano G, Regnaut S & Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611-2620.
- Giustina LD, Luz LN, Vieira FS, Rossi FS, Soares-Lopes CRA, Pereira TNS & Rossi AAB (2014) Population structure and genetic diversity in natural populations of Theobroma speciosum Willd. ex Spreng (Malvaceae). Genetics and Molecular Research 13: 3510-3519.
- Haffer J (1969) Speciation in Amazonian forest birds. Science 165: 131-137.
- Haffer J & Prance GT (2002) Impulsos climáticos da evolução na Amazônia durante o Cenozóico: sobre a teoria dos Refúgios da diferenciação biótica. Estudos Avançados 16: 175-206.
- Hamrick JL, Godt MJW, Murawski DA & Loveless MD (1991) Correlations between species traits and allozyme diversity: in implications for conservation biology. In: Falk DA & Holsinger KE (eds.) Genetic and conservation of rare plants. Oxford University Press, New York. Pp. 75-86.
- Kageyama PY, Sebbenn AM, Ribas LA, Gandara FB, Castellen M, Perecim MB & Vencovsky R (2003) Diversidade genética em espécies arbóreas tropicais de diferentes estágios sucessionais por marcadores genéticos. Scientia Florestalis 64: 93-107.
- Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A & Lagoda PJL (1999) Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology 8: 2141-2143.
- Laurance WF & Vasconcelos H (2009) Consequências ecológicas da fragmentação florestal na Amazônia. Oecologia Brasiliensis 13: 434-451.
- Liu K & Muse S (2005) Power Marker: integrated analysis environment for genetic marker data. Bioinformatics 21: 2128-2129.
- Mace GM, Smith TB, Bruford MW & Wayne RK (1996) An overview of the issues. In: Smith TB & Wayne RK (eds.) Molecular genetic approaches in conservation. Oxford University Press, New York. Pp. 3-12.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, PNAS, USA 70: 3321-3323.
- Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143-1155.
- Nybom H & Bartish I (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3: 93-114.
- Oosterhou TCV, Hutchinson WF, Wills DPM & Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535-538.
- Peakall R & Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology 6: 288-295.
- Pritchard JK & Wen W (2004) Documentation for structure software: Version 2.2. Available at <https://web.stanford.edu/group/pritchardlab/software/structure22/readme.pdf>. Access on 27 January 2021.
» https://web.stanford.edu/group/pritchardlab/software/structure22/readme.pdf - Pritchard J, Stephens M & Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
- Rivas LH, Giustina LD, Luz LN, Karsburg IV, Pereira TNS & Rossi AAB (2013) Genetic diversity in natural populations of Theobroma subincanum Mart. in the Brazilian Amazon. Genetics and Molecular Research 12: 4998-5006.
- Rossi FS, Rossi AAB, Dardengo JFE, Brauwers LR, Silva ML & Sebbenn AM (2014) Diversidade genética em populações naturais de Mauritia flexuosa L.f. (Arecaceae) com uso de marcadores ISSR. Scientia Forestalis 42: 631-639.
- Silva BM, Rossi AAB, Dardengo JFE, Araujo VAAC, Rossi FS, Oliveira LO & Clarindo WR (2016) Diversidade genética estimada com marcadores entre sequências simples repetidas em cultivos comerciais de Cupuaçuzeiro. Ciência Rural 46: 108-113.
- Silva BM, Rossi AAB, Dardengo JFE, Silva CR, Silva IV, Silva ML & Silva CJ (2015) Genetic structure of natural populations of Theobroma in the Juruena National Park, Mato Grosso state, Brazil. Genetics and Molecular Research 14: 10365-10375.
- Silva AR & Martins MB (2004) A new anthophilic species of Drosophila Fallén belonging to the bromeliae group of species (Diptera, Drosophilidae). Revista Brasileira de Zoologia 21: 435-437.
- Souza MS & Venturieri AG (2010) Floral Biology of Cacauhy (Theobroma speciosum - Malvaceae). Brazilian Archives of Biology and Technology 53: 861-872.
- Steege HT, Vaessen RW, Cárdenas-López D, Sabatier D, Antonelli A, Oliveira SM, Pitmannigel CA, Jorgensen PM & Salomão RP (2016) The discovery of the Amazonian tree flora with na updated checklist of all known tree taxa. Scientific Reports 6: 29549.
- Tamura K, Dudley J, Nei M & Kumar S (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596-1599.
- Vanzolini PE & Williams EE (1970) South American anoles: the geographic differentiation and evolution of Anolis chrysolepis species group (Sáuria, Iguanidae). Arquivos de Zoologia 19: 1-298.
- Varella TL, Rossi AAB, Dardengo JFE, Silveira GF, Souza MDA & Carvalho MLS (2016) Effect of fragmentation on the natural genetic diversity of Theobroma speciosum Willd. ex Spreng. populations. Genetics and Molecular Research 15: 1-10.
- Weir BS & Cockerham CC (1984) Estimating F-statistics for the analysis ofpopulation structure. Evolution 38: 1358-1370.
- Young A & Boyle T (2000) Forest fragmentation. In: Young A, Boshier D & Boyle T (eds.) Forest conservation genetics: principles and practice. CSIRO Publishing, Collingwood. Pp. 123-132.
- Zhang D, Martínez WJ, Johnson ES, Somarriba E, Phillips- Mora W, Astorga C, Mischke S & Meinhardt LW (2012) Genetic diversity and spatial structure in a new distinct Theobroma cacao L. population in Bolivia. Genetic Resources and Crop Evolution 59: 239-252.
Edited by
Publication Dates
-
Publication in this collection
08 Mar 2021 -
Date of issue
2021
History
-
Received
29 June 2018 -
Accepted
02 Apr 2020