Abstract
Abstract: Authentication of cell lines is of paramount importance to validate the results from their use in biomedical research. Although isoenzyme polymorphism is the standard method, molecular methods based on mitochondrial DNA (mtDNA) have been developed to replace it. The aim of this study was the improvement of our isoenzyme electrophoretic analysis and the validation of one molecular technique targeted at mtDNA for the authentication of our animal cell lines. The combined method of cellular lysing through osmotic shock, followed by freezing-thawing in N2 to obtain isoenzyme extracts, and with 42 × 106 cells maintained the best efficiency. The superior electrophoretic conditions were PAGE run at 200 V. All cell lines had isoenzymatic mobility corresponding to their species to lactate dehydrogenase, malate-dehydrogenase, and glucose-6-phosphate dehydrogenase isoenzymes, and could be distinguished from each other. Two molecular techniques based on mtDNA were tested, one on the cytochrome b gene and other on cytochrome c oxidase I subunit gene. Due to difficulties in distinguishing all cell lines using only one these techniques, we merged the primers of two methods in such a way that there was a sufficient differentiation of all DNA fragments. The sequencing of these PCR products was also performed to validate these data.
Key words
authentication; cell line; DNA barcode; isoenzyme
INTRODUCTION
Several areas of basic and applied science demand cell lines, such as viral isolation and diagnosis; production of vaccines and biopharmaceuticals; toxicity testing of materials of synthetic or natural origin; and studies of complex physiological systems (ButtlerBUTTLER M. 2005. Animal cell cultures: recent achievements and perspectives in the production of biopharmaceuticals. Appl Microbiol Biotechnol 68: 283-291. 2005, FreshneyFRESHNEY RI. 2005. Culture of animal cells: a manual of basic technique, 5th ed., New York: Wiley-Liss, 672 p. 2005, Capes-DavisCAPES-DAVIS A et al. 2010. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer 127: 1-8. et al. 2010, CreeCREE IA. 2011. Cancer cell culture, Methods and protocols, 2nd ed., Portsmouth: Human press, 493 p. 2011). Given this extensive and important applicability, techniques must be employed to confirm the identity of cell lines, as well as to monitor cross- and microbial contaminations, with the goal of validating the results (MarkovicMARKOVIC O and MARKOVIC N. 1998. Cell cross- contamination in cell cultures: the silent and neglected danger. In Vitro Cell Dev-An 34(1): 1-8. and Markovic 1998, MastersMASTERS JR et al. 2001. Short tandem repeat profiling provides an international reference standard for human cell lines. PNAS 98(1): 8012-8017. et al. 2001, Capes-Davis et al. 2010, GeraghtyGERAGHTY RJ et al. 2014. Guidelines for the use of cell lines in biomedical research. Brit J Cancer 111: 1021-1046. et al. 2014). Good laboratory practice is also fundamental to maintain quality control of these cell lines, even though there is an increase in the facilities for development and exchange of new cell lines (HartungHARTUNG T, BALLS M, BARDOUILLE C, BLANCK O, COECKE S, GSTRAUNTHALER G and LEWIS D. 2002. Good Cell Culture Practice. ATLA 30: 407-414. et al. 2002, Cree 2011, Geraghty et al. 2014). Thus, their acquisition should be from reliable sources or, preferably, from reference centers that ensure their certification and quality control; furthermore, continuous monitoring should always be performed (Hartung et al. 2002, Freshney 2005, Cree 2011, Geraghty et al. 2014).
The certification or characterization of a cell line is based on a series of determinations, such as population growth, morphology, cytogenetics, absence of adventitious agents, viral susceptibility and, mainly, species identification (HayHAY RJ. 1992. ATCC Quality control methods for cell lines, 2nd ed., Rockville: American Type Culture Collection, 132 p. https://www.amazon.com/ATCC-Quality-Control-Methods- Lines/dp/0930009460.
https://www.amazon.com/ATCC-Quality-Cont...
1992, FernandesFERNANDES MJB and SIMONI IC. 1995a. Caracterização de linhagens celulares. I- Identificação de espécies por análise isoenzimática. Arq Inst Biol 62: 59-63. and Simoni 1995a, bFERNANDES MJB and SIMONI IC. 1995b. Caracterização de linhagens celulares. II- Suscetibilidade aos vírus da doença de Aujeszky e da febre aftosa. Arq Inst Biol 62: 65-72., Freshney 2005, ATCCATCC - AMERICAN TYPE CULTURE COLLECTION. 2007. Cell line authentication test recommendations. ATCC Technical Bulletin nº 8. https://www.atcc.org/~/media/PDFs/Technical%20Bulletins/tb08.ashx.
https://www.atcc.org/~/media/PDFs/Techni...
2007). With regard to the latter item, several approaches have been used to authenticate cell lines and methodologies, such as karyotyping, immunological techniques, isoenzymes and molecular analysis, have been employed (MontesMONTES DE OCA F, MACY ML and SHANNON JE. 1969. Isoenzyme characterization of animal cell cultures. P Soc Exp Biol Med 132: 462-469. de Oca et al. 1969, StulbergSTULBERG CS, PETERSON WDJR and SIMPSON WF. 1976. Identification of cells in culture. Am J Hematol 1(2): 237-242. et al. 1976, Freshney 2005, ATCC 2007). Until now, isoenzyme analysis by electrophoresis is the technique recommended by main cell banks for the authentication of cell lines (ParodiPARODI B, ARESU O, BINI D, LORENZINI R, SCHENA F, VISCONTI P, CESARO M, FERRERA D, ANDREOTTI V and RUZZON T. 2002. Species Identification and Confirmation of Human and Animal Cell Lines: A PCR-Based Method. BioTechniques 32(2): 432-440. et al. 2002, LosiLOSI CG, FERRARI S, SOSSI E, VILLA R and FERRARI M. 2008. An alternative method to isoenzyme profile for cell line identification and interspecies cross-contaminations: cytochrome b PCR-RLFP analysis. In Vitro Cell Dev-An 44(8): 321-329. et al. 2008). The genetic polymorphism of enzymes seen through their electrophoretic mobility can discriminate cell lines from different animal species (Montes de Oca et al. 1969, SteubeSTEUBE KG, GRUNICKE D and DREXLER HG. 1995. Isoenzyme analysis as a rapid method for the examination of the species identity of cell cultures. In Vitro Cell Dev-An 31(2): 115-119. et al. 1995, ATCC 2007, RamyaRAMYA R, NAGARAJAN T, SIVAKUMAR V, SENTHILKUMAR RL, OBULAPATHI BB, THIAGARAJAN D and SRINIVASAN AA. 2009. Identification of cross-contaminated animal cells by PCR and isoenzyme analysis. Cytotechnology 61: 81-92. et al. 2009). Our initial isoenzyme studies were directed to the certification of cell lines using lactate dehydrogenase (LDH) and glucose-6-phosphate dehydrogenase (G-6-PHD) (Fernandes and Simoni 1995a). Although these enzymes are sufficient for species identification, several authors recommend the use of three or more isoenzymes (Hay 1992, NimsNIMS RW, SHOEMAKER AP, BAUERNSCHUB MA, REC LJ and HARBELL JW. 1998. Sensitivity of isoenzyme analysis for the detection of interspecies cell line cross-contamination. In Vitro Cell Dev-An 34(1): 35-39. et al. 1998). It is noted that both isoenzymes do not distinguish several species of primates, and either isoenzyme distinguishes the human from chimpanzee cell lines (HaltonHALTON DM, PETERSON WDJR and HUKKU B. 1983. Cell culture quality control by rapid enzymatic characterization. In Vitro 19(1): 16-24. et al. 1983, Steube et al. 1995). There is a commercially available kit (AuthentiKit system, Innovative Chemistry Inc.), that utilizes seven isoenzymes (Nims and Herbstritt 2005NIMS RW and HERBSTRITT CJ. 2005. Strategies for accurate speciation and case studies for the detection of cell line cross-contamination using a commercial kit. Biopharm International 18(6): 76-82.). This electrophoretic technique also detects inter- or intra-species contaminations of cell lines (Nims et al. 1998, Markovic and Markovic 1998, Ramya et al. 2009). Our further isoenzyme studies focused on experimental crosscontamination of cell lines using these two enzymes and two others, malate-dehydrogenase (MDH) and esterase (BarrosBARROS TF, SIMONI IC and FERNANDES MJB. 2008. Análise eletroforética de isoenzimas para detecção de contaminação cruzada entre linhagens celulares. Rev Biociências 14: 17-22. et al. 2008). We demonstrated a sensitivity of this technique for inter-species contamination monitoring of around 10%, also observed by Nims et al. (1998).
Several studies have introduced molecular techniques for cell line authentication and even suggested their replacement of isoenzyme technique. The targets are the mitochondrial DNA (mtDNA) (Parodi et al. 2002, CooperCOOPER JK, SYKES G, KING S, COTTRILL K, IVANOVA NV, HANNER R and IKONOMI P. 2007. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cell Dev-An 43: 344-351. et al. 2007, OnoONO K, SATOH M, YOSHIDA T, OZAWA Y, KOHARA A, TAKEUCHI M, MIZUSAWA H and SAWADA H. 2007. Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cell Dev-An 43(5): 168-175. et al. 2007, Losi et al. 2008. Ramya et al. 2009); aldolase gene (LiuLIU MY, LIN SC, LIU H, CANDAL F and VAFAI A. 2003. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev-An 39: 424-427. et al. 2003); or short tandem repeat (STR) profiling (Masters et al. 2001). DNA nucleotide sequencing for determination of species origin is widely applied in forensic studies, with mtDNA being the most frequent target (LinacreLINACRE A and TOBE SS. 2011. An overview to the investigative approach to species testing in wildlife forensic. Invest Genet 2: 1-9. and Tobe 2011). The two internationally recommended mitochondrial genes for forensic species identification are the cytochrome b gene (cyt b) and the cytochrome c oxidase I subunit gene (cox I). For this reason, we selected two methodologies based on these genes for our studies.
This study aimed to optimize the isoenzyme methodology performed in our laboratory (Fernandes and Simoni 1995a, Barros et al. 2008), as well as to validate a molecular method based on the mtDNA gene for the authentication of our animal cell lines.
MATERIALS AND METHODS
CELL LINES
Seven cell lines from seven animal species were used in the study (Table I). The LLC-PK1 cell line was cultivated in Medium 199 supplemented with 3% FBS with 5% CO2 atmosphere at 37 °C. The others were cultivated in Minimum Essential Eagle Medium (MEM) supplemented with 8% fetal bovine serum (FBS). In the case of the RK13 cell line, a 1% non-essential amino acid solution was also added.
ISOENZYME ANALYSIS OF CELL LINES
The active enzyme extracts were obtained by cellular lysis using an osmotic shock/freezing-thawing method (Fernandes and Simoni1995a). Cell suspensions were adjusted to a concentration of 42 ×106 cells, washed twice with phosphate-buffered saline (PBS; pH 7.2), and once with TNM buffer (pH 7.8; containing 10 mM Tris, 10 mM NaCl and 1.5 mM MgCl2) and centrifuged at 2500 g for 10 min at 4 °C. Pelleted cells were resuspended in 1 mL of TNM buffer and maintained on ice for 15 min, followed by three cycles of freezing in N2. The supernatants were collected by centrifugation at 2500 g for 20 min at 4 °C, and stored at -80 °C. The following three dehydrogenase isoenzymes were analyzed: lactate dehydrogenase (LDH), malate-dehydrogenase (MDH) and glucose-6-phosphate dehydrogenase (G-6-PDH). In this isoenzyme extraction methodology, we tested some changes using three cell lines and LDH isoenzyme: a) removal of the osmotic shock buffer (CRFK and RK13 cell lines); b) freezing/thawing process in ultra-freezer at -80 ºC (MDCK.2 cell line); and c) cell concentration of 21 × 106 cells (RK13 cell line). The extracts were subjected to vertical electrophoresis as described in Barros et al. (2008) and the change of current (20 mA) to voltage (200 V) was applied in all protocols. After electrophoresis, the isoenzymes were revealed on gels by the electron transfer dye staining method in an adaptation of that described by HarrisHARRIS H and HOPKINSON DA. 1976. Handbook of enzyme electrophoresis in human genetics. Amsterdam: North-Holland. and Hopkinson (1976). For this, the gels were incubated at 37ºC with a specific staining solution for each isoenzyme and kept in dark until the development of the bands (formazans). For LDH, the staining components were 0.05 M Tris/HCl buffer at pH 8.0, 8mM Lactic acid, 0.38mM nicotinamide adenine nucleotide (NAD), 0.15mM nitro blue tetrazolium (NBT), and 0.2mM phenazine methosulphate (PMS). For MDH, 0.1 M Tris/HCl buffer at pH 8.0, 50 mM Malic acid (pH again adjusted to 8.0), 0.3 mM NAD, 0,18 mM NBT and 0.33mM PMS. For G-6-PDH, 0.2 M Tris/HCl buffer at pH 8.0, 0.2 mM MgCl2, 0.6 mM Glucose-6- phosphate disodium salt and 0.13 mM NAD phosphate (NADP) were substituted for the first three components of MDH.
MOLECULAR ANALYSIS OF CELL LINES
DNA from 5 × 106 cells was extracted with the QIAamp® DNA Mini kit according manufacturer’s instructions. Two PCR approaches were used to amplify mtDNA genes using the GoTaq ® Hot Start Colorless Master Mister kit (Promega) or the KAPATaq ReadyMix DNA polymerase kit (Kapabiosystems). The first technique was a PCR performed with primers directed to the interspecific region of the cytochrome c oxidase I (cox I) subunit gene from each cell line (Parodi et al. 2002). The second technique was a nested PCR performed with primers targeting the region within the cytochrome b, d-loop, and 12S and 16S ribosomal rRNA genes. The first external primer pair was universal and the second internal primer pair was species-specific (Ono et al. 2007).
EVALUATION OF SENSITIVITY AND SPECIFICITY OF PCR
To evaluate the specificity of the identification, 10 µL aliquots of each cell line DNA were mixed and this pool of DNA templates was amplified with each specific primer separately (positive pool). The so-called negative pools contained all cell line DNAs, except that than the primer pair was used for amplification. PCR reaction with each specific primer pair at 10 µM in a 25 µL final volume was prepared using anyone of PCR kits and the primers and PCR conditions were those reported in Parodi et al. (2002). For the sensitivity assays, DNA from the MDCK.2 cell line was diluted at 1:2, 1:100, 1:1000 and 1:10 000 for use as a template in the PCR reaction as described above. Additionally, multiplex PCR was performed with species-specific primer mixes of MDBK, CRFK and RK13 cell lines following amplification with individual DNA templates and one mixed template from CRFK, and RK13 DNAs. Primers and PCR conditions used for these last two reactions were those reported in Ono et al. (2007).
RESULTS
IDENTIFICATION OF CELL LINE SPECIES BY ISOENZYME ANALYSIS
Figures 1–3 show LDH, MDH, and G-6-PDH isoenzymes of the cell lines using the cell extracts without alterations in their preparation.
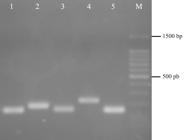
Lactate dehydrogenase (LDH) isoenzyme on PAGE. Cell lines: MDBK (1); Vero (2); CRFK (3); MDCK.2 (4); RK13 (5); and LLC-PK1 (6).
Multiple bands were obtained for LDH, with MDCK.2 cell line presenting three bands and all the others four bands (Figure 1). Regardless of same number of bands for most cell lines, it was possible to distinguish all species by the heterogeneity of electrophoretic mobility.
Both MDH and G-6-PDH presented a single band in all cell lines, but with different electrophoretic mobilities that allowed them to be distinguished (Figures 2 and 3). For G-6-PDH, a 2% agarose gel electrophoresis was used because the PAGE was not productive for this isoenzyme, and even so, the LLC-PK1 and Vero bands remained very weak and almost invisible, respectively.
The electrophoretic mobility patterns of three isoenzymes from all cell lines enabled them to be distinguished from each other. Additionally, comparative analysis of the isoenzymes confirmed their species identity and authentication.
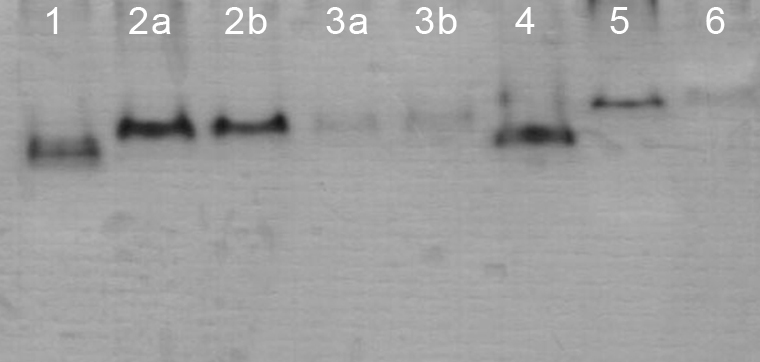
Malate dehydrogenase (MDH) isoenzyme on PAGE. Cell lines: MDBK (1); MDCK.2 with N2 (2a) and ultrafreezer (2b) freezing process; CRFK with (3a) and without (3b) osmotic shock buffer; LLC-PK1 (4); RK13 (5); and Vero (6).
Glucose-6-phosphate dehydrogenase (G-6-PDH) on agarose gel. Cell lines: CRFK (1); Vero (2): imperceptible band (arrow), MDBK (3); RK13 (4); LLC-PK1 (5); and MDCK.2 (6).
The changes applied to the isoenzyme extraction procedure were evaluated only in the MDCK.2, CRFK and RK13 cell lines. Although the number of bands of LDH for these cell lines remained the same, there was a lower resolution linked mainly to a concomitant reduction in the amount of cells. Furthermore, some bands were almost imperceptible (Figure 4). For MDH (Figure 2) and G-6-PDH, this extraction procedure was able to detect normally their unique band.
Lactate dehydrogenase (LDH) isoenzyme on PAGE with changes applied to the isoenzyme extraction procedure. Cell lines: CRFK with (1) and without (2) osmotic shock buffer; MDCK.2 with N2 (3) and ultrafreezer (4) freezing process; RK13 with (5) and without (6) osmotic shock buffer. RK13 had 21 x 106 cells while CRFK and MDCK.2 had 42 x 106 cells.
IDENTIFICATION OF CELL LINE SPECIES BY MOLECULAR ANALYSIS
In Table II is the list of primers used in PCR reactions with their respective sizes and reference.
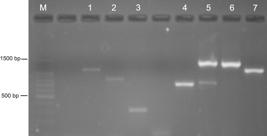
The PCR directed to cox I gene amplified fragments with expected sizes (Figure 5), but those from RK13, MDCK.2, and L929 cell lines had sizes, which were very similar, making it difficult to distinguish these three cell lines.
PCR products from species-specific primer pairs of cell lines based in Parodi et al. (2002) on 2% agarose gel electrophoresis at 80V. Lane 1: RK13 (151 bp); lane 2: CRFK (180 bp); lane 3: MDCK.2 (153 bp); lane 4: Vero (222 bp); lane 5: L929 (150 bp); lane 6: 100 bp DNA ladder (Promega).
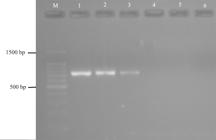
Already the first product of nested PCR with the universal primers did not present the expected size, only unspecific bands. Despite this, we tested the second reaction for CRFK and Vero cell lines by adding of 2 µL of purified amplicons. Species-specific fragments were generated for these cell lines (Table II). Based on this, nested PCR was not carried out and amplification was done by PCR using DNA from the cell lines as targets and species-specific primer pairs. There was success in the identification of the species-specific bands of the cell lines (Figure 6 and 7). For some reactions were necessary 2 µL of templates.
Gel electrophoresis of PCR (a) and multiplex-PCR products (b) from species-specific primer pairs of cell lines used in 2nd direct reaction of Ono et al. (2007) on 2% agarose gel at 80V. A: M (Marker): 100 bp DNA Ladder (Promega); lane 1: MDBK (1090 bp); lane 2: LLC-PK1 (819 bp); lane 3: Vero (301 bp). B: Lane 4 and 5: RK13 (704 bp), lane 5 and 6: CRFK (1391 bp), and lane 7: MDBK (1090 bp).
Gel electrophoresis of PCR products for sensitivity of PCR reaction using the MDCK.2 cell line (5 x 106 cells) and the 2nd direct reaction of Ono et al. (2007) on 2% agarose gel electrophoresis at 80V. M (Marker): 100 bp DNA Ladder (Promega); lane 1: pure cells; lane 2: diluted cells 1:2; lane 3-6: serially diluted cells 1:100, 1:1000, 1:10,000 and 1:100,000, respectively.
Based on the results overall, none of techniques were fully successful. In the first technique, the primers of some cell lines generated fragments of little distinguishable size and in the second technique, only the amplification of 2nd product was productive using the species-specific primers. Taking this into account, we combined these two techniques in a single one by selecting primers from these two techniques (Table II). Thus, the primers of first technique for RK13 and MDCK.2 cell lines were tested with the amplification cycle of second technique, and vice versa. There was success in the amplification of all cell line fragments using both cycles without the necessity of selecting any specific cycle for this new protocol. For the primers, the selection focused on producing DNA fragments with well distinguished sizes for all cell lines. From this approach, we selected the primers from the second technique for RK13 and MDCK.2 cell lines because their primers distinguished better these cell lines from L929 cell line. L929 primers is only available in the first technique, and primers for the MDBK and LLC-PK1 cell lines in the second technique. For the Vero and CRFK cell lines, the primers of both techniques produced differentiable fragments between them and those of other cell lines. Thus, there was no need to make a selection. After this selection of primers, complementary studies regarding the primer specificity and sensitivity of the PCR reaction were carried out. We used the second technique and the MDCK.2 cell line for the sensitivity test. The sensitivity of the PCR was up to 5 × 104 cells (Figure 7, lane 3).
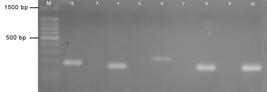
With regard to the specificity of the PCR using the first technique, in the positive pools only the DNA of the cell line corresponding to the primer used was amplified (Figure 8, even lanes), while no band was amplified in the negative pools (Figure 8, odd lanes). These results demonstrated that the presence of DNA from other cell lines did not interfere with the amplification of DNA from each cell line or contamination to the PCR reaction confirming the specificity of the primers.
Gel electrophoresis of the PCR products for specificity of PCR reaction using Parodi et al. (2002) on 2% agarose gel electrophoresis at 80V. M (Marker): 100 bp DNA ladder (Promega); Positive Pools on even lanes: CRFK-180bp (lane 2), MDCK.2-153bp (lane 4), Vero-222bp (lane 6) and L929-150bp (lane 8); Negative Pools on odd lanes: NCRFK (lane 3); NMDCK.2 (lane 5); NVero (lane 7); NL929 (lane 9).
The PCR-multiplex using the MDBK, CRFK and RK13 cell lines and second technique amplified only the band of each specific cell line used as template (Figure 6b, lanes 4, 6 and 7). When mixed DNA templates from the last two cell lines were used, two fragments corresponding to both cells them were obtained (Figure 6b, lane 5).
All PCR products were purified and sequenced, confirming the species identity of cell lines.
DISCUSSION
Isoenzyme studies on the authentication of cell lines began by employing the vertical starch-gel technique (Montes de Oca et al. 1969, RuddleRUDDLE FH and RAPOLA J. 1970. Changes in lactate dehydrogenase and esterase specific activities, isozymic patterns, and cellular distribution during the growth cycle of PK cells in vitro. Exp Cell Res 59: 399-412. and Rapola 1970). Subsequently, most of investigations used agarose electrophoresis film from a commercially available kit (Steube et al. 1995, Nims et al. 1998, Ramya et al. 2009). In contrast, a few laboratories have prepared their own electrophoresis support and staining, either in an agarose system (Stulberg et al. 1976, MachadoMACHADO JZ, MIRANDA ACS and CRUZ AS. 2016. G6PD and MDH isoenzyme electrophoresis for cell lines authentication. Virus Rev Res 21(1): 1-4. et al. 2016), or in a polyacrylamide horizontal (Fernandes and Simoni 1995a) or vertical (Barros et al. 2008) system.
To optimize our isoenzyme methodology, we incorporated some changes into the protocol: changing of current to voltage to improve resolution or detection of isoenzymes in the PAGE system; pulling out of buffer TNM, substitution of N2, and reduction of cell culture flasks to save reagents; or alternative use of other supports as agarose. However, the combined method of cellular lysis through osmotic shock and freezing-thawing in N2 and using 42 × 106 cells to obtain the isoenzyme extracts (Fernandes and Simoni 1995a) was more efficient than these modifications in protocol. Only the constant voltage (200 V) with electrophoresis yielded more benefits. Run with current of 20 mA automatically induced an increase in voltage and heating of the electrophoresis system. Although minimized under system refrigeration, heating caused the formation of sinuous bands, longer run time, and isoenzyme denaturation, which was also observed by Harris and Hopkinson (1976). However, the main problem was reaching the voltage limit (300 V), leading to interruption of the run and total loss of the experiment.
In this study, isoenzymes of cell lines representing six different species (bovine, simian, swine, feline, canine and leporine) were analyzed. All our cell lines had isoenzymatic mobility corresponding to their species and could be distinguished from each other by this comparative mobility.
In MDBK (bovine), Vero (simian), LLC-PK1 (swine) and CRFK (feline) cell lines, we detected only four of five LDH bands described in the literature for these species. Previously, we identified all five bands for the first three cell lines (Fernandes and Simoni 1995a) as did MachadoMACHADO JZ, MIRANDA ACS, GONÇALVES CR, IKEDA TI and CRUZ AS. 2014. Isoenzyme electrophoresis and karyotype techniques as an alternative for cell line identity confirmation. Virus Rev Res 19(1): 1-5. et al. (2014). Montes de Oca et al. (1969) and Hay (1992) also identified five bands for bovine and simian species and only one band for the swine species. Hay (1992) detected all five bands for feline species, while we obtained four for the CRFK cell line. Already for the RK13 (leporine) cell line, we found four bands similar to Machado et al. (2016), but Montes de Oca et al. (1969) and Hay (1992) detected three. In the case of the MDCK.2 (canine) cell line, three bands were detected such as those reported by Montes de Oca et al. (1969) and Hay (1992), while Machado et al. (2016) detected five in the same cell line.
The failure to detect all bands in certain cell lines could be associated with the low cell concentration linked to cell growth phase at the collection time. Ruddle and Rapola (1970) demonstrated that LDH activities are influenced by the growth rate of cells, increasing during the early plateau phase of growth. Collection of cell extracts in this study occurred after the confluence of the cell monolayer, generally 3–4 days subsequent to the subculture, and no necessarily coinciding with this plateau phase of growth. Another possible explanation could be the variation in amount of enzymes in different tissues of a same species (Harris and Hopkinson 1976). Regardless of the number of bands corresponding or not to those in the literature, it was possible to differentiate all cell lines through their different electrophoretic mobility and to identify their species.
Only one band for both MDH and G-6-PDH from all cell lines was detected, comparable with that described in Machado et al. (2016). Montes de Oca et al. (1969) and Hay (1992) also identified only one G-6-PDH band for the same species of these cell lines, except for bovine where they obtained two bands. The visualization of these isoenzyme bands was also maintained using the alternative isoenzyme collection protocols. Then, laboratories without N2 or the availability to specific reagents could take advantage of these alternatives proposed in the isoenzyme methodology, at least for these isoenzymes. It is emphasized that this optimization of our technique is also intended to provide an alternative methodology for use and implementation by other laboratories, since a commercial kit has not been available since 2015.
In parallel with the isoenzyme studies, identification by DNA for the monitoring of cell lines also started a long time ago. In 1977, GrossmanGROSSMAN L I, PARKER RC, WATSON RM, CHANDLER S. EW AND TEPLITZ M. 1977. Detection of a contaminant cell culture line by restriction endonuclease cleavage patterns of mitochondrial DNA. Nucleic Acids Res 4 (5): 1267-1271. et al. identified a cell line contaminated with HeLa cell line by restriction endonuclease cleavage patterns of mtDNA; from 2000, these studies became more frequent (Masters et al. 2001, Parodi et al. 2002, Liu et al. 2003, Cooper et al. 2007, Ono et al. 2007, Losi et al. 2008, Ramya et al. 2009). Hence, another focus of this study was to introduce a molecular method for authentication of species of the cell lines in our laboratory. For this, two PCR techniques directed to different target sequences of the mtDNA were tested.
The first method was a PCR based on cox I gene described in Parodi et al. (2002). This gene region has already been used in microgenomics identification, called DNA Barcode, proposed for the taxonomy of the animal kingdom (HebertHEBERT PDN, CYWINSKA A, BALL SL and WARD JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B 270: 313-319. et al. 2003). All cell lines had their species-specific fragments amplified, but in three of them (RK13, MDCK.2 and L929), the size of the fragments were very close.
In the second method, using a nested PCR based on that of Ono et al. (2007), we were not able to amplify the first product. Studies have shown that long fragment amplifications greater than 5000 bp are not always reproducible and successful, with a specificity of reaction limited to templates up to 3000 bp (OhlerOHLER LD and ROSE EA. 2014. Optimization of long-distance PCR using a transposon- based model system. Genome Res 2: 51-59. and Rose 2014). Ono et al. (2007) also reported this problem with one of their cell lines (chicken) and the requirement of a much larger amount of DNA for identification for the second PCR same in our study. This indicates that an amplification occurred in the first reaction and, although not visualized on the gel, this was sufficient to provide a template for the second PCR.
The two methods presented advantages and disadvantages. Considering this, we merged the primers of the two methods to improve the identification of the cell lines. Cooper et al. (2007) and Ramya et al. (2009) also developed methods based on the primers of these same authors, Parodi et al. (2002) and Ono et al. (2007), respectively. The sequencing of these PCR products was also made to validate these data. The choice of primers depended on the cells of interest in such a way that there was a sufficient differentiation of the DNA fragments and thereby our cell lines could be unequivocally identified with regard to their species of origin. The use this molecular methodology (mtDNA) should therefore be adopted in our laboratory to complement isoenzyme identification of the species of the cell lines, and this is consistent with what was recently proposed by NimsNIMS RW. 2015. Suitability of the CO1 Barcode assay for animal cell identity testing. www.bioreliance.com/CO1-barcode.
www.bioreliance.com/CO1-barcode...
(2015). This researcher, as a member of International Cell Line Authentication Committee (ICLAC) and of ATCC Standards Development Organization (SDO) workgroup, helped in the development of three ATCC SDO standards. One is the current standard method (ANSI/ATCC ASN-0002-2012ANSI - AMERICAN NATIONAL STANDARDS INSTITUTE. 2011. Authentication of human cell lines: Standardization of STR profiling. ANSI/ATCC ASN-0002-2011. https://webstore.ansi.org.
https://webstore.ansi.org...
) for authenticating human cell lines by STR profiling (DNA fingerprinting) and the other is a recent standard method of animal cell line authentication using the same gene as in our study (cox I), namely the DNA barcoding method (ANSI/ATCC ASN-0003ANSI - AMERICAN NATIONAL STANDARDS INSTITUTE. 2015. Species-Level identification of animal cells through mitochondrial cytochrome c oxidase subunit 1 (CO1) DNA barcodes. ANSI/ATCC ASN-0003-2015. https://webstore.ansi.org.
https://webstore.ansi.org...
).
At present, few or no researchers include in their paper a certificate of authentication of the cell lines used in the experiments. On the other hand, journal editors do not also require it. The whole scientific cell culture community should be aware of the real importance of including evidence of authentication in a study. An authentication certificate of the cell lines will not only guarantee the species identity of cell lines, but also will indicate reliability of the research data, avoiding possible future retractions due a misidentification or cross-contamination of the cell lines. It is therefore necessary and urgent to introduce cell line monitoring as routine in all cell culture laboratories.
ACKNOWLEGMENTS
We are grateful to Mr. Ricardo Harakava for the cell lines sequencing and Mrs. Simone Bacilieri for the formatting of the figures. This work was supported by a grant from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Nº2012/04164-5, and by technical training fellowships level 3 (TT3) from FAPESP to S. B. Araújo and G. F.Patrício.
REFERENCES
- ANSI - AMERICAN NATIONAL STANDARDS INSTITUTE. 2011. Authentication of human cell lines: Standardization of STR profiling. ANSI/ATCC ASN-0002-2011. https://webstore.ansi.org
» https://webstore.ansi.org - ANSI - AMERICAN NATIONAL STANDARDS INSTITUTE. 2015. Species-Level identification of animal cells through mitochondrial cytochrome c oxidase subunit 1 (CO1) DNA barcodes. ANSI/ATCC ASN-0003-2015. https://webstore.ansi.org
» https://webstore.ansi.org - ATCC - AMERICAN TYPE CULTURE COLLECTION. 2007. Cell line authentication test recommendations. ATCC Technical Bulletin nº 8. https://www.atcc.org/~/media/PDFs/Technical%20Bulletins/tb08.ashx
» https://www.atcc.org/~/media/PDFs/Technical%20Bulletins/tb08.ashx - BARROS TF, SIMONI IC and FERNANDES MJB. 2008. Análise eletroforética de isoenzimas para detecção de contaminação cruzada entre linhagens celulares. Rev Biociências 14: 17-22.
- BUTTLER M. 2005. Animal cell cultures: recent achievements and perspectives in the production of biopharmaceuticals. Appl Microbiol Biotechnol 68: 283-291.
- CAPES-DAVIS A et al. 2010. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer 127: 1-8.
- COOPER JK, SYKES G, KING S, COTTRILL K, IVANOVA NV, HANNER R and IKONOMI P. 2007. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cell Dev-An 43: 344-351.
- CREE IA. 2011. Cancer cell culture, Methods and protocols, 2nd ed., Portsmouth: Human press, 493 p.
- FERNANDES MJB and SIMONI IC. 1995a. Caracterização de linhagens celulares. I- Identificação de espécies por análise isoenzimática. Arq Inst Biol 62: 59-63.
- FERNANDES MJB and SIMONI IC. 1995b. Caracterização de linhagens celulares. II- Suscetibilidade aos vírus da doença de Aujeszky e da febre aftosa. Arq Inst Biol 62: 65-72.
- FRESHNEY RI. 2005. Culture of animal cells: a manual of basic technique, 5th ed., New York: Wiley-Liss, 672 p.
- GERAGHTY RJ et al. 2014. Guidelines for the use of cell lines in biomedical research. Brit J Cancer 111: 1021-1046.
- GROSSMAN L I, PARKER RC, WATSON RM, CHANDLER S. EW AND TEPLITZ M. 1977. Detection of a contaminant cell culture line by restriction endonuclease cleavage patterns of mitochondrial DNA. Nucleic Acids Res 4 (5): 1267-1271.
- HALTON DM, PETERSON WDJR and HUKKU B. 1983. Cell culture quality control by rapid enzymatic characterization. In Vitro 19(1): 16-24.
- HARRIS H and HOPKINSON DA. 1976. Handbook of enzyme electrophoresis in human genetics. Amsterdam: North-Holland.
- HARTUNG T, BALLS M, BARDOUILLE C, BLANCK O, COECKE S, GSTRAUNTHALER G and LEWIS D. 2002. Good Cell Culture Practice. ATLA 30: 407-414.
- HAY RJ. 1992. ATCC Quality control methods for cell lines, 2nd ed., Rockville: American Type Culture Collection, 132 p. https://www.amazon.com/ATCC-Quality-Control-Methods- Lines/dp/0930009460.
» https://www.amazon.com/ATCC-Quality-Control-Methods- - HEBERT PDN, CYWINSKA A, BALL SL and WARD JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B 270: 313-319.
- LINACRE A and TOBE SS. 2011. An overview to the investigative approach to species testing in wildlife forensic. Invest Genet 2: 1-9.
- LIU MY, LIN SC, LIU H, CANDAL F and VAFAI A. 2003. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev-An 39: 424-427.
- LOSI CG, FERRARI S, SOSSI E, VILLA R and FERRARI M. 2008. An alternative method to isoenzyme profile for cell line identification and interspecies cross-contaminations: cytochrome b PCR-RLFP analysis. In Vitro Cell Dev-An 44(8): 321-329.
- MACHADO JZ, MIRANDA ACS, GONÇALVES CR, IKEDA TI and CRUZ AS. 2014. Isoenzyme electrophoresis and karyotype techniques as an alternative for cell line identity confirmation. Virus Rev Res 19(1): 1-5.
- MACHADO JZ, MIRANDA ACS and CRUZ AS. 2016. G6PD and MDH isoenzyme electrophoresis for cell lines authentication. Virus Rev Res 21(1): 1-4.
- MARKOVIC O and MARKOVIC N. 1998. Cell cross- contamination in cell cultures: the silent and neglected danger. In Vitro Cell Dev-An 34(1): 1-8.
- MASTERS JR et al. 2001. Short tandem repeat profiling provides an international reference standard for human cell lines. PNAS 98(1): 8012-8017.
- MONTES DE OCA F, MACY ML and SHANNON JE. 1969. Isoenzyme characterization of animal cell cultures. P Soc Exp Biol Med 132: 462-469.
- NIMS RW, SHOEMAKER AP, BAUERNSCHUB MA, REC LJ and HARBELL JW. 1998. Sensitivity of isoenzyme analysis for the detection of interspecies cell line cross-contamination. In Vitro Cell Dev-An 34(1): 35-39.
- NIMS RW and HERBSTRITT CJ. 2005. Strategies for accurate speciation and case studies for the detection of cell line cross-contamination using a commercial kit. Biopharm International 18(6): 76-82.
- NIMS RW. 2015. Suitability of the CO1 Barcode assay for animal cell identity testing. www.bioreliance.com/CO1-barcode.
» www.bioreliance.com/CO1-barcode - OHLER LD and ROSE EA. 2014. Optimization of long-distance PCR using a transposon- based model system. Genome Res 2: 51-59.
- ONO K, SATOH M, YOSHIDA T, OZAWA Y, KOHARA A, TAKEUCHI M, MIZUSAWA H and SAWADA H. 2007. Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cell Dev-An 43(5): 168-175.
- PARODI B, ARESU O, BINI D, LORENZINI R, SCHENA F, VISCONTI P, CESARO M, FERRERA D, ANDREOTTI V and RUZZON T. 2002. Species Identification and Confirmation of Human and Animal Cell Lines: A PCR-Based Method. BioTechniques 32(2): 432-440.
- RAMYA R, NAGARAJAN T, SIVAKUMAR V, SENTHILKUMAR RL, OBULAPATHI BB, THIAGARAJAN D and SRINIVASAN AA. 2009. Identification of cross-contaminated animal cells by PCR and isoenzyme analysis. Cytotechnology 61: 81-92.
- RUDDLE FH and RAPOLA J. 1970. Changes in lactate dehydrogenase and esterase specific activities, isozymic patterns, and cellular distribution during the growth cycle of PK cells in vitro. Exp Cell Res 59: 399-412.
- STEUBE KG, GRUNICKE D and DREXLER HG. 1995. Isoenzyme analysis as a rapid method for the examination of the species identity of cell cultures. In Vitro Cell Dev-An 31(2): 115-119.
- STULBERG CS, PETERSON WDJR and SIMPSON WF. 1976. Identification of cells in culture. Am J Hematol 1(2): 237-242.
Publication Dates
-
Publication in this collection
14 Oct 2019 -
Date of issue
2019
History
-
Received
22 May 2018 -
Accepted
5 Feb 2019