Abstracts
OBJECTIVE: To assess the association of single nucleotide polymorphisms (SNPs) in five genes - leptin, leptin receptor (LEPR), adiponectin (APM1), peroxisome proliferator-activated receptor gamma (PPARG) and uncoupling protein 1 - with anthropometric, metabolic, and dietary parameters in a Southern Brazilian cohort of 325 children followed up from birth to 4 years old. MATERIALS AND METHODS: SNPs were analyzed using polymerase chain reaction-based procedures, and their association with phenotypes was evaluated by t-test, analysis of variance, and general linear models. RESULTS: LEPR223Arg allele (rs1137101) was associated with higher daily energy intake at 4 years of age (P = 0.002; Pcorrected = 0.024). PPARG 12Ala-carriers (rs1801282) presented higher glucose levels than Pro/Pro homozygotes (P = 0.007; Pcorrected = 0.042). CONCLUSIONS: Two of the six studied SNPs presented consistent associations, showing that it is already possible to detect the influences of genetic variants on susceptibility to overweight in 4-year-old children.
Adiponectin; leptin receptor; PPARG; polymorphism; energy intake; children
OBJETIVO: Avaliar a associação de polimorfismos de nucleotídeo único (SNPs) em cinco genes: leptina, receptor da leptina (LEPR), adiponectina (APM1), receptor ativado por proliferadores de peroxissomas gama (PPARG) e proteína desacopladora 1 com parâmetros antropométricos, metabólicos e dietéticos em uma coorte sul-brasileira composta por 325 crianças acompanhadas desde o nascimento até os 4 anos. MATERIAIS E MÉTODOS: Os SNPs foram analisados por meio da reação em cadeia da polimerase e sua associação com os fenótipos foi avaliada utilizando teste T, análise de variância e análise fatorial. RESULTADOS: O alelo LEPR223Arg (rs1137101) foi associado a uma maior ingestão energética diária aos 4 anos (P = 0,002; Pcorrigido = 0,024). Os portadores do alelo PPARG12Ala (rs1801282) apresentaram maior glicemia em relação aos homozigotos Pro/Pro (P = 0,007; Pcorrigido = 0,042). CONCLUSÕES: Dois dos seis SNPs estudados apresentaram associações consistentes, mostrando que aos 4 anos de idade já é possível detectar as influências de variantes genéticas sobre a suscetibilidade ao excesso de peso.
Adiponectina; receptor da leptina; PPARG; polimorfismo; ingestão de energia; crianças
ORIGINAL ARTICLE
Polymorphisms in LEPR, PPARG and APM1 genes: associations with energy intake and metabolic traits in young children
Polimorfismos nos genes LEPR, PPARG e APM1: associações com ingestão energética e parâmetros metabólicos em crianças em idade precoce
Marília Remuzzi ZandonáI,II; Raquel Olmedo RodriguesI; Gabriela AlbieroI; Paula Dal Bó CampagnoloII; Márcia Regina VitoloII; Silvana AlmeidaI,II; Vanessa Suñé MatteviI,II
ILaboratório de Biologia Molecular, Universidade Federal de Ciências da Saúde de Porto Alegre (UFCSPA), Porto Alegre, RS, Brazil
IIPrograma de Pós-Graduação em Ciências da Saúde, UFCSPA, Porto Alegre, RS, Brazil
Correspondence to Correspondence to: Vanessa Suñé Mattevi Universidade Federal de Ciências da Saúde de Porto Alegre Rua Sarmento Leite, 245, sala 309 90050-170 Porto Alegre, RS, Brasil vmattevi@ufcspa.edu.br
ABSTRACT
OBJECTIVE: To assess the association of single nucleotide polymorphisms (SNPs) in five genes leptin, leptin receptor (LEPR), adiponectin (APM1), peroxisome proliferator-activated receptor gamma (PPARG) and uncoupling protein 1 with anthropometric, metabolic, and dietary parameters in a Southern Brazilian cohort of 325 children followed up from birth to 4 years old.
MATERIALS AND METHODS: SNPs were analyzed using polymerase chain reaction-based procedures, and their association with phenotypes was evaluated by t-test, analysis of variance, and general linear models.
RESULTS: LEPR223Arg allele (rs1137101) was associated with higher daily energy intake at 4 years of age (P = 0.002; Pcorrected = 0.024). PPARG 12Ala-carriers (rs1801282) presented higher glucose levels than Pro/Pro homozygotes (P = 0.007; Pcorrected = 0.042).
CONCLUSIONS: Two of the six studied SNPs presented consistent associations, showing that it is already possible to detect the influences of genetic variants on susceptibility to overweight in 4-year-old children.
Keywords: Adiponectin; leptin receptor; PPARG; polymorphism; energy intake; children
RESUMO
OBJETIVO: Avaliar a associação de polimorfismos de nucleotídeo único (SNPs) em cinco genes: leptina, receptor da leptina (LEPR), adiponectina (APM1), receptor ativado por proliferadores de peroxissomas gama (PPARG) e proteína desacopladora 1 com parâmetros antropométricos, metabólicos e dietéticos em uma coorte sul-brasileira composta por 325 crianças acompanhadas desde o nascimento até os 4 anos.
MATERIAIS E MÉTODOS: Os SNPs foram analisados por meio da reação em cadeia da polimerase e sua associação com os fenótipos foi avaliada utilizando teste T, análise de variância e análise fatorial.
RESULTADOS: O alelo LEPR223Arg (rs1137101) foi associado a uma maior ingestão energética diária aos 4 anos (P = 0,002; Pcorrigido = 0,024). Os portadores do alelo PPARG12Ala (rs1801282) apresentaram maior glicemia em relação aos homozigotos Pro/Pro (P = 0,007; Pcorrigido = 0,042).
CONCLUSÕES: Dois dos seis SNPs estudados apresentaram associações consistentes, mostrando que aos 4 anos de idade já é possível detectar as influências de variantes genéticas sobre a suscetibilidade ao excesso de peso.
Descritores: Adiponectina; receptor da leptina; PPARG; polimorfismo; ingestão de energia; crianças
INTRODUCTION
The increasing prevalence of obesity in the pediatric population is a global health issue and it has become an important public health problem (1). Increased intake of high energy food and beverages is a significant risk factor for overweight and obesity. Furthermore, metabolic changes occur prior to anthropometric changes (2). Genes involved in appetite regulation, energy metabolism and fat storage may be good candidates to confer susceptibility to obesity and their identification may contribute to a potential intervention to prevent future health problems.
Leptin, the LEP gene product, is essential in the regulation of food intake and energy expenditure (3). The single nucleotide polymorphism (SNP) -2548G>A (rs7799039) has been investigated in the regulation of body weight, but findings are still inconclusive (4-6). For the leptin receptor gene (LEPR), the polymorphism Gln223Arg (rs1137101) is one of the most studied polymorphisms and has been associated with obesity and related phenotypes in Brazilian adults (7). In adolescents, this polymorphism was associated with body fat percentage (3) but, in children, no studies related to obesity were found.
Adiponectin is another adipokine exclusively produced by adipocytes. Several SNPs have already been identified in the APM1 gene and were associated with obesity, type 2 diabetes, and insulin resistance (8). The strongest associations were seen for two SNPs in the APM1 promoter: -11377C>G (rs266729) and -11391G>A (rs17300539). In a study performed with children, the same two APM1 promoter SNPs were associated with insulin resistance syndrome in overweight/obese children (9).
Peroxisome proliferator-activated receptor-gamma (PPARγ) is a nuclear transcription factor encoded by the PPARG gene. Several studies have reported an association of the Pro12Ala variant (rs1801282) with obesity, insulin sensitivity and risk of type 2 diabetes, but have yielded conflicting findings (10,11). In children, this SNP was associated with deficiency in energy storage and utilization, leading to reduced growth (12), lower BMI (13), and increased adiposity in girls (14).
Uncoupling protein-1 (UCP1), abundantly expressed in brown adipose tissue, is a mitochondrial inner membrane electron carrier. The UCP1 gene variant -3826A>G (rs1800592) has been associated with obesity and metabolic features (15-18). It has been suggested that this SNP presents functional importance since it was associated with reduced mRNA expression (19).
With increasing prevalence of childhood obesity in both developed and developing countries, it becomes noteworthy to assess the potential impact of polymorphisms in susceptibility genes in this earlier stage of life, when individuals are exposed for a shorter period to environmental factors, and interventions are still possible to prevent an obese child to become an obese adult.
The purpose of this study was to determine whether specific LEP, LEPR, PPARG, APM1, and UCP1 gene variants influence anthropometric, metabolic and dietary parameters in Brazilian children followed up from birth to 4 years old. Previous studies in the same population found an association of some of these SNPs in adults (7,10). Therefore it was our aim to investigate or confirm in children associations that have been proposed in Brazilian adults. One of the main advantages of studying such sample is that the influence of environmental factors may still not constitute a significant interference at this early age. To our knowledge, this is the first genetic study to investigate all these parameters simultaneously in children of such precocious age.
MATERIALS AND METHODS
Subjects
The population studied comprised 325 children born from October 2001 until July 2002 in the city of São Leopoldo. The mothers were recruited from the maternity ward of the city's only public hospital. This city is located in the metropolitan region of Porto Alegre, the capital of the southernmost state of Brazil.
This prospective cohort study is a continuation of a randomized controlled trial performed in the first year of life. Children were randomized in control and intervention groups at birth. The intervention group received dietary advice during home visits in the first year of life, based on the 'Ten Steps to Healthy Feeding', a Brazilian national health policy for primary care, supported by World Health Organization. A detailed account of the methodology of the first phase of the study can be found elsewhere (20). Anthropometric, dietary, socioeconomic and demographic data were collected between ages of 12 and 16 months. No intervention was performed after that. Between 3-4 years old, children were re-evaluated and anthropometric, dietary, familial, and socioeconomic data were obtained. Blood samples were also collected for DNA extraction and biochemical analyses. The children's mothers provided informed written consent and the study was approved by the Research Ethics Committee of the Federal University of Health Sciences, Porto Alegre, Brazil, protocol number 143/06, and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Race/ethnicity were self-defined by parents by skin color, as whites and nonwhites, as officially used in demographic censuses in the country.
Anthropometric measurements
At 12-16 months, children's height and weight were evaluated. At 3-4 years, height, weight, tricipital and subscapular skinfold thickness, and waist circumference were measured. The sum of the two individual skinfolds was computed. Body mass index (BMI) was calculated: [weight(kg)/height(m2)], and the values were transformed in Z-scores. Nutritional status was determined according to the International Child Growth Standards from the World Health Organization, and children with a BMI-for-age Z-score greater than +1.0 were classified as overweight (21). The detailed measurement procedure was published by Vitolo and cols. (20).
Dietary patterns
At 12 to 16 months of age, a 24-hour diet recall carried out by nutrition undergraduate students recorded the child's food intake on the day before the home visit. At 3 to 4 years of age, two 24-hour diet recalls were collected on two randomly selected and non-consecutive days; the first diet recall conducted at home and the second in the municipal health unit. The NutWin (Nutritional Support Program from the Federal University of São Paulo) and food chemical composition tables were used to calculate nutritional values of the diets (22,23), in addition to information obtained from food industries about products not included in the tables. To assess the intake of sugar-dense foods (SDF) and lipid-dense foods (LDF) at 3 to 4 years old, the foods from two dietary recalls were classified as SDF when they had 50% or more sugar in 100g of composition (e.g. candies, soft drink, sugar and honey), and as LDF when they had 30% or more fat content in 100g of composition (e.g. salty snacks, filled cookies and chocolate). The mean food intake from the two diet recalls performed at 3 to 4 years old was used for analysis.
Biochemical analyses
Blood samples were collected from children at 3-4 years after a 12-h fast. Total cholesterol (TC), high density lipoprotein (HDL), triglycerides (TG), and glucose levels were determined using standard enzymatic methods with an automatic analyzer (Cobas Integra 400®, Roche) at the Clinical Analysis Laboratory of the Cardiology Institute of Rio Grande do Sul. Low density lipoprotein (LDL) was calculated according to the standard method.
Genotyping
Genomic DNA was extracted from whole blood using a standard salting-out procedure. Single nucleotide polymorphisms (SNPs) in the five candidate genes were selected based on previous association data in adults, minor allele frequency of at least 5% in the general population and potential for changing protein function and/or expression. The regions containing the SNPs LEP -2548G>A (rs7799039) (24), LEPR Gln223Arg (rs1137101) (25), APM1 -11391G>A (rs17300539) (26), APM1 -11377C>G (rs266729) (26), PPARG Pro12Ala (rs1801282) (27) and UCP1 -3826A>G (rs1800592) (28) were amplified by polymerase chain reaction (PCR) using primer sequences and PCR cycling conditions previously described. Analysis of restriction fragment length polymorphisms was employed to determine genotypes. Appropriate controls were included to confirm the genotypes in each test.
Statistical analysis
Allele frequencies were estimated by gene counting. The agreement of genotype frequencies with Hardy-Weinberg expectations was tested using the chi-square test, as well as the comparison of genotype and allele frequencies between ethnic groups and overweight and normal-weight children.
Mean values of normally distributed continuous variables among genotypes were compared using analysis of variance (ANOVA) or Student's t-test for independent samples. Homogeneity of variances among genotype groups was previously tested for each variable by the Levene's test for unequal variances, and non-parametric tests were used when appropriate. When ANOVA provided significant results, the Tukey post-hoc test for multiple comparisons was used. For non-normally distributed variables (triglycerides and sum of skinfolds), we used the logarithmically transformed values for analyses. The non-adjusted means are presented for these two parameters. Student's t-test was used to compare differences in anthropometric, metabolic and dietary parameters between intervention and control groups, and there were no differences between groups (P > 0.05). The only variable that showed a significant difference was the total daily energy intake at 12-16 months, which was then adjusted by intervention or control group. BMI percent variation between 12-16 months and 3-4 years of age was compared between genotypes using general linear models and gender, age variation between phases and intervention or control group at 12-16 months were included as covariates. Statistical analyses were performed in SPSS 16.0 statistical package for Windows software. Nominal and Bonferroni-corrected for multiple testing P values are presented for all analyses (29).
Because of the very low number of PPARG Ala/Ala individuals (n = 2) and APM1 -11391 A/A (n = 2), all analyses for these two polymorphisms were performed under a dominant genetic model.
For APM1 -11391G>A and APM1 -11377C>G polymorphisms, haplotypes were inferred using Multiple Locus Haplotype Analysis software, version 2.0 (30), and linkage disequilibrium value (D') was calculated (31).
RESULTS
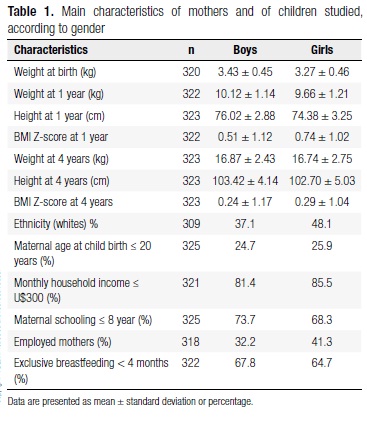
Three hundred and twenty-five children from this cohort were followed up since birth and were genotyped for the six gene polymorphisms. This sample included 186 boys (57.2%) and 139 girls (42.8%). In the intervention group, 58.7% were boys and 41.3% girls. The main features of the sample are described in Table 1. Minor allele frequencies of the LEP, LEPR, APM1-11391, APM1-11377, PPARG, and UCP1 gene variants were 0.46 (A), 0.43 (Arg), 0.09 (A), 0.24 (G), 0.08 (Ala), 0.36 (G); respectively. Genotype frequencies for all polymorphisms in the sample studied were in agreement with those expected under the Hardy-Weinberg equilibrium.
Genotype and allele frequencies were not different between ethnic groups; therefore association analyses were performed in the whole sample (whites n = 129; nonwhites n = 180).
Weight status of children studied was assessed according to BMI Z-score at 3-4 years: 261 (80.8%) had normal-weight and 62 (19.2%) were overweight. No statistically significant differences in genotype and allele frequencies were observed across the groups (data not shown).
Additional anthropometric parameters evaluated were: skinfolds, waist circumference and percent change in BMI from 12-16 months to 3-4 years. Food intake was evaluated by dietary patterns, such as total daily energy intake, SDF, and LDF. To assess the metabolic profile of children, we analyzed TC, HDL, LDL, TG, and glucose plasma levels. The means of these variables were compared among the genotypes of the six polymorphisms. These results are presented in detail below.
LEPR gene
At 3-4 years, the LEPR Gln223Arg polymorphism showed a significant association with total daily energy intake (Kruskal-Wallis test, P = 0.002; Pcorrected=0.024). Individuals with one or two Arg alleles exhibited higher total daily energy intake than wild-type homozygotes (Table 2). A nominal significant association between this polymorphism and fasting glucose levels at the same age was found; Arg/Arg genotype carriers had significantly higher fasting glucose levels in relation to Gln/Gln homozygotes (P = 0.017). However, after Bonferroni-correction for multiple testing, the Pcorrected was 0.204.
APM1 gene
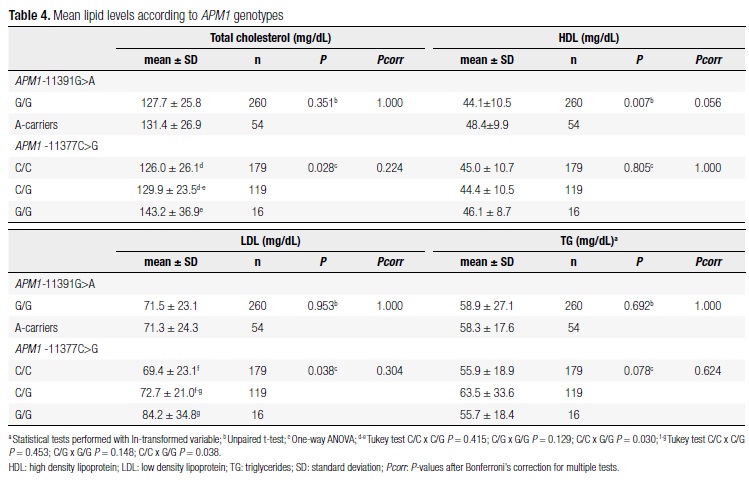
In table 3, before multiple test correction, we could note significant associations of the APM1 -11391G>A variant with daily energy intake at 12-16 months and fasting glucose levels and amount of subcutaneous fat at 3-4 years. Individuals carrying the A-allele had lower fasting glucose levels, sum of skinfolds and total energy intake/day than G/G homozygotes.
In table 4, we can observe that there seems to be an association between the APM1 -11391G>A SNP and HDL cholesterol. The A-allele showed increased HDL levels (48.4 vs. 44.1 mg/dl, P = 0.007; Pcorrected = 0.056). Besides, the APM1 -11377C>G polymorphism was significantly associated with TC and LDL before multiple test correction. Tukey test showed that these observed differences were between G/G homozygotes and C/C homozygotes.
There was strong linkage disequilibrium between APM1 -11391G>A and APM1 -11377C>G polymorphisms (D' = -1.0), with three observed haplotypes: G-C (frequency = 0.67), G-G (frequency = 0.24) and A-C (frequency = 0.09). Therefore, comparison of mean variables among carriers of these haplotypes did not provide information beyond that revealed by both SNPs separately (data not shown).
PPARG, LEP and UCP1 genes
At 3-4 years, the PPARG Pro12Ala polymorphism showed a significant association with fasting glucose levels. The Ala-carriers had significantly higher levels in relation to wild-type homozygotes (74.6 vs. 71.7 mg/dL, P=0.007, Pcorrected = 0.042). No other association was found for this SNP.
No associations were observed for LEP -2548A>G and UCP1 -3826A>G polymorphisms with anthropometric, dietary and metabolic parameters, either.
DISCUSSION
In the current study, LEPR Gln223Arg polymorphism was associated with energy intake and PPARG Pro12Ala was associated with fasting glucose levels, both associations at 4 years old. We initially observed an association between APM1 -11391G>A and daily energy intake at 1 year, subcutaneous fat, fasting glucose levels and HDL levels at aged 4, although P values did not remain significant after multiple test correction. The same occurred in the APM1 -11377C>G association with total cholesterol and LDL plasma and in the LEPR Gln223Arg with fasting glucose levels.
Despite the unquestionable relevance of environmental influence on dietary behavior, specially the mothers' choice regarding foods they offer to their children, the biological mechanisms of hunger and satiety may be active and directly affecting the child's choice about what, when, and how much to eat. Our findings suggest the genetic influences on this behavior are yet active in young children. Our data indicate a relationship between LEPR Gln223Arg and higher food consumption in childhood, as Arg-carriers presented increased total daily energy intake at ages 3 to 4 (Table 2). In accordance with our findings, adult Arg-carriers showed higher energy intake in dinner (32). This same allele was previously associated with higher BMI and abdominal adiposity in Brazilian adults from the same geographical region (7). Although we did not find an association of the Gln223Arg with BMI and other adiposity parameters in children, we believe the influence this SNP should appear progressively with age, as a result of their interaction with environmental factors. We also showed association of the Arg/Arg genotype of LEPR Gln223Arg polymorphism with higher fasting glucose levels at 3-4 years of age. Although the P value did not remain significant after multiple test correction, we suggest that this polymorphism, although exerting a small effect on glucose levels (Table 2), in interaction with environment factors may in the future increase type 2 diabetes risk. Our findings extend recently reported data in adults: Arg allele may predispose healthy individuals to develop metabolic disorders (4). It has been recently suggested that the LEPR Gln223Arg polymorphism, which results in an amino acid change, from glutamine to arginine, alters leptin-binding activity by producing a receptor with an impaired function (4). Additionally, it was reported that LEPR Gln223Arg polymorphism is associated with conversion to type 2 diabetes (33) and insulin resistance (34).
Regarding the APM1 gene, data from the present study also suggest that the two polymorphisms investigated in APM1 gene influence obesity-related traits in childhood. The APM1 -11391A allele showed better dietary, anthropometric, and metabolic profile. The APM1 -11377C allele appears to confer better lipid profile. Although P values did not remain significant after multiple test correction, our results are in accordance with previous findings in the literature (9,35,36). As the APM1 -11391A and APM1 -11377C alleles are in absolute linkage disequilibrium, we were not able to determine which one has a functional effect over the APM1 gene expression. A previous study by our group in HIV-infected individuals showed these same alleles were associated with higher adiponectin levels (37). Although we had no samples to measure circulating adiponectin in the present sample, it is our intent to perform future expression studies to clarify this issue.
Our investigation found the PPARG 12Ala variant is significantly associated with higher fasting glucose levels in children aged 4. A previous study in the same population found that the same allele contributing to higher BMI in adult males (10). There is still no consensus in the literature on the association between Pro12Ala genotypes and metabolic profile. So far, such variant has shown possible heterogeneity in the magnitude of the association among populations and with environmental factors (11). In Brazil, both studies already performed point out that the Ala-allele may be a risk marker for obesity-related phenotypes.
One of the limitations of the present study is the moderate sample size. However, it has a longitudinal design and a very detailed characterization of the sample in terms of phenotypes. Another limitation is the small number of SNPs analyzed. It is important to take into consideration that most genome-wide association (GWA) studies have been performed in populations of European descent and have focused on common variants. Rare variants might be responsible for unexplained phenotypic variation and are not analyzed in GWA studies. Risk variants (and effect sizes), allele frequencies and prevalence of disease can differ across populations (38). Therefore, studies like the current one are needed to indicate possible associations in other populations.
From all the significant nominal P-values presented herein, only the associations between LEPR Gln223Arg and total energy intake, and between PPARG Pro12Ala and glucose levels, both at 4 years of age, remained significant after correction for multiple testing. However, when the probability of the type I error (false positive) decreases, the probability of type II error (false negative) increases. The appropriate application of the multiple testing corrections is a matter of strong discussion (39) and, therefore, the two P values (before and after corrections) are shown. Although the possibility of statistical error could not be excluded, the associations found herein, in a moderate-sized sample, are all in accordance with previous associations in adults and functional evidence regarding these polymorphisms. Independent replication of significant associations in different cohorts from the same population is an important aspect of a credible genetic association result.
Children surveyed herein come from low-income families, users of the government-supported health system. This sample has been originally designed to evaluate the impact of a nutritional intervention in the first year of life on the dietary quality of 3 to 4 years old children (20). Notwithstanding, it seems also to be a good source of information for genetic studies of the general southern Brazilian population, since allele and genotype frequencies observed in the present study were in the same range of those previously reported in European and European-derived populations, including the southern Brazilian population (5,7,10,14,40).
In conclusion, this study showed that SNPs located in LEPR and PPARG genes are associated with obesity-related phenotypes in children in a very young age, and we suggest that APM1 gene polymorphisms may affect serum lipid profile. These results are in the same direction of associations found among these gene variants and obesity-related phenotypes in adults. This demonstrates that it is already possible to detect the influence of genetic variants on nutritional and metabolic characteristics at this early age, even before the manifestation of this effect on anthropometric measures. These findings may not only help in understanding the role of these genetic variants in childhood obesity, but also reinforce the notion that there is an important and precocious interaction between genes and environment in the development of excessive weight gain and its related complications. The follow-up of the present sample is ongoing and will enable us to clarify the influence of these and other polymorphisms on adiposity, metabolic, and dietary parameters over the years.
Acknowledgements: this study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (Fapergs, Brazil) and Pronex/Fapergs/CNPq.
Received on Aug/23/2012
Accepted on Apr/14/2013
Disclosure: no potential conflict of interest relevant to this article was reported.
- 1. Silventoinen K, Kaprio J. Genetics of tracking of body mass index from birth to late middle age: evidence from twin and family studies. Obes Facts. 2009;2(3):196-202.
- 2. Chang Y, Ryu S, Suh BS, Yun KE, Kim CW, Cho SI. Impact of BMI on the incidence of metabolic abnormalities in metabolically healthy men. Int J Obes (Lond). 2012;36(9):1187-94.
- 3. Guizar-Mendoza JM, Amador-Licona N, Flores-Martinez SE, Lopez-Cardona MG, Ahuatzin-Tremary R, Sanchez-Corona J. Association analysis of the Gln223Arg polymorphism in the human leptin receptor gene, and traits related to obesity in Mexican adolescents. J Hum Hypertens. 2005;19(5):341-6.
- 4. Constantin A, Costache G, Sima AV, Glavce CS, Vladica M, Popov DL. Leptin G-2548A and leptin receptor Q223R gene polymorphisms are not associated with obesity in Romanian subjects. Biochem Biophys Res Commun. 2010;391(1):282-6.
- 5. Hinuy HM, Hirata MH, Forti N, Diament J, Sampaio MF, Armaganijan D, et al. Leptin G-2548A promoter polymorphism is associated with increased plasma leptin and BMI in Brazilian women. Arq Bras Endocrinol Metabol. 2008;52(4):611-6.
- 6. Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, Nakazawa M, et al. The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum Genet. 2010;127(3):287-94.
- 7. Mattevi VS, Zembrzuski VM, Hutz MH. Association analysis of genes involved in the leptin-signaling pathway with obesity in Brazil. Int J Obes Relat Metab Disord. 2002;26(9):1179-85.
- 8. Yang WS, Yang YC, Chen CL, Wu IL, Lu JY, Lu FH, et al. Adiponectin SNP276 is associated with obesity, the metabolic syndrome, and diabetes in the elderly. Am J Clin Nutr. 2007;86(2):509-13.
- 9. Petrone A, Zavarella S, Caiazzo A, Leto G, Spoletini M, Potenziani S, et al. The promoter region of the adiponectin gene is a determinant in modulating insulin sensitivity in childhood obesity. Obesity (Silver Spring). 2006;14(9):1498-504.
- 10. Mattevi VS, Zembrzuski VM, Hutz MH. Effects of a PPARG gene variant on obesity characteristics in Brazil. Braz J Med Biol Res. 2007;40(7):927-32.
- 11. Gouda HN, Sagoo GS, Harding AH, Yates J, Sandhu MS, Higgins JP. The association between the peroxisome proliferator-activated receptor-gamma2 (PPARG2) Pro12Ala gene variant and type 2 diabetes mellitus: a HuGE review and meta-analysis. Am J Epidemiol. 2010;171(6):645-55.
- 12. Cecil JE, Fischer B, Doney AS, Hetherington M, Watt P, Wrieden W, et al. The Pro12Ala and C-681G variants of the PPARG locus are associated with opposing growth phenotypes in young schoolchildren. Diabetologia. 2005;48(8):1496-502.
- 13. Cecil JE, Palmer CN, Fischer B, Watt P, Wallis DJ, Murrie I, et al. Variants of the peroxisome proliferator-activated receptor gamma- and beta-adrenergic receptor genes are associated with measures of compensatory eating behaviors in young children. Am J Clin Nutr. 2007;86(1):167-73.
- 14. Lagou V, Scott RA, Manios Y, Chen TL, Wang G, Grammatikaki E, et al. Impact of peroxisome proliferator-activated receptors gamma and delta on adiposity in toddlers and preschoolers in the GENESIS Study. Obesity (Silver Spring). 2008;16(4):913-8.
- 15. Clement K, Ruiz J, Cassard-Doulcier AM, Bouillaud F, Ricquier D, Basdevant A, et al. Additive effect of A-->G (-3826) variant of the uncoupling protein gene and the Trp64Arg mutation of the beta 3-adrenergic receptor gene on weight gain in morbid obesity. Int J Obes Relat Metab Disord. 1996;20(12):1062-6.
- 16. Heilbronn LK, Kind KL, Pancewicz E, Morris AM, Noakes M, Clifton PM. Association of -3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia. 2000;43(2):242-4.
- 17. Kotani K, Sakane N, Saiga K, Adachi S, Shimohiro H, Mu H, et al. Relationship between A-3826G polymorphism in the promoter of the uncoupling protein-1 gene and high-density lipoprotein cholesterol in Japanese individuals: a cross-sectional study. Arch Med Res. 2008;39(1):142-6.
- 18. Oppert JM, Vohl MC, Chagnon M, Dionne FT, Cassard-Doulcier AM, Ricquier D, et al. DNA polymorphism in the uncoupling protein (UCP) gene and human body fat. Int J Obes Relat Metab Disord. 1994;18(8):526-31.
- 19. Sramkova D, Krejbichova S, Vcelak J, Vankova M, Samalikova P, Hill M, et al. The UCP1 gene polymorphism A-3826G in relation to DM2 and body composition in Czech population. Exp Clin Endocrinol Diabetes. 2007;115(5):303-7.
- 20. Vitolo MR, Rauber F, Campagnolo PD, Feldens CA, Hoffman DJ. Maternal dietary counseling in the first year of life is associated with a higher healthy eating index in childhood. J Nutr. 2010;140(11):2002-7.
-
21World Health Organization. Anthro Manual 2005, Beta version Feb. 17 2006: software for assessing growth and development of the world's children. Geneva 2006 [01/15/2007]; Available from: http://www.who.int/childgrowth/en/
- 22. Philippi S. Tabela de composição de alimentos: suporte para decisão nutricional. São Paulo: Metha; 2002.
-
23Unicamp. Tabela Brasileira de Composição de Alimentos (TACO): versão 2. São Paulo: Núcleo de Estudos e Pesquisas em Alimentação; 2006.
- 24. Mammes O, Betoulle D, Aubert R, Giraud V, Tuzet S, Petiet A, et al. Novel polymorphisms in the 5' region of the LEP gene: association with leptin levels and response to low-calorie diet in human obesity. Diabetes. 1998;47(3):487-9.
- 25. Matsuoka N, Ogawa Y, Hosoda K, Matsuda J, Masuzaki H, Miyawaki T, et al. Human leptin receptor gene in obese Japanese subjects: evidence against either obesity-causing mutations or association of sequence variants with obesity. Diabetologia. 1997;40(10):1204-10.
- 26. Poitou C, Lacorte JM, Coupaye M, Bertrais S, Bedel JF, Lafon N, et al. Relationship between single nucleotide polymorphisms in leptin, IL6 and adiponectin genes and their circulating product in morbidly obese subjects before and after gastric banding surgery. Obes Surg. 2005;15(1):11-23.
- 27. Yen CJ, Beamer BA, Negri C, Silver K, Brown KA, Yarnall DP, et al. Molecular scanning of the human peroxisome proliferator activated receptor gamma (hPPAR gamma) gene in diabetic Caucasians: identification of a Pro12Ala PPAR gamma 2 missense mutation. Biochem Biophys Res Commun. 1997;241(2):270-4.
- 28. Valve R, Heikkinen S, Rissanen A, Laakso M, Uusitupa M. Synergistic effect of polymorphisms in uncoupling protein 1 and beta3-adrenergic receptor genes on basal metabolic rate in obese Finns. Diabetologia. 1998;41(3):357-61.
- 29. Brown BW, Russell K. Methods of correcting for multiple testing: operating characteristics. Stat Med. 1997;16(22):2511-28.
- 30. Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56(3):799-810.
- 31. Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120(3):849-52.
- 32. Bienertova-Vasku J, Bienert P, Forejt M, Tomandl J, Brazdova Z, Vasku A. Genotype x nutrient association of common polymorphisms in obesity-related genes with food preferences and time structure of energy intake. Br J Nutr. 2010;103(3):352-9.
- 33. Salopuro T, Pulkkinen L, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, et al. Genetic variation in leptin receptor gene is associated with type 2 diabetes and body weight: The Finnish Diabetes Prevention Study. Int J Obes (Lond). 2005;29(10):1245-51.
- 34. Chiu KC, Chu A, Chuang LM, Saad MF. Association of leptin receptor polymorphism with insulin resistance. Eur J Endocrinol. 2004;150(5):725-9.
- 35. Goyenechea E, Collins LJ, Parra D, Abete I, Crujeiras AB, O'Dell SD, et al. The - 11391 G/A polymorphism of the adiponectin gene promoter is associated with metabolic syndrome traits and the outcome of an energy-restricted diet in obese subjects. Horm Metab Res. 2009;41(1):55-61.
- 36. Warodomwichit D, Shen J, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring). 2009;17(3):510-7.
- 37. Trinca JR, Sprinz E, Lazzaretti RK, Hutz MH, Kuhmmer R, de Almeida S, et al. SNPs in the APM1 gene promoter are associated with adiponectin levels in HIV-infected individuals receiving HAART. J Acquir Immune Defic Syndr. 2010;55(3):299-305.
- 38. Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356-66.
- 39. Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;316(7139):1236-8.
- 40. Bouatia-Naji N, Meyre D, Lobbens S, Seron K, Fumeron F, Balkau B, et al. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes. 2006;55(2):545-50.
Correspondence to:
Publication Dates
-
Publication in this collection
11 Dec 2013 -
Date of issue
Nov 2013
History
-
Received
23 Aug 2012 -
Accepted
14 Apr 2013