Abstract
A possible role for the indole-3-acetamide (IAM) pathway in the indole-3-acetic acid (IAA) production was investigated in developing rice grains. IAM-hydrolase proposed to convert IAM to IAA primarily through the identification of IAM and IAM-hydrolase activity in some plant species. Expression profiles of the two putative rice IAM-hydrolase genes, OsAMI1&2, were compared to the previously quantified IAA level. The abrupt increase in IAA level between 4 and 7 days after anthesis (DAF) was not found to correlate with changes in the expression of OsAMI1 or OsAMID2 suggesting that the IAM pathway may not contribute significantly to IAA pool in rice grains. Production of a biological compound other than IAA may explain the high activity of OsAMI1&2 in developing rice grains. OsAMI1 that reported to be conserved across the plant kingdom showed higher expression level in most analyzed reproductive rice tissues whereas OsAMID2 showed more fluctuation in expression comparing to OsAMI1. Role of the IAM pathway in IAA production was also discussed in other plant systems and Arabidopsis seed was recommended as an ideal tissue to identify enzyme(s) convert(s) tryptophan to IAM as well as physiological effects of IAA produced via this pathway.
indole-3-acetamide; IAM-hydrolase; Auxin, Indole-3-acetic acid; Tryptophan
INTROCUCTION
The phytohormone auxin is structurally a versatile group of chemicals. IAA is the predominant naturally occurring auxin. IAA acts as a regulator for many aspects of plant growth and development including cell division, elongation and differentiation as well as organ patterning, shoot architecture, vascular development, root growth, leaf expansion, and fruit development (reviewed by 11. Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Annals of Botany. 2005 Apr;95(5):707-35., 22. Mockaitis K, Estelle M. Auxin Receptors and Plant Development: A New Signaling Paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55-80.). Several pathways of IAA synthesis have been proposed to be operated in plants including a Tryptophan (Trp)-independent as well as Trp-dependent pathways (Figure 1). The indole-3-acetaldoxime (IAOx), the tryptamine (TAM), the indole-3-pyruvic acid (IPA), and the indole-3-acetamide pathways were reported to synthesis IAA from Trp (Fig. 1). So far only the IPA pathway of IAA synthesis in plants has been completely elucidated in Arabidopsis (33. Kriechbaumer V, Wang P, Hawes C, Abell BM. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 2012;70(2):292-302.
4. Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. PNAS. 2011;108(45):18512-7.
5. Stepanova NA, Yun J, Robles ML, Novak O, He W, Guo H, et al. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. The Plant Cell. 2011;23:3961-73.-66. Won C, Shen X, Mashiguchi K, Zheng Z, Daia X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. PNAS. 2011;108(45):18518-23.), maize (77. Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, et al. Vanishing tassel2 Encodes a Grass-Specific Tryptophan Aminotransferase Required for Vegetative and Reproductive Development in Maize The Plant Cell. 2011;23:550-66) as well as Pisum sativum (88. Tivendale ND, Davidson SE, Davies NW, Smith JA, Dalmais M, Bendahmane AI, et al. Biosynthesis of the Halogenated Auxin, 4-Chloroindole-3-Acetic Acid. Plant Physiology 2012;159:1055-63.). In this pathway, Trp aminotransferase (TAA) converts Trp to IPA whereas YUCCA, a flavin monooxygenase catalyses the conversion of IPA to IAA. The mutants in TAA or YUCCA of Arabidopsis and other plants are not lethal suggesting that other pathways together with the IPA are expected. The mutant in Vanishing tassel2, which encode grass-specific TAA, was reported to account for 60% reduction in IAA level (77. Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, et al. Vanishing tassel2 Encodes a Grass-Specific Tryptophan Aminotransferase Required for Vegetative and Reproductive Development in Maize The Plant Cell. 2011;23:550-66). This hormone in plant is crucial and two pathways of IAA production or more may be needed to ensure the efflux of enough amount of IAA. Moreover, multiple IAA biosynthetic pathways may contribute to regulation of IAA production.
Proposed routes from tryptophan to IAA in plants. Bold lines refer to the first complete dissected pathway of IAA synthesis in plants. Dashed lines indicate that neither a gene nor enzyme activity has been identified in any member of plants. Trp: tryptophan; IAOx: Indole-3-acetaldoxime; IAM: indole-3-acetamide; IAA: indole-3-acetic acid; IPA: indole-3-pyruvic acid; TAM: tryptamine; HTAM: N-hydroxyl tryptamine; IAAld: indole-3-acetaldehyde; IAN: indole-3-acetonitrile; TAA: tryptophan aminotransferase; AMI: amidase; NIT: nitrilase; AO: aldehyde oxidase; YUCCA: a flavin monooxygenase. References for mutant phenotypes are described in (Stepanova, Robertson-Hoyt et al. 2008) wei8; (Tao, Ferrer et al. 2008)(sav3); (Yamada, Greenham et al. 2009)tir2; (Zhao, Hull et al. 2002)cytp79b2/3; (Nafisi, Goregaoker et al. 2007)pad4; and (Normanly, Grisafi et al. 1997)nit1. Gene abbreviations are given in upper case italics. Mutant alleles are given in lower case italics.
As in the IPA pathway, the IAM pathway has been completely elucidated in bacteria and proposed to be operated in plants. In this pathway the IAM-hydrolase was proposed to catalyze the conversion of IAM to IAA whereas the enzyme or enzymes that convert Trp to IAM has not been identified so far in any plant members. The occurrence of IAM pathway in plants has been suggested due to firstly, the identification of endogenous IAM in a wide range of monocot and dicot plants including Poncirus trifoliate (99. Kawaguchi M, Fujioka S, Sakurai A, Yamaki YT, Syono K. Presence of a Pathway for the Biosynthesis of Auxin via Indole-3-Acetamide in Trifoliata Orange. Plant Cell Physiol 1993;34 (1): 121-8. .), aseptically grown Cucurbita maxima (1010. Rajagopal R, Tsurusaki K-i, Kannangara G, Kuraishi S, Sakurai N. Natural Occurrence of Indoleacetamide and Amidohydrolase Activity in Etiolated Aseptically-Grown Squash Seedlings. Plant Cell Physiol. 1994;35(3):329-39.), young fruit of Citrus unshiu (1111. Takahashi N, Yamaguchi I, Kono T, Igoshil M, Hirose K, Suzuki K. Characterization of plant growth substances in Citrus unshiu and their change in fruit development. Plant & Cell Physiol. 1975;16:1101-11.), hypocotyls of Prunus jamasakura (1212. Saotome M, Shirahata K, Nishimura R, Yahaba M, Kawaguchi M, Syono K, et al. The Identification of Indole-3-Acetic Acid and Indole-3-Acetamide in the Hypocotyls of Japanese Cherry. Plant and Cell Physiology. 1993;34(1):157-9.), and epicotyls of Pisum sativum (1313. Prinsen E, Bercetche J, Chriqui D, Van Onckelen H. Pisum sativum epicotyls inoculated with Agrobacterium rhizogenes agropine strains harbouring various T-DNA fragments: Morphology, histology and endogenous indole-3-acetic acid and indole-3-acetamide content. Plant Physiol. 1992;140:75-83.). Recently, the IAM has been detected in the model dicot plant Arabidopsis (1414. Pollmann S, Muller A, Piotrowski M, Weiler EW. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta. 2002 Nov;216(1):155-61., 1515. Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, et al. Biochemical analyses of indole-3-acetaldoximedependent auxin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar;106(13):5430-5.), Oryza sativa, Zea mays (1515. Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, et al. Biochemical analyses of indole-3-acetaldoximedependent auxin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar;106(13):5430-5.), as well as Nicotiana tabacum (1515. Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, et al. Biochemical analyses of indole-3-acetaldoximedependent auxin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar;106(13):5430-5.
16. Nemoto K, Hara M, Goto S, Kasai K, Seki H, Suzuki M, et al. The aux1 gene of the Ri plasmid is sufficient to confer auxin autotrophy in tobacco BY-2 cells. Plant Physiology. 2009;166:729-38.-1717. Lemcke K, Prinsen E, van Onckelen H, Schmülling T. The ORF8 gene product of Agrobacterium rhizogenes TL-DNA has tryptophan 2-monooxygenase activity. Mol Plant Microbe Interact 2000;13(7):787-90.). Secondly, the IAM-hydrolase activity was detected in rice calli (1818. Kawaguchi M, Kobayashi M, Sakurai A, Syono K. The Presence of an Enzyme that Converts IndoIe-3-acetamide into IAA in Wild and Cultivated Rice. Plant Cell Physiol. 1991;32(2):143-9., 1919. Arai Y, Kawaguchi M, Syono K, Ikuta A. Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J Plant Res. 2004;117:191-8.), crude extract from young fruits of Poncirus trifoliate (99. Kawaguchi M, Fujioka S, Sakurai A, Yamaki YT, Syono K. Presence of a Pathway for the Biosynthesis of Auxin via Indole-3-Acetamide in Trifoliata Orange. Plant Cell Physiol 1993;34 (1): 121-8. .), Arabidopsis (2020. Pollmann S, Neu D, Weiler E. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry. 2003; 62(3):293-300.), Nicotiana (2121. Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Letters 2009;583:487-92.). The importance of IAM as an intermediate in IAA synthesis was also stressed by the observation that IAM interferes with the conversion of [2H]5-Trp to [2H]5- IAA by the IAA synthase complex extracted from 14 different plant species (2222. Pollmann S, Düchting P, Weiler E. Tryptophan-dependent indole-3-acetic acid biosynthesis by IAA-synthase proceeds via indole-3-acetamide. Phytochemistry. 2009;70(4):523-31.).
In transgenic tobacco infected with Agrobacterium rhizogenes, Trp can be converted to IAM and then to IAA by expression of the 2 integrated genes AUX1 and 2 respectively (1616. Nemoto K, Hara M, Goto S, Kasai K, Seki H, Suzuki M, et al. The aux1 gene of the Ri plasmid is sufficient to confer auxin autotrophy in tobacco BY-2 cells. Plant Physiology. 2009;166:729-38.). Interestingly, overexpression of the AUX1 gene alone is sufficient to allow the rapid growth of tobacco cell lines in the absence of auxin and at lower concentration of IAM (10-5 M) whereas growth of these cell lines were found to be completely inhibited in the RNAi-mediated suppression of NtAMI1 in IAM-containing medium (2121. Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Letters 2009;583:487-92.). Moreover, phylogenetic analysis showed that that the IAM-hydrolase is a common component of cells in a wide array of monocot and dicot plants as well as the non-seed plant, Selaginella moellendorffii, and the moss Physcomitrella patens (2323. Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. Journal of Experimental Botany. 2012;doi:10.1093/jxb/ers091:1-20.
https://doi.org/10.1093/jxb/ers091:1-20...
, 2424. Abu-Zaitoon YM. IAA synthesis by a conserved IAM -hydrolase in developing rice grains. 2000. In Press.). Arabidopsis genome was reported to have 4 putative IAM-hydrolase genes, one of them Arabidopsis amidase 1 (AtAMI1, At1g08980) was found to be the only Arabidopsis sequence to specifically converts IAM to IAA (2020. Pollmann S, Neu D, Weiler E. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry. 2003; 62(3):293-300.). In (2121. Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Letters 2009;583:487-92.) Nemoto, et al. isolated the AtAMI1 homologue in Nicotiana tabacum (NtAMI1) that showed IAM-hydrolase activity.
Rice has been chosen as a plant of study because it is the most popular crop in the world. The completion of sequencing rice genome is considered the first step to explore and develop traits of this crucial crop plant and ultimately all cereals. The abundance of genetic information derived from microarrays, expressed sequence tags, massively parallel signature sequencing make rice an ideal monocot plant to explore genes involved in the IAM pathway of IAA synthesis. Rice was reported to accumulate large amount of IAA during grain's development (25). The largest increase in IAA was detected between 4 and 7 DAF. In a previous study (24), rice proteome was found to have 2 putative IAM-hydrolase enzymes Os04g02780 (OsAMI1) and Os04g02754 (OsAMI2). OsAMI1 was reported to be conserved across the plant kingdom and found in the same clade as AtAMI1 and NtAMI, the two isolated and well characterized IAM-hydrolase enzymes in plants (2020. Pollmann S, Neu D, Weiler E. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry. 2003; 62(3):293-300., 2121. Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Letters 2009;583:487-92.). Moreover, All IAM-hydrolase sequences of the conserved clade were reported to encode amidase with significant sequence similarity to the IAM-hydrolyzing bacterial ones. Therefore OsAMI1 was concluded to have a major role in IAA synthesis or other primary process in plant growth and development (2424. Abu-Zaitoon YM. IAA synthesis by a conserved IAM -hydrolase in developing rice grains. 2000. In Press.).
The importance of IAM pathway in IAA synthesis has been highlighted primarily by the identification of IAM as well as IAM-hydrolase activity in species across the plant kingdom. Nevertheless, major questions regarding the IAM pathway need to be answered. The enzyme or enzymes converting Trp to IAM has not been identified in any plant. Contribution of the IAM pathway to IAA pool and associated phenotypes as well as interaction with other pathways of IAA synthesis was not determined. Physiological effects of IAA produced by the IAM pathway in plants remains elusive. The major aim of this research was to find out whether the IAM pathway is operated in developing rice grains. Therefore, correlation if any between the expression of OsAMI1 and OsAMI2 genes and changes in IAA level in the developing rice grains was explored. Expression profiles of IAM-hydrolase genes were also investigated in a group of reproductive rice organs.
MATERIALS AND METHODS
Plant Materials
Data were downloaded from the following experiments: OS5 (2626. Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology. 2007;143(4):1467-83.), OS8, OS16 (2727. Li M, Xu W, Yang W, Kong Z, Y X. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 2007;144(4):1797-812.), and OS89 (2828. Gao L, Xue H. Global analysis of expression profiles of rice receptor-like kinase genes. Mol Plant Microbe Interact. 2012;5(1):143-53.). Data analysed in this study were obtained from various rice tissues including seedling root, leaf, panicle and seeds (Os5 experiment), root, seedling leaf, and seeds (Os8 experiment), seedling shoot and root, anther, stigma, ovary, embryo and endosperm (Os16 experiment), as well as ovary, embryo and seed (experiment 89)Rice (subsp. indica var IR64) was used in Os5 experiment whereas Nipponbare and japonica cultivar Zhonghua11 were used in Os16 and Os89 experiments respectively.
Microarray gene hybridization
Expression profiles of the OsAMI1&2 were analyzed using Public microarray data downloaded from PLEXdb data base (2929. Dash S, Van Hemert J, Hong L, Wise RP, JA D. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Research. 2012;40(D1):D1194-D201.) (http://www.plexdb.org). Total RNA of each sample from Os5, Os16, and Os89 was isolated and purified as described by Jain, et al.,(3030. Jain M, Nijhawan A, Tyagi A, Khurana J. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2006;345(2):646-51.), Qiagen, TRIzol reagent and protocols respectively. Five µg RNA was then hybridized on Affymetrix GeneChip Rice Genome Arrays. In most experiments at least 2 biologically replicates were used. Preparation, hybridization to arrays, washing, staining, and scanning was performed according to manufacturer's protocol (Affymetrix).
Data authentication and gene annotations
Validity of microarray data was confirmed by real time PCR analysis and then by RNA in situ hybridization. Real-Time PCR was conducted using gene-specific primers as recommended by Lan (3131. Lan L, W C, Y L, J S, Z K, C L, et al. Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice ( Oryza sativa L.) with a 10K cDNA microarray. Plant Mol Biol. 2004;54(4):471-87.) and Lai (3232. Lai Z, W M, B H, L L, Y Z, G H, et al. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 2002;50(1):29-42.). At least two biological and three technical replicates were used. PCR data was normalized through the amplification of 18S rRNA.
Gene annotations were obtained from PlaNet platform (http://aranet.mpimp-golm.mpg.de/ricenet).
RESULTS AND DISCUSSION
In this study the expression profiles of OsAMI1 and OsAMI2 were studied in various reproductive rice tissues (Figure 2). The highest expression level of OsAMI1 was found in Y leaf, inflorescence (6 DAF), seedling shoot, endosperm (9 DAF), and embryo (3 DAF) whereas peaks of expression level for OsAMI2 were detected in Y leaf, inflorescence (6 DAF), seedling root, embryo (12 DAF), and endosperm (16 DAF). The lowest expression level of the 2 IAM-hydrolase genes was detected in the first 3 developmental stages of inflorescence. Generally, expression of OsAMI1is high in most tested tissues and changes in the expression is very limited comparing to that of OsAMI2. In Arabidopsis, transcripts for AtAMI1 were detected in all analyzed tissues. The highest expression levels were detected in inflorescence, young leaves, and germinating seeds (3333. Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta. 2006;224:1241-53.). High amount of the Arabidopsis amidase (at the translational level, protein) was only detected in young leaves. Discrepancy in the expression level of amidase from the same tissue in different plants suggests the multiphysiological effects of this enzyme in plant life.
Expression profiles ofOsAMI(Os04g02780)and OsAMI2 (Os04g02754) as downloaded from PLEXdb data base (http://www.plexdb.org). Each point represents the mean ±standard error of 3 biological replicates for (A & stigma and ovary for C), 2 biological replicates for (B & D). All data were normalized by RMA from Rice 57k genechip at the experiment OS5 (A), OS8 (B), OS16 (C), OS89 (D). (A) Inflorescence 1 (1-3 cm), inflorescence 2 (3-5 cm), inflorescence 3 (5-10 cm), inflorescence 4 (10-15 cm), inflorescence 5 (15-22 cm), inflorescence 6 (22-30 cm), SAM (shoot apical meristem), seedling root (7-days old). (B) Embryo and endosperm (6 DAF), seedling. (D) Ovary (0 DAF).
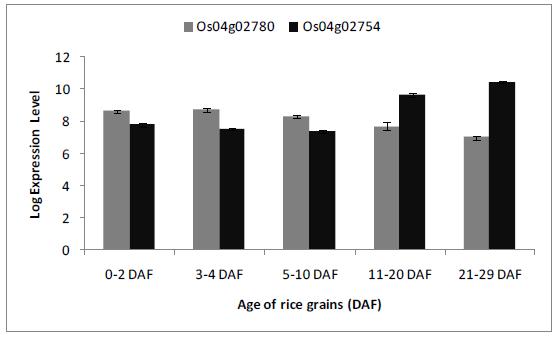
It has been reported that IAA is produced in high amount in rice kernels compared to vegetative tissue. Abu-Zaitoon, et al. (2525. Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiologia Plantarum. 2012;146(4):487-99.) showed that IAA levels in kernels increase from undetectable limit at 4 DAF to 2µg/g FW during 14 days from anthesis. The greatest change in auxin synthesis occurs from 4 to 7 DAF. This period of grain development was investigated to find out whether the expression of OsAMI1 and OsAMID2 correlate with this change in IAA level and therefore to suggest a role for amidase in IAA synthesis.
Figure (3) shows change in expression level of OsAMI1 and OsAMI2. The expression of the two IAM-hydrolase genes was not found to change significantly (p=0.04 and 0.3 respectively) between 3-4 and 5-10 DAF. Due to lack of correlation between change in the expression level of rice amidases and IAA level during the active period of IAA synthesis (4-7 DAF), a significant role if any for OsAMIs and ultimately the IAM pathway in IAA synthesis in the developing rice grains is not expected. High expression levels of OsAMI1 and OsAMI2 in developing rice grains may be linked to a biological activity other than IAA synthesis such as hydrolysis of IAA conjugates. The same conclusion was drawn by Arai, et al., (19) who partially purified IAM-hydrolase from rice calli. The Km value of the isolated rice enzyme (0.96 mM) was found to be considerably higher for IAM comparing to that of the Agrobacterium tumefaciens (1.2 µM) (3434. Kemper E, Wafenschmidt S, Weiler EW, Rausch T, Schoder J. T-DNA-encoded auxin formation in crown-gall cells. Planta. 1985;163( 2 ):257-62.). Additionally, no endogenous IAM was detected from various rice organs including shoots, roots, calli and young fruits (19).
Expression profiles ofOs04g02780 (OSAMI) and OsAMI2 (Os04g02754) as downloaded from PLEXdb data base (http://www.plexdb.org). Each point represents the mean ±standard error of 3 biological replicates normalized by RMA from Rice57k genechip at the experiment OS5(Jain, Nijhawan et al. 2007; Wise, Caldo et al. 2007).
Unlike the IAM pathway, the IPA pathway was reported by our research group to have a pivot role in IAA synthesis in rice grains (25). Changes in the expression of OsYUCCA9, OsYUCCA11 as well as OsTAA1were found to significantly correlate with the abrupt and large change in IAA synthesis in the active period of IAA accumulation (between 4 and 7 DAF). A minor role if any for the IAM pathway is also expected in the maize endosperm. A 95% reduction in IAA level was reported in the de-18 mutant that results from a mutation in the YUCCA1 gene of maize endosperm (35). As in the developing rice grains, most IAA in the maize endosperm was reported to be obtained through the IPA pathway (35). IAM was also excluded as an intermediate in IAA synthesis in seeds of pea (Pisum sativum). Labeled IAM was not detected after injection the liquid endosperm of pea seed with deuterated Trp and the endogenous IAM was below the detection limit (8). In Brassica rapa (turnip), correlation between amidase activity and free IAA level were not detected in a group of studied tissues including leaves, hypocotyls and roots (36).
However, this may not be the case in other systems including Arabidopsis thaliana. Correlation between free IAA and IAM levels were found in imbibed seeds and sterile-grown seedlings. The amount of IAA and IAM were found to be 270 and 40-fold higher in imbibed seeds comparing to the 2-week old seedlings (14). Interestingly, IAM were found to be parallel to IAA level in this system. The 2 metabolites were also found to largely decrease and at the same rate during the 2 weeks after seed imbibition. This system is ideal to follow the expression level of AtAMI1 and to generate knock out as well as overexpression amidase mutants to detect effects on de novo IAA level as well as auxin phenotypes. After that, Microarray or real-time PCR analysis of any changes in the expression level of genes in these mutants may be enough to identify gene (s) catalyze(s) the conversion of Trp to IAM. The 2 weeks interval after seed imbibition will be also valuable to screen AtYUCCAs expression level and therefore the contribution of both pathways in IAA level. To sum up unlike the IPA, the IAM is not expected to be a ubiquitous pathway and seems to have a very limited role in IAA synthesis in plant tissues. Even though IAM and IAM-hydrolase activity has been identified in a group of monocot and dicot plants, correlation between IAA level and the expression of IAM-hydrolase genes were not found most studied plants.
CONCLUSION
Expression patterns of OsAMI1 and OsAMI2 were studied using public microarray data from Os5, Os8, Os16, and Os89 experiments. Data were obtained from a wide variety of vegetative and reproductive rice tissues. Changes in the expression of these two genes were then compared to the very rapidly period of IAA accumulation in developing rice grains to identify a possible role for the gene products in IAA production. Significant correlation between the expression profiles of OsAMI1 &2 were not detected in developing rice grain suggesting that their products are unlikely to be involved in IAA synthesis. However, a possible role for amidase in IAA synthesis in other rice tissues or plants is not excluded. The high and constant expression levels of both genes in this system may be linked to a biochemical role other than IAA synthesis. It will be interesting to knockout or overexpress one or both rice amidase genes and observe any changes in IAA level or phenotypes.
ACKNOWLEDGMENT
I would like to highly appreciate and thank my superior supervisor Dr. Heather M. Nonhebel from the University of New England; Australia, and my colleagues Ahmad Khlifat, Aram Al-Kwaldeh and Lina Kraishan for their help in analyzing data.
REFERENCES
-
1Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Annals of Botany. 2005 Apr;95(5):707-35.
-
2Mockaitis K, Estelle M. Auxin Receptors and Plant Development: A New Signaling Paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55-80.
-
3Kriechbaumer V, Wang P, Hawes C, Abell BM. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 2012;70(2):292-302.
-
4Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. PNAS. 2011;108(45):18512-7.
-
5Stepanova NA, Yun J, Robles ML, Novak O, He W, Guo H, et al. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. The Plant Cell. 2011;23:3961-73.
-
6Won C, Shen X, Mashiguchi K, Zheng Z, Daia X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. PNAS. 2011;108(45):18518-23.
-
7Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, et al. Vanishing tassel2 Encodes a Grass-Specific Tryptophan Aminotransferase Required for Vegetative and Reproductive Development in Maize The Plant Cell. 2011;23:550-66
-
8Tivendale ND, Davidson SE, Davies NW, Smith JA, Dalmais M, Bendahmane AI, et al. Biosynthesis of the Halogenated Auxin, 4-Chloroindole-3-Acetic Acid. Plant Physiology 2012;159:1055-63.
-
9Kawaguchi M, Fujioka S, Sakurai A, Yamaki YT, Syono K. Presence of a Pathway for the Biosynthesis of Auxin via Indole-3-Acetamide in Trifoliata Orange. Plant Cell Physiol 1993;34 (1): 121-8. .
-
10Rajagopal R, Tsurusaki K-i, Kannangara G, Kuraishi S, Sakurai N. Natural Occurrence of Indoleacetamide and Amidohydrolase Activity in Etiolated Aseptically-Grown Squash Seedlings. Plant Cell Physiol. 1994;35(3):329-39.
-
11Takahashi N, Yamaguchi I, Kono T, Igoshil M, Hirose K, Suzuki K. Characterization of plant growth substances in Citrus unshiu and their change in fruit development. Plant & Cell Physiol. 1975;16:1101-11.
-
12Saotome M, Shirahata K, Nishimura R, Yahaba M, Kawaguchi M, Syono K, et al. The Identification of Indole-3-Acetic Acid and Indole-3-Acetamide in the Hypocotyls of Japanese Cherry. Plant and Cell Physiology. 1993;34(1):157-9.
-
13Prinsen E, Bercetche J, Chriqui D, Van Onckelen H. Pisum sativum epicotyls inoculated with Agrobacterium rhizogenes agropine strains harbouring various T-DNA fragments: Morphology, histology and endogenous indole-3-acetic acid and indole-3-acetamide content. Plant Physiol. 1992;140:75-83.
-
14Pollmann S, Muller A, Piotrowski M, Weiler EW. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta. 2002 Nov;216(1):155-61.
-
15Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, et al. Biochemical analyses of indole-3-acetaldoximedependent auxin biosynthesis in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar;106(13):5430-5.
-
16Nemoto K, Hara M, Goto S, Kasai K, Seki H, Suzuki M, et al. The aux1 gene of the Ri plasmid is sufficient to confer auxin autotrophy in tobacco BY-2 cells. Plant Physiology. 2009;166:729-38.
-
17Lemcke K, Prinsen E, van Onckelen H, Schmülling T. The ORF8 gene product of Agrobacterium rhizogenes TL-DNA has tryptophan 2-monooxygenase activity. Mol Plant Microbe Interact 2000;13(7):787-90.
-
18Kawaguchi M, Kobayashi M, Sakurai A, Syono K. The Presence of an Enzyme that Converts IndoIe-3-acetamide into IAA in Wild and Cultivated Rice. Plant Cell Physiol. 1991;32(2):143-9.
-
19Arai Y, Kawaguchi M, Syono K, Ikuta A. Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J Plant Res. 2004;117:191-8.
-
20Pollmann S, Neu D, Weiler E. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry. 2003; 62(3):293-300.
-
21Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Letters 2009;583:487-92.
-
22Pollmann S, Düchting P, Weiler E. Tryptophan-dependent indole-3-acetic acid biosynthesis by IAA-synthase proceeds via indole-3-acetamide. Phytochemistry. 2009;70(4):523-31.
-
23Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. Journal of Experimental Botany. 2012;doi:10.1093/jxb/ers091:1-20.
» https://doi.org/10.1093/jxb/ers091:1-20 -
24Abu-Zaitoon YM. IAA synthesis by a conserved IAM -hydrolase in developing rice grains. 2000. In Press.
-
25Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiologia Plantarum. 2012;146(4):487-99.
-
26Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology. 2007;143(4):1467-83.
-
27Li M, Xu W, Yang W, Kong Z, Y X. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 2007;144(4):1797-812.
-
28Gao L, Xue H. Global analysis of expression profiles of rice receptor-like kinase genes. Mol Plant Microbe Interact. 2012;5(1):143-53.
-
29Dash S, Van Hemert J, Hong L, Wise RP, JA D. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Research. 2012;40(D1):D1194-D201.
-
30Jain M, Nijhawan A, Tyagi A, Khurana J. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2006;345(2):646-51.
-
31Lan L, W C, Y L, J S, Z K, C L, et al. Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice ( Oryza sativa L.) with a 10K cDNA microarray. Plant Mol Biol. 2004;54(4):471-87.
-
32Lai Z, W M, B H, L L, Y Z, G H, et al. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 2002;50(1):29-42.
-
33Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta. 2006;224:1241-53.
-
34Kemper E, Wafenschmidt S, Weiler EW, Rausch T, Schoder J. T-DNA-encoded auxin formation in crown-gall cells. Planta. 1985;163( 2 ):257-62.
-
35Bernardi J, Lanubile A, Li Q-B, Kumar D, Kladnik A, Cook SD, et al. Impaired Auxin Biosynthesis in the defective endosperm18 Mutant Is Due to Mutational Loss of Expression in the ZmYuc1 Gene Encoding Endosperm-Specific YUCCA1 Protein in Maize. Plant Physiology November 2012;160:1318-28.
-
36Ishikawa T, Kuroda H, Okazaki K, Itoh K, Mitsui T, Hori H. Evaluation of roles of amidase which converts indole-3-acetamide to indole-3-acetic acid, in formation of clubroot in turnip Bulletin of the Faculty of Agriculture. 2007;60 (1):53-60.
-
ErratumIn Article “Contribution of the IAM Pathway to IAA Pool in Developing Rice Grains ”, with the number of DOI: http://dx.doi.org/10.1590/1678-4324-2016150677, published in journal Brazilian Archives of Biology and Technology, vol. 59, the 01 page.That read:“Twenty four fungal species were screened for their ability to produce alkaline L-methioninase on methionine-glucose liquid medium. Aspergillus ustus AUMC 10151 displayed the highest yield of enzyme (10.8 U/mg protein), followed by A. ochraceus and Fusarium proliferatum. Upon optimization of the submerged fermentation (SmF)conditions, the maximum enzyme yield (18.23 U/mg protein) was obtained on a medium containing L-methionine (0.5%), sucrose (0.95%), KH2PO4 (0.1%) and 175 rpm. Seven agro-industrial by-products were screened as substrates for L-methioninase production under solid-state fermentation (SSF). Wheat bran resulted 38.1 U/mg protein, followed by rice bran (27.6 U/mg protein) and soya bean meal (26.6 U/mg protein). Maximum alkaline L-methioninase (99.56U/mg protein) was achieved at initial moisture content of 71.5%, inoculum size of 2.0 mL of spore suspension, initial pH 8.5, incubation period eight days at 30°C and supplementation of the salt basal medium with pyridoxine(100 μg/mL) and beet molasses (20% v/v). The productivity of L-methioninase by A. ustus under SSF was higher than that of SmF about 5.45 fold under optimum conditions.Key words: L-methioninase, Aspergillus ustus, Submerged fermentation, Solid state fermentation, Wheat bran.”Read:“A possible role for the indole-3-acetamide (IAM) pathway in the indole-3-acetic acid (IAA) production was investigated in developing rice grains. IAM-hydrolase proposed to convert IAM to IAA primarily through the identification of IAM and IAM-hydrolase activity in some plant species. Expression profiles of the two putative rice IAM-hydrolase genes, OsAMI1&2, were compared to the previously quantified IAA level. The abrupt increase in IAA level between 4 and 7 days after anthesis (DAF) was not found to correlate with changes in the expression of OsAMI1 or OsAMID2 suggesting that the IAM pathway may not contribute significantly to IAA pool in rice grains. Production of a biological compound other than IAA may explain the high activity of OsAMI1&2 in developing rice grains. OsAMI1 that reported to be conserved across the plant kingdom showed higher expression level in most analyzed reproductive rice tissues whereas OsAMID2 showed more fluctuation in expression comparing to OsAMI1. Role of the IAM pathway in IAA production was also discussed in other plant systems and Arabidopsis seed was recommended as an ideal tissue to identify enzyme(s) convert(s) tryptophan to IAM as well as physiological effects of IAA produced via this pathway.Key words: indole-3-acetamide, IAM-hydrolase, Auxin, Indole-3-acetic acid, Tryptophan.”
Publication Dates
-
Publication in this collection
2016
History
-
Received
04 Nov 2015 -
Accepted
16 Mar 2016