ABSTRACT
The genus Planktothricoides Suda & Watanabe is considered as a 2-methylisoborneol (MIB) producer, affecting water quality and aquatic animal products worldwide. To date, there is limited information about the diversity of this genus from Thailand. In this study, Thai Planktothricoides strains were isolated from fish ponds and reservoirs in North, Northeast and Central regions for morphological examination, phylogenetic analyses based on 16S rRNA, rbcLX and MIB synthase genes as well as GC/MS/MS analyses. The morphological results and the 16S rRNA and rbcLX phylogenies of Thai Planktothricodes strains enabled them to be designated as Planktothricoides raciborskii. Cell dimensions of Thai strains tested were in 1.86 to 5.96 µm length (L), 2.83 to 13.70 µm width (W), and the L/W ratio ranged from 1:6 to 1:1. Among Planktothricoides strains, the 16S rRNA phylogenies demonstrated that three subclades (A, B and C groups) were apparently divided. The similarity of 16S rRNA genes between subclades were 96-98%. From the detection of MIB synthase genes and GC/MS/MS analyses, some strains grouped into A group were considered as MIB-producers. In this study, most Thai Planktothricoides strains belonging to the A group were found in all three regions, while the strains forming the B and C groups were not distributed in the North region. To our knowledge, the present study is the first report investigation and characterization of the potential MIB-producing Planktothricoides from Thailand. Therefore, providing a valuable tool as a model for the early prediction and detection of taste and odor event is necessary.
Key word:
2-methylisoborneol; filamentous cyanobacteria; 16S rRNA; MIB synthase; Thailand
INTRODUCTION
Increasing eutrophication of water resources with changing climate are making cyanobacterial blooms more common, which can produce taste and odor episodes 11 Li L, Wan N, Gan N, Xiao BD, Song LR. Annual dynamics and origins of the odorous compounds in the pilot experimental area of Lake Dianchi, China. Water Sci Technol. 2007; 55(5):43-50.
2 Watson SB, Charlton M, Rao YR, Howell T, Ridal J, Brownlee B, Marvin C, Millard S. Off flavours in large water bodies: physics, chemistry and biology in synchrony. Water Sci Technol . 2007; 55(5):1-8.
3 Su M, Gaget V, Giglio S, Burch M, An W, Yang M. Establishment of quantitative PCR methods for the quantification of geosmin-producing potential and Anabaena sp. in freshwater systems. Water Res. 2013; 47(10): 3444-3454.-44 Wang Z, Xiao P, Song G, Li Y, Li R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ Sci Pollut R. 2015; 22(16):12133-12140.. These undesirable effects are not only restricted to drinking water sources but are also significant causes of economic losses in the aquaculture industry due to their partitioning into fish flesh 55 Lu J, Wills PS, Wilson PC. Trace analysis of off-flavor/odor compounds in water using liquid-liquid microextraction coupled with gas chromatography- positive chemical ionization-tandem mass spectrometry. Front Environ Sci Eng. 2016; 10(3):477-481.. The two most common compounds known to cause earthy and musty off-flavors are the organic chemicals geosmin and 2-methylisoborneol (MIB) 66 Guttman L, Van Rijn J. Identification of conditions underlying production of geosmin and 2-methylisoborneol in a recirculating system. Aquaculture. 2008; 279: 85-91.
7 Schrader KK, Summerfelt ST. Distribution of off-flavor compounds and isolation of geosmin-producing bacteria in a series of water recirculating systems for rainbow trout culture. N Am J Aquacult. 2010; 72(1): 1-9.-88 Houle S, Schrader K, Lefrancois NR, Comeau Y, Kharoune M, Summerfelt ST, Savoie A, Vandenberg GW. Geosmin causes off-flavor in Arctic Charr raised in recirculating aquaculture systems. Aquaculture Res. 2011; 42:360-365., which are secondary metabolic products of certain species of cyanobacteria and actinomycete bacteria. In Thailand, the occurrences of off-flavor episodes were frequently found in aquaculture 99 Yamprayoon J, Noomhorm A. Geosmin and off-flavor in Nile tilapia (Oreochromis niloticus). J Aqua Food Prod T. 2000; 9(2):29-41.
10 Gutierrez R, Whangchai N, Sompong U, Prarom W, Iwami N, Itayama T, et al. Off-flavour in Nile tilapia (Oreochromis niloticus) cultured in an integrated pond-cage culture system. Maejo Int J Sci Technol. 2013; 7:1-13.
11 Yarnpakdee S, Benjakul S, Penjamras P, Kristinsson HG. Chemical compositions and muddy flavor/odour of protein hydrolysate from Nile tilapia and broadhead catfish mince and protein isolate. Food Chem. 2013; 142:210-216.
12 Pimolrat P, Whangchai N, Chitmanat C, Lebel L. Musty/earthy off-flavor in aquatic animal: impact, causes and solutions. KKU Research Journal. 2014; 19(6):927-938.
13 Pimolrat P, Whangchai N, Chitmanat C, Itayama T, Lebel L. Off-flavor characterization in high-nutrient-load Tilapia ponds in Northern Thailand. Turkish J Fish Aquat Sci. 2015; 15(2):273-281.-1414 Suwanpakdee S, Gutierrez R, Pithakpol S, Jampeetong A, Pathom-aree W, Nomura N, et al. Earthy-musty odour and off-flavour taints in Phayao Lake, Thailand. Chiang Mai J Sci. 2016; 43(1):1076-1085.. These episodes erode consumer confidence in the quality and safety of production. Currently, accurate diagnostic tools for the detection of taste and odor producers are important. It would be invaluable to use this information for early detection of potentially odor-producing cyanobacterial blooms and thus leading to management solutions.
The cyanobacterial genus Planktothricoides Suda and Watanabe has also been reported as one of the most important MIB-producing genera and water bloom-forming cyanobacteria that could be widely distributed in aquatic and terrestrial environments, including fish ponds, rivers and reservoirs 1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.
16 Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665.-1717 Suurnӓkki S, Gomez-Saez GV, Rantala-Ylinen A, Jokela J, Fewer DP, Sivonen K. Identification of geosmin and 2-methylisoborneol in cyanobacteria and molecular detection methods for the producers of these compounds. Water Res . 2015; 68:56-66.. This genus includes two species: P. raciborskii Suda and Watanabe and P. attenuate Komárek and Komáarkova-Legnerová as currently accepted taxonomically. Nevertheless, the morphological characters of the genus Planktothricoides are frequently similar to those of the genus Planktothrix. One of the most important dissimilarities is the fact that Planktothricoides have solitary trichomes attenuated towards their ends and are sometimes slightly bent near the apex 1818 Komàrek J, Komárková J. Taxonomic review of the cyanoprokaryotic genera Planktothrix and Planktothricoides. Czech Phycology. 2004; 4:1-18.,1919 Komàrek J, Anagnostidis K. Cyanoprokaryota 2. Teil: Oscillatoriales. Süsswasserflora von Mitteleuropa 19/2. München: Elsevier GmbH; 2005.. Though both genera are morphologically similar, the genus Planktothricoides comprises an independent phylogenetic cluster from the genus Planktothrix based on 16S rDNA sequence analysis 1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.,1919 Komàrek J, Anagnostidis K. Cyanoprokaryota 2. Teil: Oscillatoriales. Süsswasserflora von Mitteleuropa 19/2. München: Elsevier GmbH; 2005..
Molecular analysis is recommended to clarify ambiguity in taxonomy based upon morphology, and cyanotoxin investigations are also widely used 2020 Padisák J, Vasas G, Borics G. Phycogenography of freshwater phytoplankton: traditional knowledge and new molecular tools. Hydrobiologia. 2016; 76(1):3-27.. Several genetic markers such as the small-subunit rRNA (16S rRNA) gene, phycocyanin encoding locus and the conserved gene encoding D-ribulose 1,5-bisphosphate carboxylase-oxygenase large subunit (rbcLX ) have been used for assessing cyanobacterial diversity and phylogenetic relationship 1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.,2121 Neilan BA, Burns BP, Relman DA, Lowe DR. Molecular identification of cyanobacteria associated with stromatolites from distinct geographical locations. Astrobiology. 2002; 2(3):271-280.
22 Lin S, Wu Z, Yu G, Zhu M, Yu B, Li R. Genetic diversity and molecular phylogeny of Planktothrix (Oscillatoriales, Cyanobacteria) strains from China. Harmful Algae. 2010; 9(1):87-97.
23 Komàrek J, Kaštovský J, Mareš J, Johansen JR. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 2014; 86:295-335.-2424 Singh P, Fatma A, Mishra AK. Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann Microbiol. 2015; 65(2):799-807.. Particularly, the 16S rRNA gene is highly conserved between different species of cyanobacteria and has become an important molecular marker for phylogenetic analysis 2525 Nübel U, Garcia-Pichel F, Muyzer G. PCR Primers to amplifiy 16S rRNA genes from Cyanobacteria. Appl Environ Microbiol. 1997; 63(8): 3327-3332.. Recently, Giglio et al.2626 Giglio S, Chou WKW, Ikeda H, Cane DE, Monis PT. Biosynthesis of 2-methylisoborneol in cyanobacteria. Environ Sci Technol. 2011; 45(3): 992-998. discovered the MIB biosynthetic genes in cyanobacteria from Pseudanaaena limnetica (Castaic Lake) and was further confirmed in P. limnetica NIVA- CYA 111 (Lake Biwa) and Oscillatoria limosa LBD 305b. More recently, Wang et al.1616 Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665. reported that Pseudoanabaena sp. dqh15 and P. raciborskii CHAB 3331 from China were confirmed to be 2-MIB producing organisms by using the 2-MIB synthesis-associated genes and GC-MS analyses. Afterwards, there were several cyanobacterial strains (e.g., Oscillatoria,Planktothrixand Letolyngbya bijugata) reported as the MIB producers 44 Wang Z, Xiao P, Song G, Li Y, Li R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ Sci Pollut R. 2015; 22(16):12133-12140.,1717 Suurnӓkki S, Gomez-Saez GV, Rantala-Ylinen A, Jokela J, Fewer DP, Sivonen K. Identification of geosmin and 2-methylisoborneol in cyanobacteria and molecular detection methods for the producers of these compounds. Water Res . 2015; 68:56-66.. These studies provided fundamental information for the development of molecular technologies to monitor odor-producing microorganisms and investigate the relationship between odor production and the corresponding synthesis genes. Several protocols for the detection of geosmin have been reported along with the elucidated genetic background. At present, real-time PCR assays have been established for quantification of geosmin-producing Streptomyces2727 Auffret M, Pilote A, Proulx E, Proulx D, Vandenberg G, Villemur R. Establishment of a real-time PCR method for quantification of geosmin-producing Streptomyces spp. in recirculating aquaculture systems. Water Res . 2011; 45(20):6753-6762. and Anabaena species 33 Su M, Gaget V, Giglio S, Burch M, An W, Yang M. Establishment of quantitative PCR methods for the quantification of geosmin-producing potential and Anabaena sp. in freshwater systems. Water Res. 2013; 47(10): 3444-3454.,2828 Tsao, H.W., Michinaka, A., Yen, H.K., Giglio, S., Hobson, P., Monis, P. et al. Monitoring of geosmin producing Anabaena circinalis using quantitative PCR. Water Res . 2014; 49(1):416-425.. However, no molecular technique for MIB-producing cyanobacteria has been established.
Understanding the diversity of off-odor-producing groups is essential for the monitoring and management of off-flavor events 33 Su M, Gaget V, Giglio S, Burch M, An W, Yang M. Establishment of quantitative PCR methods for the quantification of geosmin-producing potential and Anabaena sp. in freshwater systems. Water Res. 2013; 47(10): 3444-3454.,44 Wang Z, Xiao P, Song G, Li Y, Li R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ Sci Pollut R. 2015; 22(16):12133-12140.,2828 Tsao, H.W., Michinaka, A., Yen, H.K., Giglio, S., Hobson, P., Monis, P. et al. Monitoring of geosmin producing Anabaena circinalis using quantitative PCR. Water Res . 2014; 49(1):416-425.,2929 Zuo Y, Li L, Wu Z, Song L. Isolation, identification and odour-producing abilities of geosmin/2-MIB in actinomycetes from sediments in Lake Lotus, China. J. Water Supply Res T. 2009; 58(8):552-561.. Although off-odor has been described as a common problem for cultured fish, there are very few molecular genetic studies of cyanobacterial species producing off-odor from Thailand because of difficulties in the isolation and purification of strains. To evaluate diversity based on morphological and molecular analyses as well as ability of MIB production, Thai Planktothricoides strains were isolated in this study from several fishponds and reservoirs in Thailand. Then, microscopic morphological examination, pigment analysis and molecular diversity analysis based on 16S rRNA, rcbLX and MIB synthase genes between MIB-producing and non-MIB-producing strains were performed. Furthermore, GC/MS/MS analysis was also conducted for confirmation.
MATERIALS AND METHODS
Isolation, culturing condition and morphological characterization of Thai Planktothricoides
The Planktothricoides filaments were isolated from several freshwater ponds, most of which are used for fisheries or reservoirs from ten provinces of Thailand (Fig. 1). The strains were picked under a BX51 light microscope (Olympus, Tokyo, Japan) using the Pasteur Micropipette method 3030 Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1998; 167:1572-1574.. The filaments were subsequently transferred to double concave sides and washed twice with distilled water. Then, these strains were maintained in capped tubes containing 10 ml of BG11 medium3131 Andersen R. Algal Culturing Techniques. Burlington: Elsevier Academic Press; 2005.. The cultural condition was kept under temperature of 28º C, a light intensity of 20 µmol photons m-2s-1 and a light-dark cycle of 16:8 h. During the exponential growth phase (around 28 days old), successfully grown Planktothricoides strains were observed for their morphological characters under a BX51 light microscope (Olympus, Tokyo, Japan) with digital camera for species identification according to the descriptions of Suda et al.15 and Komárek and Komárková-Legnerová 3232 Komàrek J, Komárková-Legnerová J. Several rare freshwater planktic Cyanobacteria (Cyanoprokaryotes) from reservoirs in South America. Hoehnea. 2007; 34(1):49-58.. At least 50 filaments of strain tested, morphometric information as length (L) and width (W) were measured using the AxioVision software (Carl Zeiss, Jena, German). The L/W ratio was also calculated. Aberrant cell morphology was discarded for morphometric analyses.
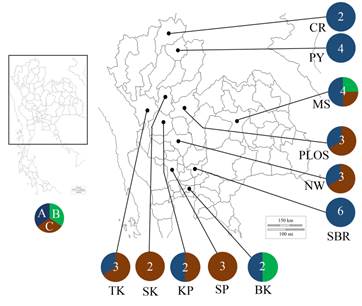
Geographic distributions based on the 16S rRNA gene sequences of Planktothricoides plotted on map showing upper part of Thailand. The numbers in each pie indicated the number of strains used for phylogenetic analyses. Each color in pies corresponds to a subclade in 16S rRNA phylogenetic trees, i.e. dark blue: Group A, green: Group B, brown: Group C. Each province is marked as CR: Chiang Rai, PY: Phayao, SK: Sukhothai, TK: Tak, KP: Kamphaeng Phet, PLOS: Phisanulok, MS: Maha Sarakham, NW: Nakhon Sawan, SP: Suphanburi, SBR: Saraburi, BKK: Bangkok (reported by Suda et al.1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.).
In vivo and phycobilin pigment absorption spectra determination
Thai Planktothricoides strains were used for determination of in vivo absorption and phycobilin pigment absorption following the method described by Suda et al. 15. An aliquot of culture was collected and the in vivo absorption spectra was measured from 350 to 850 nm by using UV-VIS HACH DR/4000 spectrophotometer (HACH, Loveland, USA). Then, the cells were harvested by centrifuging at 1,500 × g for 15 min at room temperatures. Then, the pellet cell was suspended in 0.1 M of phosphate buffer (pH 7), followed by five successive freezing and thawing cycles in order to disrupt the cell wall and consequently to isolate the phycocyanin (PC). The broken cell suspension was centrifuged at 1,500 × g for 15 min to remove unbroken cells. The absorbance of the supernatant was measured from 350 to 750 nm as explained above.
DNA extractions, PCR and sequencing
The cells were harvested by centrifugation for 10 min at 1,500 × g. DNA extraction was performed using a modified CTAB (cetyl-trimethyl-ammonium bromide)-based extraction method adapted for cyanobacteria33. The partial 16S rRNA, rbcLX and MIB synthase genes were amplified and sequenced by using the primers set showed in Table 1. The PCR reaction mixture (25 µl) for all genes consisted of 10 µl of nanopure water, 0.25 µl of each primer (0.10 µM), 2 µl of genomic DNA (20 ng), 12.5 µl of 2× Go Taq Green Master Mix (Promega, Madison, WI, USA) containing Taq DNA polymerase, magnesium chloride (3 mM), dNTPs (400 µM each) and reaction buffers (pH 8.5). The PCR conditions for 16S rRNA, rbcLX and MIB synthase genes were carried out using a Bio-Rad MyCycler (Bio-Rad, Hercules, CA, USA) with an initial denaturation at 94 °C for 2 min followed by 30 cycles of 94° C for 30s, 50° C for 30s and 72° C for 1 min, followed by the extension at 72° C for 5 min with a final hold at 4° C until needed. The PCR product were purified using the PCR purification kit (Omega, Norcross, GA, USA) and directly sequenced using an automated ABI Prism 3730XL DNA sequencer (Applied Biosystem, Foster, CA, USA). All sequences were submitted to GenBank and accession numbers (LC157916 - LC157948 for the partial16S rRNA genes, LC157949 - LC157985 for the rbcLX genes and LC157986 - LC157992 for the MIB synthase genes) are shown in Figs 3A-C.
Alignments and Phylogenetic analyses
The obtained sequences of 16S rRNA, rbcLX and MIB synthase genes in this study were separately aligned together with reference sequences of each gene retrieved from GenBank. The 16S rRNA alignment was made using MUSCEL implemented in MEGA V5.23434 Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011; 28(10):2731-2739., while the rbcLX and MIB synthase alignments were performed with ClustalW in BioEdit3535 Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999; 41:95-98.. The hypervariable intergenic region between rbcL and rbcX genes 3636 Rudi K, Skulberg OM, Jakobsen KS. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol. 1998; 180(3):3453-3461. was removed from the analyses.
The phylogenetic trees were generated according to the Neighbor-Joining (NJ), Maximum Likelihood (ML) and Bayesian Inference (BI) methods. The phylogenetic trees according to the NJ analyses 3737 Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol . 1987; 4(4): 406-425. were constructed with Tamura-Nei 3838 Tamura K, Kumar S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol Biol Evol . 2002; 19(10):1727-1736. as a genetic distance model implemented in program MEGA 5.2 3434 Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011; 28(10):2731-2739.. The support values for the internal branches of NJ trees were estimated using the bootstrap method with 1000 replicates3939 Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985; 39(4):783-791.. Substitution models as the best-fit model for ML and BI analyses were estimated with the Akaike Information Criterion (AIC) using MrModeltest v. 2.34040 Nylander JAA. Mrmodeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2008.. The best-fit models of 16S rRNA, rbcLX and MIB synthase gene sequences were the GTR+ I + G. All parameters of selected modes are summarized in Table 2.
The ML analysis was conducted using PhyML 3.0 4141 Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biolo. 2010; 59(3):307-321. with 100-fold bootstrap analysis. The BI was performed with MrBaye 3.1.2 4242 Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19(12):1572-1574. using Markov chain Monte Carlo algorithm. The program was run 12,000,000 generations for the partial16S rRNA data set, 4,000,000 generations for the rbcLX data set and 2,000,000 generations for the MIB synthase gene data sets, sampling trees every 100 generations until the standard deviation of split frequencies reached 0.01. To calculate Bayesian posterior probabilities (pp), the burn-in periods were determined at 105,000 trees for the partial 16S rRNA data set, 29,000 trees for the rbcLX data set and 5,000 trees for the MIB synthase gene sequence.
Gas Chromatography-Mass Spectrometry
The pellet cells of Thai Planktothricoides strains tested were taken up in 1 ml of methanol and vortexed following the method of Kakimoto et al.4343 Kakimoto, M., Ishikawa, T., Miyagi, A., Saito, K., Miyazaki, M., Asaeda, T, et al. Culture temperature affects gene expression and metabolic pathways in the 2-methylisoborneol-producing cyanobacterium Pseudanabaena galeata. J Plant Physiol. 2014; 171(3-4):292-300. with a slight modification. The aliquot of each sample was sonicated at 30 °C for 20 min and then centrifuged at 2,000 × g for 5 min. The supernatant was transferred to a 1.5 ml of glass tube and directly used for gas chromatography (GC) analysis. A Varian (Varian Inc. CA, USA) Saturn 2200 tandem mass spectrometer (MS/MS) connected to a Varian 3800 GC was utilized to identify the geosmin and MIB compounds from the Thai strains as described by Kakimoto et al4343 Kakimoto, M., Ishikawa, T., Miyagi, A., Saito, K., Miyazaki, M., Asaeda, T, et al. Culture temperature affects gene expression and metabolic pathways in the 2-methylisoborneol-producing cyanobacterium Pseudanabaena galeata. J Plant Physiol. 2014; 171(3-4):292-300.. Full scan electron impact mass spectra were recorded at a range of 40 to 200 m/z in a 0.55 total scan time. Peak identifications were aided by computerized mass spectra library and by interpretation of mass fragments. Furthermore, the mass spectrometers were also run in Selected-Ion Monitoring (SIM) in order to confirm the target compounds. The selected ions at m/z 95, 107, 121, 135, 150 were monitored for MIB, whilst m/z 112, 125, 126, 149, 182 were monitored for geosmin.
RESULTS
Morphological examination
The isolated Planktothricoides cells from cultures were morphologically examined using light microscopy. Most of the isolates in this study belong to the genus Planktothricoides based on the descriptions of Suda et al.1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595. and presented typical characteristics of the Planktothricoides raciborskii Suda and Watanabe (Fig. 2). The trichomes of Thai strains were solitary, planktonic, or sometimes formed mats, settled to the bottom of the cultures tubes (Fig. 2). Most of the trichomes were unsheathed, straight, consisting of cylindrical cells, in terminal parts sometimes slightly curved, apical cells conical-rounded or rounded and without calyptra. They were constricted or indistinctly constricted at cross-walls. The color of the trichome was dark blue-green or yellow-green. The thick sections were also shown in a single trichome. These filaments were similar in cells that were much greater in width than in length.
When measuring cell dimension, Thai Planktothricoides strains showed high morphometric variability, 1.86 to 5.96 µm in length and 2.83 to 13.70 µm in width. The length and width ratio (L/W ratio) ranged from 1:6 to 1:1. The average cell width (mean ± standard deviation) ranged from an average of 5.14±0.74 µm to 11.18±1.16 µm. The average cell length (mean ± standard deviation) ranged from 2.62±0.31 µm to 3.67±0.70 µm (data not shown).
In vivo and phycobilin pigment absorption spectra
The results indicated that the Thai strains used in this study followed somewhat the same patterns. In vivo absorption spectra had a large valley around 550 nm and two peaks at about 615 nm and 680 nm, respectively (data not shown). The first peaks were considered as phycocyanin (PC). These results were confirmed by the phycobiliprotein absorption spectrum presenting the maximum peak of PC at 615 nm (data not shown).
Phylogenetic analyses
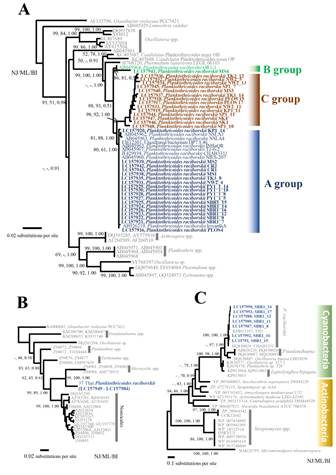
Thirty-three 16S rRNA gene sequences from this study, 9 previous 16S rRNA sequences of Planktothricoides strains and other genera in GenBank were used to construct the phylogenetic trees using the NJ, ML, and BI methods. The final alignment of partial 16S rRNA sequences (746 bp) produced 173 variable sites and 127 parsimony informative sites. The phylogenetic trees derived from NJ, ML and BI analyses were nearly identical in topology; therefore, only the NJ tree is shown for clarity of illustration (Fig. 3A). In the partial 16S rRNA phylogeny, all Thai Planktothricoides strains formed a clade with moderate support values from the three analyses (NJ=93, ML= 88 and BI=0.51). Among Planktothricoides strains, phylogenetic trees based on the 16S rRNA sequences revealed that the P. raciborskii clade comprised two previously well-supported subclades, designated as A and B groups in this study. The B group was the earliest diverging species of the genus Planktothricoides clade containing two strains from Thailand (OR1-1 and MS4) with the well support value (NJ=86, ML= 81 and BI=1.00). The A group comprised with the published P. raciboiskii strains from Japan (type strain NIES-207, koyamaA and INBaOR), China (DP7.5.46 and CHAB3331), Australia (NSLA3 and NSLA4) and Thailand (T1-6-2) together with 18 new Thai strains (NJ=80, ML=61, BI=1.00) (Fig. 3A). Interestingly, there was a novel subclade containing 14 Thai strains (KP1_11, KP1_14, SK1, SP1_10, SK4, PLOS17, PLOS19, MS3, SP1_7, NW2_5, NW2_11, NW2_13, TK3_12, and TK1_14). The significantly supported values of this subclade obtained from NJ, ML and BI analyses were 98, 92 and 1.00, respectively. This subclade belonged to A group and was thus designed as a new subclade, termed as C group (Fig. 3A). Among the three different groups of P. raciborskii based on the partial 16S rRNA gene sequences, the similarity ranged from 96.8% to 98.4% and the genetic distances ranged from 0.016 to 0.031. In contrast, at the intraspecific level within each group, the similarity and genetic distances were around 98.4% to 99.9% and 0.001 to 0.016.
Phylogenetic analyses of Thai Planktothricoides strains. Node robustness was assessed by performing bootstrap values from NJ and ML analyses and posterior probability from BI analysis, respectively. Values below 50 aren't shown. A: NJ trees based on the alignment of partial 16S rRNA gene sequence (746 bp) of Planktothricoides strains with other cyanobacterial taxa. Gloeobacter violaceus PCC7421 was used as out group. Each color corresponds to a subclade of Planktothricoides in 16S rRNA phylogenetic trees, i.e. dark blue: Group A, green: Group B, brown: Group C.; B: ML trees based on the alignment of rbcLX gene sequence (757 bp) of Planktothricoides strains with other cyanobacterial taxa. Gloeobacter violaceus PCC7421 was used as out group. Thai strains used in this study are marked in dark blue and bold; C: ML based MIB synthase gene from cyanobacteria included Thai strains and closest actinobacteria. Micromonospora olivasterospora NRRL 8178 strain was used as an out group.
For phylogenetic analyses based on the rbcLX genes, 37 rbcLX gene sequences obtained from the Thai strains were obtained and aligned with reference sequences from GenBank. The final alignment containing the regions for about 256 bp rbcL and 500 bp rbcX produced 500 variable sites and 467 parsimony informative sites. The rbcLX trees made by NJ, ML and BI methods revealed that almost all topology positions were the same; therefore, only the ML tree is presented in Fig. 3B. The phylogenetic trees based on rbcLX sequences indicated that all Thai Planktothricoides strains formed a monophyletic clade with significantly supported values by NJ (99), ML (100) and BI (1.00) methods, respectively. This Planktothricoides clade was phylogenetically positioned between Nostocales and Tychonema borurrellyi clades with strong support values (NJ= 99, ML = 89 and BI = 1.00).
An approximately 988 bp of new MIB synthase gene sequence in the strains from Saraburi province was successfully amplified and subsequently aligned with MIB synthase gene sequence of other MIB-producing cyanobacteria and actinomyces. The final alignment produced 848 variable sites and 740 parsimony informative sites. The MIB synthase phylogenetic trees determined from NJ, ML and BI methods revealed that almost all topology positions were the same; therefore, only the ML tree is presented in Fig. 3C. In the MIB synthase phylogeny, the monophyletic cyanobacteria branch formed a clade with other actinobacteria group. Among MIB-producing cyanobacteria strains, Oscillatoria, Planktothrix and Leptolyngbya branch was a basal sister branch, respectively. Subsequently, seven Thai Planktothricoides strains from Saraburi and two previous Planktothricoides sequences from Genbank (TH3 and CHAB3331) formed a clade with the robustly supported values from NJ=100, ML=100 and BI=1.00. This clade was the sister to the branch containing three strains of Pseudanabaena spp. and a strain of Oscillatoria limosa LBD305b. The MIB synthase gene sequences of Thai Planktothricoides strains had similarity of 99% compared with other MIB-producing Planktothricoides sequences.
Chemical analysis
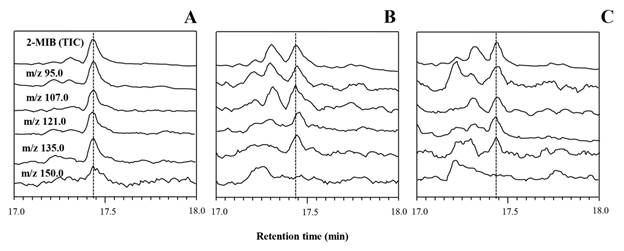
To confirm the results of olfactory and MIB synthesis gene detections, three strains (SBR1_11, SBR1_15 and SBR1_16) representing the potential MIB-producing strains from Saraburi were selected for GC/MS/MS analysis. The odorous compounds were identified and confirmed according to the retention time and the ion chromatogram in the scan mode and SIM mode. Their total ion chromatography and mass spectrum revealed the compound peak with a retention time of 17.42 min according to MIB (Fig. 4). The specific mass spectrum ion for MIB from all three tested strains monitored by SIM mode revealed the molecular mass of 95, 107, 121, and 135, while the molecular mass for the geosmin did not show. As the results of GC/MS/MS, it can conclude that Thai Planktothricoides strains from Saraburi can produce the MIB compound.
Total Ion Chromatograms (TIC) and Selected-Ion Monitoring (SIM) chromatogram according to MIB (m/z 95.0, 107.0, 121.0, 135.0 and 150) of Thai Planktothricoides strains SBR1_11 (A), SBR1_15 (B) and SBR1_15 (C) detected at around 17.42 min.
Distribution
All Planktothricoides strains used in this study were collected from fish ponds and fresh water resources in Northern (Chiang Rai, Phayao, Sukhothai, Tak, Kamphaeng Phet and Phisanulok), Northeastern (Maha Sarakham) and Central (Nakhon Sawan, Suphanburi and Saraburi) areas of Thailand (Fig. 1). The generic composition of Planktothricoides sp. based on 16S rRNA gene sequence was plotted onto a map of Thailand. Furthermore, two previous strains isolated from a reservoir in Bangkok1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595. were also included. During sampling, the temperature and pH in each sampling sites ranged from 26° C to 29° C and 7.5 to 9.0, respectively. Based on the 16S rRNA gene phylogenetic trees, most Thai Planktothricoides strains belonging to the A group were distributed in all three areas of Thailand. In this study, the strains forming the B and C groups were distributed in the Northeast and Central regions. The Planktothricoides strains isolated from Chiang Rai, Phayao and Saraburi only fell in the A group, while strains isolated from Sukhothai and Suphanburi belonged only to the C group.
DISCUSSION
In this study, during the period of cultivation, all strains (SBR1_6, SBR1_7, SBR1_8, SBR1_11, SBR1_12, SBR1_14, SBR1_15, SBR1_16 and SBR1_17) from Saraburi province were considered as potential odor-producing strains after olfaction. However, these strains do not differ morphologically from non-odor-producing strains. The Planktothricoides strains from Thailand were morphologically identical to the Planktothricoides
racborskii corresponding to original description by Suda et al.1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.. Trichomes of Thai Planktothricoides strains appeared generally straight or slightly bent near the apex and attenuated towards the ends. These characteristics are different from P. attenuate
Komárek and Komárková-Legnerová of which trichomes are always straight, wide in the middle and very gradually tapered towards ends3232 Komàrek J, Komárková-Legnerová J. Several rare freshwater planktic Cyanobacteria (Cyanoprokaryotes) from reservoirs in South America. Hoehnea. 2007; 34(1):49-58.. Meanwhile, the genus Planktothrix showed trichomes that were not attenuated or attenuated towards the ends3232 Komàrek J, Komárková-Legnerová J. Several rare freshwater planktic Cyanobacteria (Cyanoprokaryotes) from reservoirs in South America. Hoehnea. 2007; 34(1):49-58.. The type of phycobilin pigment composition is the morphological character corresponding to the original description of the genus Planktothricoides. Phycocyanin is a blue-colored pigment protein complex with the absorption maxima for phycocyanins found between 610 and 635 nm4444 Mimuro M. Photon capture, exciton migration and trapping and fluorescence emission in Cyanobacteria and red algae. In: Papageorgiou GC, Govindjee, editors. Chlorophyll a Fluorescence: A signature of photosynthesis. Netherland: Springer; 2004. p. 173-195.. As the results of the pigment analyses, this study has demonstrated that Thai strains contained phycocyanin as dominant pigments belong to a group I-pc as with the designation of Suda et al.1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.. Meanwhile, cell dimensions are largely heritable across a range of organisms, including prokaryotes and eukaryotes 4545 Chien AC, Hill NS, Levin PA. Cell size control in bacteria. Curr Biol. 2012; 22(9):R340-R349.
46 Lloyd, A. C. 2013. The regulation of cell size. Cell. 2012; 154(64):1194-1205.
47 Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. A constant size extension drives bacterial cell size homeostasis. Cell. 2014; 159(6):1433-46.-4848 Montgomery BL. Light-dependent governance of cell shape dimensions in Cyanobacteria. Front Microbiol. 2015; 6(514):1-8.. The results of morphometric information indicated that cell length, cell width and length to width ratio of Thai strains tested mostly overlapped with those of P. raciborskii strain NIES207, a type strain1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.. However, using cell size as a taxonomic criterion for cultured isolated of cyanobacteria can be problematic, since the character may vary according to the growth condition. This makes it difficult to define and to establish taxonomic limits for its identification 4949 Bittencourt-Oliveira MC. Development of Microcystis aeruginosa (Kützing) Kützing (Cyanophyceae/Cyanobacteria) under cultivation and its taxonomic implications. Algol Stud. 2000; 99:29-37.
50 Otsuka S, Suda S, Li R, Matsumoto S, Watanabe MM. Morphological variability of colonies of Microcystis morphospecies in culture. J Gen Appl Microbiol. 2000; 46(1):39-50.-5151 Otsuka S, Suda S, Shibata A, Oyaizu H, Matsumoto S, Watanabe MM. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kützing ex Lemmermann 1907 under the Rules of the Bacteriological Code. Int Syst Evol Microbiol. 2001; 51(Pt3):873-879.. Considering all results of morphological analyses, all Thai Planktothricoides strains tested were identified as P. raciborskii. Furthermore, this study suggests that both non-odor-producing and odor-producing Planktothricoides strain cannot be distinguished clearly using the basic information of morphological characters. Thus, in order to confirm these observations, it is necessary to increase the number of morphological analyses in the genus Planktothricoides.
The partial 16S rRNA genes cover approximately the first half of the gene, where most of the variable sites reside 5252 Lohtander K, Oksanen I, Rikkinen J. Genetic diversity of green algal and cyanobacterial photobionts in Nephroma (Peltigerales). Lichenologist. 2003; 35(4):325-339.. The results, based on the partial 16S rRNA gene sequences trees using the cyano-specific primers 5353 Neilan BA, Jacobs D, DelDot T, Blackall LL, Hawkins PR, Cox PT, et al. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int J Syst Bacteriol. 1997; 47(3):693-697., are in accord with the earlier studies 1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.,5454 Guidi-Rontani C, Jean MRN, Gonzalez-Rizzo S, Bolte-Kluge S, Gros O. Description of new filamentous toxic Cyanobacteria (Oscillatoriales) colonizing the sulfidic periphyton mat in marine mangroves. FEMS Microbiol Lett. 2014; 359(2):173-181., indicating a monophyletic clade of the genus Planktothricoides. Corresponding to the 16S rRNA phylogenetic result of Suda et al. 1515 Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595., the previous two subclades, named in this study as A and B groups, were also obtained from Thai Planktothricoides strains. Interestingly, an outcome of the present study was the discovery of a novel subclade and designated as C group. However, the 16S rRNA gene phylogeny results have revealed inconsistencies with the morphological classification of each group. This may be found in several cyanobacterial species in which differences in morphology are not reflected in analyses of 16S rRNA gene 5555 Garcia-Pichel F, Nübel U, Muyzer G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol. 1998; 169(6):469-482.,5656 Gugger M, Lyra C, Henriksen P, Couté A, Humbert JF, Sivonen K. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int J Syst Evol Microbiol. 2002; 52(Pt5):1867-1880.,5757 Lyra C, Suomalainen S, Gugger M, Vezie C, Sundman P, Paulin L, et al. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int J Syst Evol Microbiol. 2001; 51(Pt2):513-526.. The cut-off points of 97.5% and 95% 16S rRNA gene sequence similarity have been suggested for bacterial species and genus definition, respectively (Ludwig et al., 1998). Bosshard et al. 5959 Bosshard PP, Zbinden R, Abels S, Böddinghaus B, Altwegg M, Böttger EC. 16S rRNA gene sequencing versus the API 20 NE system and the Vitek 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J Clin Microbiolo. 2006; 44(4):1359-1366. and Janda and Abbott 6060 Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, Perils, and Pitfalls. J Clin Microbiol. 2007; 45(9):2761-2764. mentioned that the given ≥99% similarity of 16S rRNA gene sequence is regarded to be the type strain or reference strain of species. According to those definitions, the existence of cryptic species was also suspected from Thai strains in this study when 16S rRNA gene similarity of < 97.5% was observed. This finding suggests that the Planktothricoides strains in three subclades within the genus Planktothricoides probably evolved separately. Moreover, cryptic diversity can be expected as still limited sequences from a few geographical areas are available for this species. Thus, the examination based on the morphological features, ecology and secondary metabolite profile of each subclade in detail are necessary.
In this study, the result of the rbcLX phylogeny was incongruity compared with that the partial 16S rRNA phylogeny with the absence of three subclades. This difference may be because the rbcLX genes are too conservative to determine at the subclass level 6161 Doebley J, Durbin ML, Golenberg EM, Clegg MT, MA DP. Evolutionary analysis of the large subunit of carboxylase (rbcL) nucleotide sequence among the grasses (Graminae). Evolution. 1990; 44(4):1097-1108.,6262 Myllys L, Stenroos S, Thell A, Kuusinen M. High cyanobiont selectivity of epiphytic lichens in old growth boreal forest of Finland. New Phytol. 2007; 173(3):621-629., while the 16S rRNA genes can provide a much higher degree of resolution among cyanobacterial taxa than either morphological or chemical traits 6363 Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Dordrecht: Kluwer Academic Publishers; 1994. p. 1-25.. Rudi et al.3636 Rudi K, Skulberg OM, Jakobsen KS. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol. 1998; 180(3):3453-3461. also found that the topologies of the 16S rRNA gene and rbcLX trees were not congruent for genetically closely related Nostoc and Anabaena strains. Different regions of the genome may have experienced different evolutionary pressure, promoting an incomplete lineage sorting, leading to a divergence in the fixation rates among the locus and the taxon itself 6464 Gogarten JP, Zhaxybayeva O. Horizontal gene transfer: its detection and role in microbial evolution. In: Xu Y, Gogarten JP, editors. Computational Methods for Understanding Bacterial and Archaeal Genomes. Singapore: Imperial College Press; 2008. p. 137-152.. Considering the phylogenetic analysis results in this study, the 16S rRNA genes are more suitable for distinguishing genetic diversity within the genus Planktothricoides at the subclade level.
Genes (MIB synthase) amplified in this study showed sequence homology with other MIB-producing cyanobacterial species from previous studies 1616 Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665.,2626 Giglio S, Chou WKW, Ikeda H, Cane DE, Monis PT. Biosynthesis of 2-methylisoborneol in cyanobacteria. Environ Sci Technol. 2011; 45(3): 992-998.,6565 Ludwig F, Medger A, Börnick H., Opitz M, Lang K, Göttfert M, et al. Identification and expression analyses of putative sesquiterpene synthase genes in Phormidium sp. and prevalence of geoA-like genes in a drinking water reservoir. Appl Environ Microbiol . 2007; 73(21):6988-6993.
66 Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J Bacteriol . 2008; 190(18):6084-6096.-6767 Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008; 105(21):7422-7427.. Corresponding to the suggestion of Wang et al.1616 Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665., all 2-MIB-producing genes in cyanobacteria may have a common origin showing that homologous genes can be found between Pseudanabaena sp. dqh15 and P. raciborskii CHAB 3331. Moreover, the results of 16S rRNA gene phylogenies revealed that all MIB-producing P. raciborskii strains from Thailand and China (CHAB3331)1616 Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665. were grouped together with non-MIB-producing strains into the A group. This present study demonstrated that a single distinct group of MIB-producing strains cannot be supported as analyzed by 16S rRNA genes including rbcLX genes. The results can only indicate that the Planktothriocoides strain grouped in the A group should probably be considered as a potential MIB producer. On other hand, this study has hypothesized that MIB-producing strains within the genus Planktothricoides can coexist and distribute the ability of MIB production with non-MIB-producing strains due to horizontal gene transfer (HGT); for example, Nakasugi et al. 6868 Nakasugi K, Alexova R, Svensonand CJ, Neilan BA. Functional analysis of pilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J Bacteriol . 2007; 189(5):1689-1697.. Previous studies implied that the process of horizontal gene transfer may play a role in the distribution of hepatoxic strains within the genus Microcystis6969 Neilan BA., Dittmann E, Rouhiainen L, Bass RA, Schaub V, Sivonen K, et al. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J Bacteriol . 1999; 181(13):4089-4097.
70 Tillett D, Parker DL, Neilan BA. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyaninoperon (phycocyanin intergenic spacer) phylogenies. Appl Environ Microb. 2001; 67(6):2810-2818.-7171 Mikalsen B, Boison G, Skulberg OM, Fastner J, Davies W, Gabrielsen TM, et al. Natural variation in the microcystin synthetase operon mcy ABC and impact on microcystin production in Microcystis strain. J Bacteriol . 2003; 185(9):2774-2785.. However, there is no clear experimental evidence which favors horizontal gene transfer as a mechanism to explain the distribution of MIB-producing strains in cyanobacteria. Alternatively, the lack of the MIB synthase genes of non-MIB-producing Thai Planktothricoides strains may consequently be due to losses of this gene during evolution. Similarly, preliminary data suggest that the loss of mcy genes seem to be a continuous process starting with a loss of function by mutation7272 Kurmayer R, Christiansen G, Fastner J, Börner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004; 6(8):831-841.. Thus, future research should also be focused on finding appropriate genetic markers to conclusively investigate evolutionary relationships of MIB-producers within the genus Planktothricoides.
The qualitative confirmation of MIB compounds in GC/MS/MS analyses is needed with a minimum of four corresponding Selected-Ions Monitoring (SIM) analysis. The results of SIM mode in this study showed that peaks of m/z values according to MIB compound7373 Ding Z, Peng S, Xia W, Zheng H, Chen X, Yin L. Analysis of five earthy-musty odorants in environmental water by HS-SPME/GC-MS. International Journal of Analytical Chemistry. 2014; 2014:1-11. were detected from Thai Planktothricoides strains isolated from Saraburi province. Thus, the results of the olfaction in the initial cultivation and GC/MS/MS analysis corresponded with those of the MIB synthase gene phylogenetic analyses, considering that Thai Planktothricoides strains isolated from Saraburi province were MIB producers. These results are useful to provide genetic information from Thai Planktothricoides species in order to prevent the risk of taste-odor events caused by the cyanobacterial origin of MIB. Future research could use molecular technologies to monitor odor-producing cyanobacterial species and investigate the relationship between odor production and the corresponding synthesis gene.
An interesting characteristic of cyanobacteria is their broad geographical distribution, which reflects the group’s genotypic and phenotypic variation. The dispersal capability of the genus Planktothricoides in each region of Thailand is still poorly understood. Based on the 16S rRNA genes, the present study expected that the Planktothricoides strains belonging in the A group is the cosmopolitan species which can be found in all areas of Thailand. Depending on the aquatic environmental factors associating to chemical and physical parameters, the concentration of cyanobacteria can increase and produce unpleasant odor-causing compound in drinking water and aquaculture production. Addition research is needed to determine the distribution, abundance as well as environmental factors affecting growth of Planktothricoides species, in particular with MIB-producing strains, in several freshwater resources.
CONCLUSIONS
In conclusion, the results of morphological and phylogenetic (16S rRNA and rbcLX gene sequences) analyses revealed that all Thai Planktothricoides strains used in this study could be designated as Planktothricoides raciborskii (Woloszynska) Suda & Watanabe. MIB-producing and non-MIB-producing strains could not be distinguished using morphological features. The 16S rRNA gene sequences of Thai Planktothricoides strains fell into three different subclades (A, B and novel C groups), indicating the genetic variation into the genus Planktothricoides. From the detection of MIB synthase genes and GC/MS/MS analysis, strains from Saraburi were considered as MIB-producing strains. Moreover, the 16S rRNA phylogeny showed that the MIB-producing strains including Thai strains from Saraburi fell entirely into the A group. This information is provided for the development of quantitative molecular technologies to monitor odor-producing cyanobacterial species in environmental samples. In-depth studies on the environmental factors affecting the growth characterization and MIB bioaccumulation will also be required.
ACKNOWLEDGMENT
The author wishes to thank Dr.Pongsanat Pongcharoen for her valuable suggestion, and Mr. Cliff Sloane for editing the English in the manuscript. This research (R2558C226) was supported financially by Naresuan University.
REFERNCES
-
1Li L, Wan N, Gan N, Xiao BD, Song LR. Annual dynamics and origins of the odorous compounds in the pilot experimental area of Lake Dianchi, China. Water Sci Technol. 2007; 55(5):43-50.
-
2Watson SB, Charlton M, Rao YR, Howell T, Ridal J, Brownlee B, Marvin C, Millard S. Off flavours in large water bodies: physics, chemistry and biology in synchrony. Water Sci Technol . 2007; 55(5):1-8.
-
3Su M, Gaget V, Giglio S, Burch M, An W, Yang M. Establishment of quantitative PCR methods for the quantification of geosmin-producing potential and Anabaena sp. in freshwater systems. Water Res. 2013; 47(10): 3444-3454.
-
4Wang Z, Xiao P, Song G, Li Y, Li R. Isolation and characterization of a new reported cyanobacterium Leptolyngbya bijugata coproducing odorous geosmin and 2-methylisoborneol. Environ Sci Pollut R. 2015; 22(16):12133-12140.
-
5Lu J, Wills PS, Wilson PC. Trace analysis of off-flavor/odor compounds in water using liquid-liquid microextraction coupled with gas chromatography- positive chemical ionization-tandem mass spectrometry. Front Environ Sci Eng. 2016; 10(3):477-481.
-
6Guttman L, Van Rijn J. Identification of conditions underlying production of geosmin and 2-methylisoborneol in a recirculating system. Aquaculture. 2008; 279: 85-91.
-
7Schrader KK, Summerfelt ST. Distribution of off-flavor compounds and isolation of geosmin-producing bacteria in a series of water recirculating systems for rainbow trout culture. N Am J Aquacult. 2010; 72(1): 1-9.
-
8Houle S, Schrader K, Lefrancois NR, Comeau Y, Kharoune M, Summerfelt ST, Savoie A, Vandenberg GW. Geosmin causes off-flavor in Arctic Charr raised in recirculating aquaculture systems. Aquaculture Res. 2011; 42:360-365.
-
9Yamprayoon J, Noomhorm A. Geosmin and off-flavor in Nile tilapia (Oreochromis niloticus). J Aqua Food Prod T. 2000; 9(2):29-41.
-
10Gutierrez R, Whangchai N, Sompong U, Prarom W, Iwami N, Itayama T, et al. Off-flavour in Nile tilapia (Oreochromis niloticus) cultured in an integrated pond-cage culture system. Maejo Int J Sci Technol. 2013; 7:1-13.
-
11Yarnpakdee S, Benjakul S, Penjamras P, Kristinsson HG. Chemical compositions and muddy flavor/odour of protein hydrolysate from Nile tilapia and broadhead catfish mince and protein isolate. Food Chem. 2013; 142:210-216.
-
12Pimolrat P, Whangchai N, Chitmanat C, Lebel L. Musty/earthy off-flavor in aquatic animal: impact, causes and solutions. KKU Research Journal. 2014; 19(6):927-938.
-
13Pimolrat P, Whangchai N, Chitmanat C, Itayama T, Lebel L. Off-flavor characterization in high-nutrient-load Tilapia ponds in Northern Thailand. Turkish J Fish Aquat Sci. 2015; 15(2):273-281.
-
14Suwanpakdee S, Gutierrez R, Pithakpol S, Jampeetong A, Pathom-aree W, Nomura N, et al. Earthy-musty odour and off-flavour taints in Phayao Lake, Thailand. Chiang Mai J Sci. 2016; 43(1):1076-1085.
-
15Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, et al. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Micr. 2002; 52(Pt5):1577-1595.
-
16Wang Z, Xu Y, Shao J, Wang J, Li R. Genes associated with 2-Methylisoborneol biosynthesis in cyanobacteria: isolation, characterization, and expression in response to light. PLoS ONE. 2011; 6(4): e18665.
-
17Suurnӓkki S, Gomez-Saez GV, Rantala-Ylinen A, Jokela J, Fewer DP, Sivonen K. Identification of geosmin and 2-methylisoborneol in cyanobacteria and molecular detection methods for the producers of these compounds. Water Res . 2015; 68:56-66.
-
18Komàrek J, Komárková J. Taxonomic review of the cyanoprokaryotic genera Planktothrix and Planktothricoides. Czech Phycology. 2004; 4:1-18.
-
19Komàrek J, Anagnostidis K. Cyanoprokaryota 2. Teil: Oscillatoriales. Süsswasserflora von Mitteleuropa 19/2. München: Elsevier GmbH; 2005.
-
20Padisák J, Vasas G, Borics G. Phycogenography of freshwater phytoplankton: traditional knowledge and new molecular tools. Hydrobiologia. 2016; 76(1):3-27.
-
21Neilan BA, Burns BP, Relman DA, Lowe DR. Molecular identification of cyanobacteria associated with stromatolites from distinct geographical locations. Astrobiology. 2002; 2(3):271-280.
-
22Lin S, Wu Z, Yu G, Zhu M, Yu B, Li R. Genetic diversity and molecular phylogeny of Planktothrix (Oscillatoriales, Cyanobacteria) strains from China. Harmful Algae. 2010; 9(1):87-97.
-
23Komàrek J, Kaštovský J, Mareš J, Johansen JR. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 2014; 86:295-335.
-
24Singh P, Fatma A, Mishra AK. Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann Microbiol. 2015; 65(2):799-807.
-
25Nübel U, Garcia-Pichel F, Muyzer G. PCR Primers to amplifiy 16S rRNA genes from Cyanobacteria. Appl Environ Microbiol. 1997; 63(8): 3327-3332.
-
26Giglio S, Chou WKW, Ikeda H, Cane DE, Monis PT. Biosynthesis of 2-methylisoborneol in cyanobacteria. Environ Sci Technol. 2011; 45(3): 992-998.
-
27Auffret M, Pilote A, Proulx E, Proulx D, Vandenberg G, Villemur R. Establishment of a real-time PCR method for quantification of geosmin-producing Streptomyces spp. in recirculating aquaculture systems. Water Res . 2011; 45(20):6753-6762.
-
28Tsao, H.W., Michinaka, A., Yen, H.K., Giglio, S., Hobson, P., Monis, P. et al. Monitoring of geosmin producing Anabaena circinalis using quantitative PCR. Water Res . 2014; 49(1):416-425.
-
29Zuo Y, Li L, Wu Z, Song L. Isolation, identification and odour-producing abilities of geosmin/2-MIB in actinomycetes from sediments in Lake Lotus, China. J. Water Supply Res T. 2009; 58(8):552-561.
-
30Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1998; 167:1572-1574.
-
31Andersen R. Algal Culturing Techniques. Burlington: Elsevier Academic Press; 2005.
-
32Komàrek J, Komárková-Legnerová J. Several rare freshwater planktic Cyanobacteria (Cyanoprokaryotes) from reservoirs in South America. Hoehnea. 2007; 34(1):49-58.
-
33Fiore MF, Moon DH, Tsai SM, Lee H, Trevors JT. Miniprep DNA isolation from unicellular and filamentous cyanobacteria. J Microbiolo Methods. 2000; 39(2):159-169.
-
34Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011; 28(10):2731-2739.
-
35Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999; 41:95-98.
-
36Rudi K, Skulberg OM, Jakobsen KS. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol. 1998; 180(3):3453-3461.
-
37Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol . 1987; 4(4): 406-425.
-
38Tamura K, Kumar S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol Biol Evol . 2002; 19(10):1727-1736.
-
39Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985; 39(4):783-791.
-
40Nylander JAA. Mrmodeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2008.
-
41Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biolo. 2010; 59(3):307-321.
-
42Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19(12):1572-1574.
-
43Kakimoto, M., Ishikawa, T., Miyagi, A., Saito, K., Miyazaki, M., Asaeda, T, et al. Culture temperature affects gene expression and metabolic pathways in the 2-methylisoborneol-producing cyanobacterium Pseudanabaena galeata. J Plant Physiol. 2014; 171(3-4):292-300.
-
44Mimuro M. Photon capture, exciton migration and trapping and fluorescence emission in Cyanobacteria and red algae. In: Papageorgiou GC, Govindjee, editors. Chlorophyll a Fluorescence: A signature of photosynthesis. Netherland: Springer; 2004. p. 173-195.
-
45Chien AC, Hill NS, Levin PA. Cell size control in bacteria. Curr Biol. 2012; 22(9):R340-R349.
-
46Lloyd, A. C. 2013. The regulation of cell size. Cell. 2012; 154(64):1194-1205.
-
47Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. A constant size extension drives bacterial cell size homeostasis. Cell. 2014; 159(6):1433-46.
-
48Montgomery BL. Light-dependent governance of cell shape dimensions in Cyanobacteria. Front Microbiol. 2015; 6(514):1-8.
-
49Bittencourt-Oliveira MC. Development of Microcystis aeruginosa (Kützing) Kützing (Cyanophyceae/Cyanobacteria) under cultivation and its taxonomic implications. Algol Stud. 2000; 99:29-37.
-
50Otsuka S, Suda S, Li R, Matsumoto S, Watanabe MM. Morphological variability of colonies of Microcystis morphospecies in culture. J Gen Appl Microbiol. 2000; 46(1):39-50.
-
51Otsuka S, Suda S, Shibata A, Oyaizu H, Matsumoto S, Watanabe MM. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kützing ex Lemmermann 1907 under the Rules of the Bacteriological Code. Int Syst Evol Microbiol. 2001; 51(Pt3):873-879.
-
52Lohtander K, Oksanen I, Rikkinen J. Genetic diversity of green algal and cyanobacterial photobionts in Nephroma (Peltigerales). Lichenologist. 2003; 35(4):325-339.
-
53Neilan BA, Jacobs D, DelDot T, Blackall LL, Hawkins PR, Cox PT, et al. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int J Syst Bacteriol. 1997; 47(3):693-697.
-
54Guidi-Rontani C, Jean MRN, Gonzalez-Rizzo S, Bolte-Kluge S, Gros O. Description of new filamentous toxic Cyanobacteria (Oscillatoriales) colonizing the sulfidic periphyton mat in marine mangroves. FEMS Microbiol Lett. 2014; 359(2):173-181.
-
55Garcia-Pichel F, Nübel U, Muyzer G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol. 1998; 169(6):469-482.
-
56Gugger M, Lyra C, Henriksen P, Couté A, Humbert JF, Sivonen K. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int J Syst Evol Microbiol. 2002; 52(Pt5):1867-1880.
-
57Lyra C, Suomalainen S, Gugger M, Vezie C, Sundman P, Paulin L, et al. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int J Syst Evol Microbiol. 2001; 51(Pt2):513-526.
-
58Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, et al. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998; 19(4):554-568.
-
59Bosshard PP, Zbinden R, Abels S, Böddinghaus B, Altwegg M, Böttger EC. 16S rRNA gene sequencing versus the API 20 NE system and the Vitek 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J Clin Microbiolo. 2006; 44(4):1359-1366.
-
60Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, Perils, and Pitfalls. J Clin Microbiol. 2007; 45(9):2761-2764.
-
61Doebley J, Durbin ML, Golenberg EM, Clegg MT, MA DP. Evolutionary analysis of the large subunit of carboxylase (rbcL) nucleotide sequence among the grasses (Graminae). Evolution. 1990; 44(4):1097-1108.
-
62Myllys L, Stenroos S, Thell A, Kuusinen M. High cyanobiont selectivity of epiphytic lichens in old growth boreal forest of Finland. New Phytol. 2007; 173(3):621-629.
-
63Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Dordrecht: Kluwer Academic Publishers; 1994. p. 1-25.
-
64Gogarten JP, Zhaxybayeva O. Horizontal gene transfer: its detection and role in microbial evolution. In: Xu Y, Gogarten JP, editors. Computational Methods for Understanding Bacterial and Archaeal Genomes. Singapore: Imperial College Press; 2008. p. 137-152.
-
65Ludwig F, Medger A, Börnick H., Opitz M, Lang K, Göttfert M, et al. Identification and expression analyses of putative sesquiterpene synthase genes in Phormidium sp. and prevalence of geoA-like genes in a drinking water reservoir. Appl Environ Microbiol . 2007; 73(21):6988-6993.
-
66Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J Bacteriol . 2008; 190(18):6084-6096.
-
67Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008; 105(21):7422-7427.
-
68Nakasugi K, Alexova R, Svensonand CJ, Neilan BA. Functional analysis of pilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J Bacteriol . 2007; 189(5):1689-1697.
-
69Neilan BA., Dittmann E, Rouhiainen L, Bass RA, Schaub V, Sivonen K, et al. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J Bacteriol . 1999; 181(13):4089-4097.
-
70Tillett D, Parker DL, Neilan BA. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyaninoperon (phycocyanin intergenic spacer) phylogenies. Appl Environ Microb. 2001; 67(6):2810-2818.
-
71Mikalsen B, Boison G, Skulberg OM, Fastner J, Davies W, Gabrielsen TM, et al. Natural variation in the microcystin synthetase operon mcy ABC and impact on microcystin production in Microcystis strain. J Bacteriol . 2003; 185(9):2774-2785.
-
72Kurmayer R, Christiansen G, Fastner J, Börner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004; 6(8):831-841.
-
73Ding Z, Peng S, Xia W, Zheng H, Chen X, Yin L. Analysis of five earthy-musty odorants in environmental water by HS-SPME/GC-MS. International Journal of Analytical Chemistry. 2014; 2014:1-11.
-
Erratum
In Article “Diversity of the Potential 2-Methylisoborneol-Producing Genotypes in Thai Strains of Planktothricoides (Cyanobacteria)”, with the number of DOI: http://dx.doi.org/10.1590/1678-4324-2017160567, published in journal Brazilian Archives of Biology and Technology, vol. 60, the 01 page.That read:“http://dx.doi.org/10.190/1678-4324-2017160567”Read:“http://dx.doi.org/10.1590/1678-4324-2017160567”
Publication Dates
-
Publication in this collection
2017
History
-
Received
01 July 2016 -
Accepted
28 Nov 2016