ABSTRACT
Studies were conducted to characterize Klebsiella pneumoniae isolates from urinary tract infection (UTI) patients in Sylhet city of Bangladesh. At the same time, all isolates were screened for some common virulence genes and four significant isolates were searched for plasmid number and sizes by mini alkaline-lysis method. Among five tested isolates from female UTI patients, gyrase subunit B2 (gyrb2) amplified in all isolates, lipase and nuclease detected in three isolates and serine protease amplifies in two isolates and gave the expected band of 1130 bp, 517 bp, 1055 bp and 211 bp respectively. Two of four isolates showed 9.82 kb plasmid band on agarose gel. Isolates bearing 9.82 kb plasmid were found to be resistant to multiple commercial antibiotics. At the same time all isolates were screened for in-vitro plate assay for proteolytic, lypolytic and hemolytic activity. Isolates with positive plasmid and more than one virulent gene with gyrB2 showed positive result in in-vitro culture plate with clear zone of proteolysis, hemolysis or lipolysis. This study will be helpful for further study in finding correlation or pattern of virulence properties for K. pneumoniae associated UTI in Bangladesh.
Keywords:
K. pneumoniae; urinary tract infection; virulence factors; plasmid profiling; correlation study
INTRODUCTION
Urinary tract infection (UTI) is the most commonly encountered hospital-acquired infection. Klebsiella pneumoniae is the second leading cause of urinary tract infection (UTI) after E. coli [11 Farajnia S, Alikhani MY, Ghotaslou R, Naghili B, Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. International Journal of Infectious Diseases. 2009 Mar 31; 13(2):140-4.]. However, although quite low prevalence, the pathogenicity of K. pneumoniae associated UTI is much higher than E. coli [11 Farajnia S, Alikhani MY, Ghotaslou R, Naghili B, Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. International Journal of Infectious Diseases. 2009 Mar 31; 13(2):140-4.,22 Foysal MJ, Rahman MM, Prodhan MS. PCR Based Molecular Detection of the Gyr-B-2 Gene from the Klebsiella Sp. Isolates from Patients who were Suffering with Pneumonia and Urinary Tract Infections (UTIs). Journal of clinical and diagnostic research: JCDR. 2013 Jan;7(1):23-5]. It is estimated that 150 million people affected by UTI each year in which approximately 12% of UTI caused by K. pneumoniae and the number is increasing alarmingly in East Asia [22 Foysal MJ, Rahman MM, Prodhan MS. PCR Based Molecular Detection of the Gyr-B-2 Gene from the Klebsiella Sp. Isolates from Patients who were Suffering with Pneumonia and Urinary Tract Infections (UTIs). Journal of clinical and diagnostic research: JCDR. 2013 Jan;7(1):23-5,33 Kucheria R, Dasgupta P, Sacks S, Khan M, Sheerin N. Urinary tract infections: new insights into a common problem. Postgraduate medical journal. 2005 Feb; 81(952):83-6]. In Bangladesh, same trend for UTI infection also observed and significant numbers of isolates became antibiotic resistante [44 Kamrul L, Eliza R, Habibura R, Ranjith R. Urinary Tract Infection in Children: An Update Bangaladesh. J Child Health. 2012; 36(2):90-7.]. Yet any study conducted on gene-virulence factors and plasmid profiling for K. pneumoniae in order to find out molecular mechanism in disease progression and correlation among virulence factors [55 Farshad S, Ranijbar R, Japoni A, Hosseini M, Anvarinejad M, Mohammadzadegan R. Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Archives of Iranian Medicine (AIM). 2012 May 1; 15(5):312-6.,22 Foysal MJ, Rahman MM, Prodhan MS. PCR Based Molecular Detection of the Gyr-B-2 Gene from the Klebsiella Sp. Isolates from Patients who were Suffering with Pneumonia and Urinary Tract Infections (UTIs). Journal of clinical and diagnostic research: JCDR. 2013 Jan;7(1):23-5]. Targeting of virulence gene is the key factor in determining the pathogenicity and disease causing ability of any isolate because this gene act multi-functionally and multi-factorially. In recent time, molecular detection of virulence genes provide a promising tool for understanding pathogenicity of an isolates as well as in disease diagnosis and prophylaxis [66 Bugarel M, Granier SA, Weill FX, Fach P, Brisabois A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC microbiology. 2011 Jun 27; 11(1):1-51.]. Several virulence genes i.e. nuclease, protease, lipase, hemolysin, gyrase etc. encode products that indulge/influence the organism in expressing its virulence properties in the host cell [77 Murugkar HV, Rahman H, Dutta PK. Distribution of virulence genes in Salmonella serovars isolated from man & animals. Indian Journal of Medical Research. 2003 Feb 1; 117:66-70]. DNA gyrase (gyrB) is an ATP requiring enzyme help in replication by unwinding DNA super coil by an unusual mechanism [88 Breuer T, Benkel DH, Shapiro RL, Hall WN, Winnett MM, Linn MJ, Neimann J, Barrett TJ, Dietrich S, Downes FP, Toney DM. A multistate outbreak of Escherichia coli O157: H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerging infectious diseases. 2001 Nov; 7(6):977-82.,99 Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proceedings of the National Academy of Sciences. 1976 Nov 1; 73(11):3872-6.]. DNA gyrase subunit two (gyrB2) is essential for cell viability and poses beta hemolytic activity: ability to lyse blood agar might contribute to the pathogenicity of any isolates [1010 Han XY, Han FS, Segal J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. International journal of systematic and evolutionary microbiology. 2008 Jun 1; 58(6):1398-403.]. Lipase (lipA) shows a wide variety of substrate and reaction specificity which catalyze hydrolysis of triglycerol. Increasing serum lipase activity results in higher chance of pancreatic inflammation and other disorders [1111 Rosenblum JL. Serum lipase activity is increased in disease states other than acute pancreatitis: amylase revisited. Clinical chemistry. 1991 Mar 1; 37(3):315-6.]. Nuclease (nucl) is capable of cleaving phosphodiester bonds between the nucleotide subunit of nucleic acids. Some of these are sequence specific endonuclease that recognizes and cut specific sequences within a particular point of base pairs. Nuclease showed to be very important virulence factors in many clinical isolates i.e. Strepcoccus sp. [1212 Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003 Nov 1; 84(4):269-78.]. Serine protease (serP) is an enzyme that cleaves peptide bonds in protein in which serine serve as the nucleophilic amino acid at the enzyme’s active site [1313 Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cellular and molecular life sciences. 2014 Mar 1; 71(5):745-70.]. Plasmid profiling involves the study of sizes and numbers of plasmid in a particular organism. After the cell lyses, the nucleic acids are subjected to electrophoresis to visualize the size and copy number plasmid [1414 Smalla K, Jechalke S, Top EM. Plasmid detection, characterization and ecology. Microbiology spectrum. 2015 Feb; 3(1):1-21.]. Plasmid profiling sometimes may not provide useful information because some species may contain variable number of plasmids or even unrelated bacteria may harbor similar number of plasmids. However, plasmid profile analysis is very important in analysis of gene-virulence relationship study [1515 Victor LY. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerging Infect Dis. 2007; 13(7): 123-128.,1616 Koga VL, Tomazetto G, Cyoia PS, Neves MS, Vidotto MC, Nakazato G, Kobayashi RK. Molecular screening of virulence genes in extraintestinal pathogenic Escherichia coli isolated from human blood culture in Brazil. BioMed research international. 2014 Apr 15; 2014:id465054.].
Antibiotic sensitivity assay is generally done concurrently with plasmid profiling in order to correlate resistance pattern with their amplified plasmid [1717 Akingbade OA, Ogiogwa IJ, Okonko IO, Okerentugba PO, Innocent-Adiele HC, Nwanze JC, Onoh CC. Plasmid Profile of Isolated Klebsiella Species in a Tertiary Hospital in Abeokuta, Ogun State, Nigeria. World Applied Science Journal. 2012. Mar; 3:371-8]. Large plasmid confer broad antibiotic resistance pattern including extended spectrum beta-lactamase (ESBL) which significantly associated with virulence properties including a number of virulence functions in a human infection [1717 Akingbade OA, Ogiogwa IJ, Okonko IO, Okerentugba PO, Innocent-Adiele HC, Nwanze JC, Onoh CC. Plasmid Profile of Isolated Klebsiella Species in a Tertiary Hospital in Abeokuta, Ogun State, Nigeria. World Applied Science Journal. 2012. Mar; 3:371-8,1818 Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences. 2015 Jul 7; 112(27):E3574-81.]. In-vitro plate assay to analyze proteolytic, hemolytic and lypolytic activity have been shown to be a promising approach to indentify co-relation between amplified gene and corresponding virulence factors [1919 Foysal MJ, Rahman MM, Rabbee MF, Nazmul M, Hossain RM, Rahman MJ, Miah MF, Islam K. Identification and assay of putative virulence properties of Eschericha coli gyrase subunit A and B among hospitalized UTI patients in Bangladesh. Inov Pharm Pharmacother. 2013; 1(1):54-9.,2020 Pinto JB, Rossatto FC, Martins PD, Frazzon AP. Genetic relationships and virulence factors in Staphylococcus aureus isolated from raw poultry in South Brazil. Annals of Microbiology. 2015 Dec 1; 65(4):1933-40.]. Thus the present study was conducted in order to i) detect the major putative virulent genes, size and copy number of plasmid in K. pneumoniae causing UTI in Eastern part of Bangladesh; ii) to find out correlations among virulence factors and plasmid of UTI causing K. pneumoniae.
MATERIALS AND METHODS
Collection of bacterial isolates
Six bacterial isolates were collected from two different hospitals and diagnostic centre of Sylhet city, Bangladesh, Popular Hospital and Diagnostic Centre of Bangladesh (isolates K1, K5 and K8) and M. A. G. Osmani Medical College and Hospital (isolates K10, K11 and K12). Isolates were cultured in ESBL medium overnight and transported immediately after culture to USDA-project laboratory of Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh by maintaining cool chain. All isolates were collected from UTI patients of different aged persons, older than 40 years except K10, which collected from 12 years old girl.
Bacterial culture condition
In laboratory, individual bacterial isolates were streaked in nutrient agar plate for pure culture and incubated at 37 ºC overnight. After incubation, single colony was picked for different biochemical tests for K. pneumoniae. For the isolation of DNA, bacterial isolates were cultured in nutrient broth (NB) at 37ºC in a shaker incubator at 150 rpm speed. For profiling of plasmid DNA, five isolates were cultured in Luria broth (LB) medium and incubated overnight at 37ºC in a shaker incubator with same speed used for DNA extraction.
Biochemical characterization
Freshly cultured bacterial isolates were tested for further confirmations belong to K. pneumoniae species. A series of biochemical test were done according to Burgey’s manual of determinative bacteriology [2121 Breed, RS, Murray, EGD, Smith, NB. Burgey's manual of determinative bacteriology. 7th Edn. Baltimore: Waverly Press Inc; 1957.]. Major tests were; gram’s test, gram staining, catalase, oxidase, oxidative-fermentative, indole, hydrogen sulfide, methyl red, voges-proskauer and, motility and so forth. All results were matched with Burgey’s manual and only sole characteristics belong to K. pneumoniae species were taken for further study.
Bacterial DNA extraction
A total of five bacterial isolates were cultured in nutrient broth overnight at 37 ºC in a shaker incubator. Bacterial genomic DNA was extracted by following instructions of commercial genomic DNA extraction kit (Bio Basic Inc., 160 Torbay Road, Markham Ontario, Canada) and extracted DNA stored initially at - 20 ºC for further use. DNA of five isolates was quantified by gel electrophoresis with lambda DNA as well as in spectrophotometer as a ratio of DNA-protein absorbance.
PCR reaction mixture set-up
The PCR was performed in 25 µl master mixtures containing DNA template (150 µg of genomic DNA of K. pneumoniae) of 1.2 µl, 1 µl of 25 mM MgCl2, 5 µl of 10x colorless reaction buffer, 0.5 µl concentration of deoxynucleotide triphosphate (dNTP), 1.2 µl of each forward and reverse primer with the concentration of 0.5 µM (Table 2) and 0.15 µl Taq DNA polymerase. The amplifications were carried out in a MultiGene gradient thermal cycler (Labnet International Inc. USA).
PCR amplification condition
PCR amplification condition was optimized after several laboratory trials with the following parameters: an initial denaturation run of 94°C for 4 min; denaturation step of 94°C for 1 min, annealing 1 min at 64 °C for nucl, lipA and serP, 62°C for gyr B2, and an extension at 72°C for 90 s; and a final extension step of 72°C for 10 min. For all genes, 35 serial cycles of amplification reaction was performed.
Profiling of bacterial plasmid DNA
Five bacterial isolates were inoculated in freshly prepared LB medium and incubated at 37ºC in a shaker incubator at 150 rpm for overnight. Plasmid DNA was isolated and purified by alkaline lyses method by using following reagents: solution 1 (50 mM glucose, 25 mM Tris HCL, and 10m MEDTA), solution 2 (0.2% NaOH, 1% SDS) and phenol-chloroform [2222 Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 4th Edn. New York: Cold Spring Harbor Laboratory Press; 2012.].
Antibiogram profile of the isolates
Resistance or sensitivity of the K. pneumoniae isolates to 12 commercial antibiotic discs was determined by disk diffusion assay [2323 Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966 Apr; 45(4):493-6.]. The antibiotic disks used in this study were ampicilin (10 µg/disk), kanamycin (30 µg/disk), erythromycin (15 µg/disk), amoxycillin (30 µg/disk), chloramphenicol (30 µg/disk), levofloxacin (5 µg/disk), ciprofloxacin (30 µg/disk), azithromycin (30 µg/disk), cefradine (25 µg/disk), gentamicin (10 µg/disk), streptomycin (10 µg/disk) and sulphamethoxazole (25 µg/disk). Overnight bacterial culture (30 µl) was spread inoculated on Isosensei Test Agar plates (Micromaster, India) and disks were aseptically placed on the culture. After 24h of incubation at 37oC, the zone of inhibitions were measured and resistance or sensitivity of the strains were estimated according to the disk manufacturers instruction.
In -vitro plate assay for study of virulence factors
Bacterial isolates were cultured in-vitro in culture plate with casein, tween20 and sheep blood agar supplementation for analyzing proteolysis, lipolysis and hemolysis. Proteolytic activity was determined by using casein as a protein source and positive isolates can hydrolyze casein and produce zone of hemolysis [2424 Rodarte MP, Dias DR, Vilela DM, Schwan RF. Proteolytic activities of bacteria, yeasts and filamentous fungi isolated from coffee fruit (Coffea arabica L.). ActaScientiarum. Agronomy. 2011 Sep; 33(3):457-64.]. Casein agar media was prepared as follows: 0.5% of casein, 0.5% of glucose, and 2% of agar (w/v), pH 7.0. Bacterial isolates were streaked into tween20 agar containing 10 (g/L) peptone; 5 (g/L) NaCl, 0.1 (g/L) CaCl2, 2% agar and 1 (ml/L) tween20 for measuring the halos of precipitation for determine lipolytic activity [2525 Michelim L, Lahude M, Araújo PR, Giovanaz DS, Müller G, Delamare AP, Costa SO, Echeverrigaray S. Pathogenic factors and antimicrobial resistance of Staphylococcus epidermidis associated with nosocomial infections occurring in intensive care units. Brazilian Journal of Microbiology. 2005 Mar; 36(1):17-23.]. Hemolytic activity was calculated by culturing isolates on TSA supplemented with 5% (v/v) sheep’s blood [2525 Michelim L, Lahude M, Araújo PR, Giovanaz DS, Müller G, Delamare AP, Costa SO, Echeverrigaray S. Pathogenic factors and antimicrobial resistance of Staphylococcus epidermidis associated with nosocomial infections occurring in intensive care units. Brazilian Journal of Microbiology. 2005 Mar; 36(1):17-23.] and the diameter of the halos of hemolysis was measured [1919 Foysal MJ, Rahman MM, Rabbee MF, Nazmul M, Hossain RM, Rahman MJ, Miah MF, Islam K. Identification and assay of putative virulence properties of Eschericha coli gyrase subunit A and B among hospitalized UTI patients in Bangladesh. Inov Pharm Pharmacother. 2013; 1(1):54-9.].
RESULTS
Identification of K. pneumoniae isolates
Although all bacterial isolates were initially supplied as K. pneumoniae culture on selective agar media supplemented with ornithine, raffinose and koser citrate [2626 Bruce SK, Schick DG, Tanaka L, Jimenez EM, Montgomerie JZ. Selective medium for isolation of Klebsiella pneumonia. J of ClinMicrobiol. 1981; 13(6): 1114-6.]; however, all bacterial isolates were assayed for their morphological, physiological and biochemical properties for further confirmation. Among six isolates, five isolates were confirmed belong to K. pneumoniae species according to Burgey’s manual [2121 Breed, RS, Murray, EGD, Smith, NB. Burgey's manual of determinative bacteriology. 7th Edn. Baltimore: Waverly Press Inc; 1957.] for identification of bacteria (Table 1). Isolates were numbered as K1, K5, K8, K10 and K11. Isolate K12 showed confusing result in oxidative-fermentative test and this was excluded from this study.
PCR amplification of virulence genes
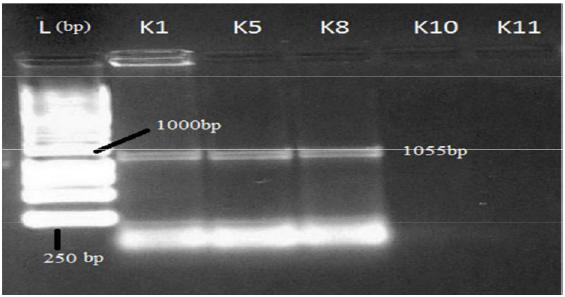
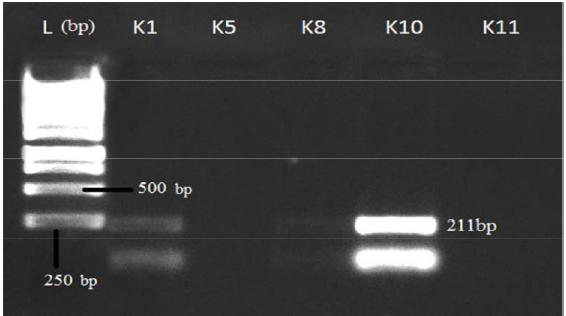
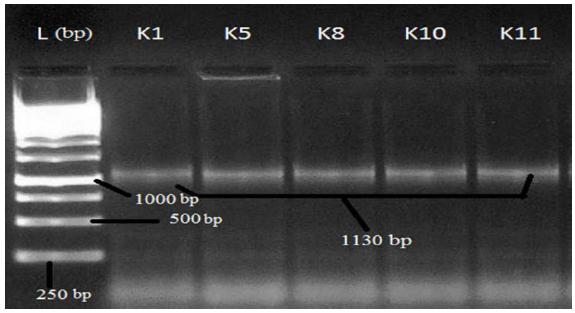
After PCR run, products were electrophoresed in 1.5% agarose gel in TBE buffer for visualization of amplified gene(s) in tested isolates. Gel stained with ethidium bromide solution (10 mg/mL) for 40 min and visualized under gel documentation system. Among five tested isolates, nulc and lipA amplified in K1, K5 and K8 whereas serP found in K1 and K10 and gave expected product length of 517 bp, 1055 bp and 211 bp respectively. Gyrase subunit B2 (gyrB2) amplified in all five isolates with PCR product size of 1130 bp in agarose gel (Figures 1- 4).
Antibiotic sensitivity of K. pneumoniae isolates
To know the antibiogram profile of the isolates, we screened 12 antibiotics representing different antibiotic groups. All of the isolates exhibited 80-100% resistant to ampicillin, erythromycin, and chloramphenicol and also for cephradine, kanamycin and sulphamethaxazole. Sensitivity was observed for only four antibiotics viz., streptomycin, gentamycin, ciprofloxacin and levofloxacin except for isolates K1 and K5 which showed sensitivity to only one and resistant to three other antibiotics. Sensitivity results for other antibiotics varied but neither one showed 100% sensitivity alone to all tested isolates. All of the isolates exhibits resistance to multiple antibiotics tested (Table 2).
Profiling of plasmid DNA
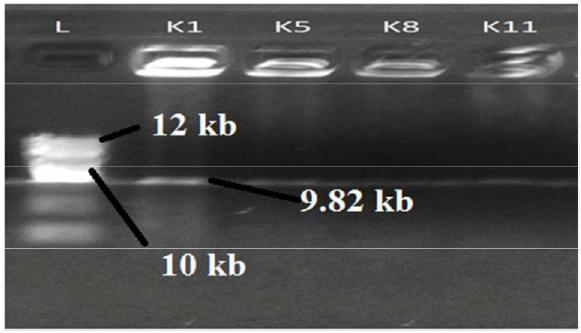
Plasmid isolation and analysis was done for isolates K1, K5, K10 and K11, those showed positive result in virulent gene amplification for nulc, lipA and serP. Plasmid DNA was amplified in two isolates (K1 and K5) and showed 9.82 kb size on 1.2% agarose gel under gel documentation system while others showed negative result for plasmid DNA (Figure 5).
In -vitro assay for virulence factors
In-vitro plate assay showed divergence pattern of activity on culture plate. Better In -vitro proteolysis and lipolysis activity was observed for isolates K1, K5 and K10 isolates while other isolates hardly hydrolyze casein and tween20 on culture plate. No significant the halo of hemolysis was noticed for four isolates of K. pneumoniae except for K1, which, slightly hydrolyze blood agar only around the center (Table 3).
DISCUSSION
K. pneumoniae reported to cause wide range of infections in human and other organisms and the pathogenicity depends on a variable number of factors directed by an array of genes and virulence factors [2727 Clegg S, Murphy CN. Epidemiology and Virulence of Klebsiella pneumoniae. Microbiology spectrum. 2016 Feb 5; 4(1).,2828 Dashti AA, Jadaon MM, Gomaa HH, Noronha B, Udo EE. Transmission of a Klebsiella pneumoniae clone harbouring genes for CTX-M-15-like and SHV-112 enzymes in a neonatal intensive care unit of a Kuwaiti hospital. Journal of medical microbiology. 2010 Jun 1; 59(6):687-92.,2929 Shenoy S, Hegde A, Dominic SR, Kamath S, Arvind N. An outbreak of extended spectrum beta-lactamase producing Klebsiella pneumoniae in a neonatal intensive care unit. Indian journal of pathology & microbiology. 2007 Jul; 50(3):669-70.]. Among those, urinary tract infection (UTI) is one of the most common diseases that cause potential threat to human health; especially for pregnant women and aged persons [1212 Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003 Nov 1; 84(4):269-78.,3030 Cortés G, Borrell N, de Astorza B, Gómez C, Sauleda J, Albertí S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infection and immunity. 2002 May 1; 70(5):2583-90.]. Present study also reveals that, women’s during pregnancy and people after age of 50 generally more vulnerable to UTI causing K. pneumoniae. Six out of ten isolates were collected from adult patients of age range between 35 and 65, in which three were pregnant women. Nuclease (nulc) and lipase (lipA) gene amplified in three isolates both of which encode plasmid of same molecular weight. In a most recent study, nuclease like protein, colibacin toxin, a putative cytotoxin in the outer membrane identified as one of the virulence factors of K. pneumonia [3131 Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, Tournebize R. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC biology. 2014 May 29; 12(1):1.]. In addition, a protein from phospholipase D family (PLD1) was also reported as a virulent gene in in-vivo K. pneumoniae mouse model after expression of its mRNA [3131 Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, Tournebize R. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC biology. 2014 May 29; 12(1):1.]. Increasing serum lipase activity also responsible for inflammation and other disorders by altering the structure of cytoplasmic membrane of host and thus exacerbates its pathogenicity [1111 Rosenblum JL. Serum lipase activity is increased in disease states other than acute pancreatitis: amylase revisited. Clinical chemistry. 1991 Mar 1; 37(3):315-6.,3232 Nawaz M, Khan SA, Khan AA, Sung K, Tran Q, Kerdahi K, Steele R. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food microbiology. 2010 May 31; 27(3):327-31.]. Serine protease, a protein that triggers proteolysis by binding and inhibiting the enzymatic activity of elastase and cathepsin G in in-vitro, has been reported as a virulence factors for K. pneumoniae in recent times [3333 Zhao Y, Olonisakin TF, Xiong Z, Hulver M, Sayeed S, Yu MT, Gregory AD, Kochman EJ, Chen BB, Mallampalli RK, Sun M. Thrombospondin-1 restrains neutrophil granule serine protease function and regulates the innate immune response during Klebsiella pneumoniae infection. Mucosal immunology. 2015 Jul 1; 8(4):896-905.]. DNA gyrase subunit B on the other hand, encode sequences essential for DNA replication, exhibits beta-hemolytic activity in mouse model and contribute to the progression of enterobacteriaceae associated disease in human [1010 Han XY, Han FS, Segal J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. International journal of systematic and evolutionary microbiology. 2008 Jun 1; 58(6):1398-403.,3434 Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical microbiology reviews. 1998 Oct 1; 11(4):589-603.]. In our present study, two isolates (K1 and K2) of K.pneumoniae had plasmid molecular weight of 982 kb. This plasmid likely to bear high antibiotic resistance encoding beta-lactamse enzyme confer resistance to ampicilin, kanamycin, chloramphenicol [3535 Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, MacDonald A, Brudney D, Sompolinsky D, Navon-Venezia S, Livermore DM. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. Journal of antimicrobial chemotherapy. 2008 Dec 1; 62(6):1261-4.,3636 Ghafourian S, Sekawi Z, Neela V, Khosravi A, Rahbar M, Sadeghifard N. Incidence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in patients with urinary tract infection. Sao Paulo Medical Journal. 2012; 130(1):37-43.]. Our present study found majority of isolates (80%) resistant to antibiotic ampicilin, 70% resistant to chloramphenicol and kanamycin. Data results expose a correlation in between 9.8 kb plasmid and antibiotic resistant pattern which suggest involvement of this plasmid in pathogenicity of K. pneumoniae. We conducted in-vitro plate assay of respective gene viz. nuclease, lipase, protease and gyrase to analyze their activity (proteolysis, lipolysis and hemolysis) extracellularly. This assay proved to be very useful in determining virulence associated factors, analyzing virulence level and predicting the correlation of these two with the amount of enzymes and toxins produced by the pathogens. Our present study showed a unique correlations: isolate K1 bearing nucl, lipA and serP, exhibited strong activity to hydrolyze casein, tween20 and moderate activity to blood agar whereas, K5 which had nucl, lipA, hydrolyze tween20 effectively but not casein and sheep blood. Finally, isolate K10 amplified with serP gene was powerful hydrolyser of casein in extracellular environment but barely utilize tween20 and blood agar. Therefore, a strong correlation was revealed among putative virulence factors and bacterial plasmid in our study that contribute to the pathogenicity of K. pneumoniae associated urinary tract infection (UTI) in Bangladesh.
CONCLUSION
K. pneumoniae pathogenicity depends on three factors: number and type of virulent genes, size of the plasmid and result of in -vitro hydrolysis assay for virulence factors. In this study, we found a strong correlation among these factors in the pathogenicity of K. pneumoniae associated urinary tract infection (UTI) in Bangladesh. Further studies i.e. sequencing of genes, analysis of the sequence for any mutation are currently going on in our research laboratory. Investigation need to develop animal model for further analysis of in-vivo relationship of these three factors.
ACKNOWLEDGEMENT
The present study was conducted under a research project titled “Virulent gene targeting and analysis of virulence factors of K. pneumoniae causing pneumonia and urinary tract infection (UTI) in Bangladesh” funded by Ministry of Science and Technology, Bangladesh (MOST).
REFERENCES
-
1Farajnia S, Alikhani MY, Ghotaslou R, Naghili B, Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. International Journal of Infectious Diseases. 2009 Mar 31; 13(2):140-4.
-
2Foysal MJ, Rahman MM, Prodhan MS. PCR Based Molecular Detection of the Gyr-B-2 Gene from the Klebsiella Sp. Isolates from Patients who were Suffering with Pneumonia and Urinary Tract Infections (UTIs). Journal of clinical and diagnostic research: JCDR. 2013 Jan;7(1):23-5
-
3Kucheria R, Dasgupta P, Sacks S, Khan M, Sheerin N. Urinary tract infections: new insights into a common problem. Postgraduate medical journal. 2005 Feb; 81(952):83-6
-
4Kamrul L, Eliza R, Habibura R, Ranjith R. Urinary Tract Infection in Children: An Update Bangaladesh. J Child Health. 2012; 36(2):90-7.
-
5Farshad S, Ranijbar R, Japoni A, Hosseini M, Anvarinejad M, Mohammadzadegan R. Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Archives of Iranian Medicine (AIM). 2012 May 1; 15(5):312-6.
-
6Bugarel M, Granier SA, Weill FX, Fach P, Brisabois A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC microbiology. 2011 Jun 27; 11(1):1-51.
-
7Murugkar HV, Rahman H, Dutta PK. Distribution of virulence genes in Salmonella serovars isolated from man & animals. Indian Journal of Medical Research. 2003 Feb 1; 117:66-70
-
8Breuer T, Benkel DH, Shapiro RL, Hall WN, Winnett MM, Linn MJ, Neimann J, Barrett TJ, Dietrich S, Downes FP, Toney DM. A multistate outbreak of Escherichia coli O157: H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerging infectious diseases. 2001 Nov; 7(6):977-82.
-
9Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proceedings of the National Academy of Sciences. 1976 Nov 1; 73(11):3872-6.
-
10Han XY, Han FS, Segal J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. International journal of systematic and evolutionary microbiology. 2008 Jun 1; 58(6):1398-403.
-
11Rosenblum JL. Serum lipase activity is increased in disease states other than acute pancreatitis: amylase revisited. Clinical chemistry. 1991 Mar 1; 37(3):315-6.
-
12Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003 Nov 1; 84(4):269-78.
-
13Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cellular and molecular life sciences. 2014 Mar 1; 71(5):745-70.
-
14Smalla K, Jechalke S, Top EM. Plasmid detection, characterization and ecology. Microbiology spectrum. 2015 Feb; 3(1):1-21.
-
15Victor LY. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerging Infect Dis. 2007; 13(7): 123-128.
-
16Koga VL, Tomazetto G, Cyoia PS, Neves MS, Vidotto MC, Nakazato G, Kobayashi RK. Molecular screening of virulence genes in extraintestinal pathogenic Escherichia coli isolated from human blood culture in Brazil. BioMed research international. 2014 Apr 15; 2014:id465054.
-
17Akingbade OA, Ogiogwa IJ, Okonko IO, Okerentugba PO, Innocent-Adiele HC, Nwanze JC, Onoh CC. Plasmid Profile of Isolated Klebsiella Species in a Tertiary Hospital in Abeokuta, Ogun State, Nigeria. World Applied Science Journal. 2012. Mar; 3:371-8
-
18Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences. 2015 Jul 7; 112(27):E3574-81.
-
19Foysal MJ, Rahman MM, Rabbee MF, Nazmul M, Hossain RM, Rahman MJ, Miah MF, Islam K. Identification and assay of putative virulence properties of Eschericha coli gyrase subunit A and B among hospitalized UTI patients in Bangladesh. Inov Pharm Pharmacother. 2013; 1(1):54-9.
-
20Pinto JB, Rossatto FC, Martins PD, Frazzon AP. Genetic relationships and virulence factors in Staphylococcus aureus isolated from raw poultry in South Brazil. Annals of Microbiology. 2015 Dec 1; 65(4):1933-40.
-
21Breed, RS, Murray, EGD, Smith, NB. Burgey's manual of determinative bacteriology. 7th Edn. Baltimore: Waverly Press Inc; 1957.
-
22Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 4th Edn. New York: Cold Spring Harbor Laboratory Press; 2012.
-
23Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966 Apr; 45(4):493-6.
-
24Rodarte MP, Dias DR, Vilela DM, Schwan RF. Proteolytic activities of bacteria, yeasts and filamentous fungi isolated from coffee fruit (Coffea arabica L.). ActaScientiarum. Agronomy. 2011 Sep; 33(3):457-64.
-
25Michelim L, Lahude M, Araújo PR, Giovanaz DS, Müller G, Delamare AP, Costa SO, Echeverrigaray S. Pathogenic factors and antimicrobial resistance of Staphylococcus epidermidis associated with nosocomial infections occurring in intensive care units. Brazilian Journal of Microbiology. 2005 Mar; 36(1):17-23.
-
26Bruce SK, Schick DG, Tanaka L, Jimenez EM, Montgomerie JZ. Selective medium for isolation of Klebsiella pneumonia. J of ClinMicrobiol. 1981; 13(6): 1114-6.
-
27Clegg S, Murphy CN. Epidemiology and Virulence of Klebsiella pneumoniae. Microbiology spectrum. 2016 Feb 5; 4(1).
-
28Dashti AA, Jadaon MM, Gomaa HH, Noronha B, Udo EE. Transmission of a Klebsiella pneumoniae clone harbouring genes for CTX-M-15-like and SHV-112 enzymes in a neonatal intensive care unit of a Kuwaiti hospital. Journal of medical microbiology. 2010 Jun 1; 59(6):687-92.
-
29Shenoy S, Hegde A, Dominic SR, Kamath S, Arvind N. An outbreak of extended spectrum beta-lactamase producing Klebsiella pneumoniae in a neonatal intensive care unit. Indian journal of pathology & microbiology. 2007 Jul; 50(3):669-70.
-
30Cortés G, Borrell N, de Astorza B, Gómez C, Sauleda J, Albertí S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infection and immunity. 2002 May 1; 70(5):2583-90.
-
31Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, Tournebize R. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC biology. 2014 May 29; 12(1):1.
-
32Nawaz M, Khan SA, Khan AA, Sung K, Tran Q, Kerdahi K, Steele R. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food microbiology. 2010 May 31; 27(3):327-31.
-
33Zhao Y, Olonisakin TF, Xiong Z, Hulver M, Sayeed S, Yu MT, Gregory AD, Kochman EJ, Chen BB, Mallampalli RK, Sun M. Thrombospondin-1 restrains neutrophil granule serine protease function and regulates the innate immune response during Klebsiella pneumoniae infection. Mucosal immunology. 2015 Jul 1; 8(4):896-905.
-
34Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical microbiology reviews. 1998 Oct 1; 11(4):589-603.
-
35Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, MacDonald A, Brudney D, Sompolinsky D, Navon-Venezia S, Livermore DM. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. Journal of antimicrobial chemotherapy. 2008 Dec 1; 62(6):1261-4.
-
36Ghafourian S, Sekawi Z, Neela V, Khosravi A, Rahbar M, Sadeghifard N. Incidence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in patients with urinary tract infection. Sao Paulo Medical Journal. 2012; 130(1):37-43.
Publication Dates
-
Publication in this collection
2018
History
-
Received
10 Aug 2016 -
Accepted
20 June 2017