Abstract
Rapid diagnosis of rabies in suspected human cases influences post-exposure prophylaxis for potential contacts of the patient and ensures appropriate patient management. Apart from the central nervous system (CNS), rabies virus (RABV) is usually present in small sensory nerves adjacent to hair follicles of infected humans. We used an RT-PCR, with primers targeted to the 3' terminal portion of the nucleoprotein gene (N), to test neck-skin samples of nine patients who had rabies in order to validate a diagnostic method that could serve as an additional tool for rabies diagnosis, particularly in antemortem samples. Six of eight postmortem samples were found to be positive for rabies by RT-PCR, and one of two samples collected antemortem was positive with this same technique. Results were confirmed by DNA sequencing; this validates RT-PCR and neck-skin as a suitable technique and type of sample, respectively, for use in the diagnosis of human rabies. RT-PCR applied to neck-skin biopsies could allow early diagnosis and lead to more effective rabies treatment.
Human rabies; neck-skin; diagnosis; RT-PCR; DNA sequencing
ORIGINAL PAPERS
Diagnosis of human rabies cases by polymerase chain reaction of neck-skin samples
Carla Isabel MacedoI; Pedro Carnieli JrI; Paulo Eduardo BrandãoI; Elizabeth S. Travassos da RosaII; Rafael de Novaes OliveiraI; Juliana Galera CastilhoI; Rita MedeirosIII; Rosangela Rocha MachadoIV; Rosely Cerqueira de OliveiraIV; Maria Luiza CarrieriI; Ivanete KotaitI
IInstituto Pasteur, SP, Brazil
IIInstituto Evandro Chagas, PA, Brazil
IIIHospital Universitário João de Barros Barreto, PA, Brazil
IVSVS/ Ministério da Saúde, Brasília, DF, Brazil
Address for correspondence Address for correspondence: Dr. Pedro Carnieli Junior Av. Paulista, 393 Zip code: 01311-001 São Paulo, SP, Brazil Tel. 55.11.3288-0088 E-mail: pecarniel@yahoo.com.br.
ABSTRACT
Rapid diagnosis of rabies in suspected human cases influences post-exposure prophylaxis for potential contacts of the patient and ensures appropriate patient management. Apart from the central nervous system (CNS), rabies virus (RABV) is usually present in small sensory nerves adjacent to hair follicles of infected humans. We used an RT-PCR, with primers targeted to the 3' terminal portion of the nucleoprotein gene (N), to test neck-skin samples of nine patients who had rabies in order to validate a diagnostic method that could serve as an additional tool for rabies diagnosis, particularly in antemortem samples. Six of eight postmortem samples were found to be positive for rabies by RT-PCR, and one of two samples collected antemortem was positive with this same technique. Results were confirmed by DNA sequencing; this validates RT-PCR and neck-skin as a suitable technique and type of sample, respectively, for use in the diagnosis of human rabies. RT-PCR applied to neck-skin biopsies could allow early diagnosis and lead to more effective rabies treatment.
Key Words: Human rabies, neck-skin, diagnosis, RT-PCR, DNA sequencing.
Rabies is an acute, progressive and fatal disease caused by an RNA virus of the Lyssavirus genus of the Rhabdoviridae family [1]; it is the 11th cause of human death by infectious disease [2]. Human mortality due to rabies is estimated to be 55,000 deaths per year worldwide; most rabies cases have been reported in Asia and Africa [3]. In the states of Pará and Maranhão in Brazil, outbreaks of rabies transmitted by vampire bats caused 21 human deaths in 2004 and 42 human deaths in 2005 [4].
The Brazilian budget for rabies prevention in 2004 was US$ 28 million. This figure included the cost of vaccines for humans and dogs, immunoglobulin, laboratory diagnosis, medical and veterinary staff, training of staff and dog vaccination campaigns [3].
Diagnosis of rabies based on clinical symptoms alone is difficult and unreliable, except when there are specific clinical signs of hydrophobia or aerophobia [3]. A diagnosis of rabies should be considered in any patient who presents encephalitis of unknown cause [5]. The first clinical signs of rabies are nonspecific, and the diagnosis is often confirmed later in the course of the disease or at postmortem. Delay in diagnosis can result in dissemination of the disease to contacts and unnecessary postexposure treatment of people at risk [6-8]. Early diagnosis can eliminate the expense of unnecessary diagnostic tests and inappropriate medical treatment [9]. It also significantly reduces the number of potential exposures to the virus during contact with the patient and allows early identification of people who are candidates for prophylactic treatment [6,10].
Furthermore, the great advances in antiviral production and intensive care treatment in hospitals make treatment of many viral diseases possible. Indeed, the first successful treatment of human rabies was recently reported in the United States, indicating an optimistic outlook for the coming years [11], highlighting the importance of early diagnosis if rabies treatment is to be effective.
Laboratory specimens used for antemortem diagnosis include serum, cerebrospinal fluid, saliva, corneal impressions and biopsy of highly innervated regions such as neck-skin [6,7,12]. The rabies virus (RABV) is usually present in nerve cells surrounding the bases of hair follicles. Neck-skin biopsies are analyzed by a fluorescent antibody test (FAT) in frozen sections of the biopsy and by reverse transcriptionpolymerase chain reaction (RT-PCR) on RNA extracted from the biopsy [7].
We examined a molecular method for diagnosing human rabies based on RT-PCR of neck-skin biopsies. This method could become an important tool for rabies diagnosis, particularly in antemortem samples.
Material and Methods
Samples
Ten approximately 1 cm2 samples of neck-skin biopsy collected in 2005 from nine patients from Augusto Correa, Pará, suspected of having rabies were analyzed. Two samples, registered as 4793a and 4795, were collected before the patient's death, and samples registered as 5446, 5447, 5448, 5449, 5450, 5451, 5452 and 5453 were collected postmortem. Samples 4795 and 5446 were collected antemortem and postmortem, respectively, from the same patient.
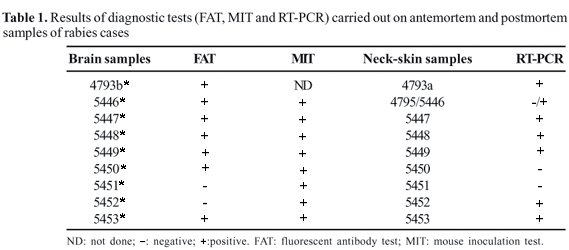
Brain samples were collected postmortem from the same nine patients from Augusto Correa suspected of having rabies and were registered as 4793b*, 5446*, 5447*, 5448*, 5449*, 5450*, 5451*, 5452* and 5453*.
Fluorescent antibody test (FAT) and mouse inoculation test (MIT)
All the brain samples were diagnosed as positive for rabies by FAT [13] and MIT [14] in the Instituto Evandro Chagas, Pará, Brazil.
RT-PCR and DNA sequencing
Neck-skin samples were cut into small pieces with a scalp and submitted to total RNA extraction with Trizol® (Invitrogen), according to the manufacturer's instructions. A reference strain (Challenge Virus Standard-CVS) was used as a positive control; RNA extracted from uninfected mouse brain tissue and ultra-pure water were used as negative controls. Reverse Transcription (RT) and Polymerase Chain Reaction (PCR) were performed using sense primer 504 (5'-TATACTCGAATCATGATGAATGGAGGTCGACT-3') and antisense primer 304 (5'-TTGACGAAGATCTTGCTCAT-3') targeted to the region corresponding to the nucleoprotein (N) gene, as previously described [15]. An aliquot of 5 mL of extracted RNA was included in the RT mixture containing 1X first-strand buffer, 40U RNAseOUT, 1.3 mM dNTP, 1 pmol/µL of primers 504 and 304, 8.5 mM DTT, 200 U M-MLV reverse transcriptase (InvitrogenTM), and RNase-free water to a final volume of 47µL. For PCR amplification, 10µL of cDNA generated in the RT reaction was added to a PCR mixture containing 1X PCR buffer, 1pmol/µL of primers 504 and 304, 0.2 mM dNTP, 2.4 mM MgCl2, 2.5 U Taq DNA recombinant polymerase (InvitrogenTM), and RNase-free water to a final volume of 102µL. Reactions were performed in an EppendorfTM thermocycler with a cycle of 60 min at 42ºC for RT and a cycle of 1 min at 94ºC followed by 40 cycles of 30 seconds at 94ºC, 30 seconds at 37ºC and 90 seconds at 72ºC, and a final cycle of 7 min at 72ºC for the PCR reactions. The RT-PCR product was visualized under UV light after gel electrophoresis on 1% agarose gel containing ethidium bromide in TBE buffer.
A second-round amplification was carried out, as previously described, with 5µL of the first-round amplification. Amplified DNA fragments were purified with GFX PCR DNA and a Gel Band Purification Kit (Amersham BiosciencesTM) and submitted to DNA sequencing reactions using sense and antisense primers (504 and 304, respectively) with DYEnamic ET Dye Terminator (Amersham BiosciencesTM) according to the manufacturer's instructions. The sequences were resolved on a MegaBACE DNA (Amersham BiosciencesTM).
Phylogenetic analysis
A 249 bp region of the nucleoprotein gene located between nucleotides 1287 and 1534 of the Pasteur Virus (PV) strain (GeneBank accession number M13215.1) was analyzed. First, data from raw sequencing were edited using CHROMAS software (Version 2.24 Copyright© 1998-2004 Technelysium Pty Ltd). The final sequence of each virus strain was aligned with homologous sequences retrieved from Genbank using the CLUSTAL W method and Bioedit software [16]. The alignments were then used to build a neighbor-joining DNA-distance phylogenetic tree, with the Kimura 2-parameter evolutionary model and 1000 bootstrap repetitions for statistical support using Mega 2.1 [17].
Results
FAT and MIT
Seven brain samples were positive by FAT and eight positive by MIT (Table 1).
RT-PCR
Seven samples of neck skin (one antemortem biopsy and six postmortem samples) were positive by RT-PCR (Table 1).
Phylogenetic analysis
For each of the seven neck-skin samples positive by RT-PCR, 249 genome nucleotides located between nucleotides 1287 and 1534 were sequenced using standard strain PV as a reference. One hundred and fifty-six nucleotides from the 3'-termini of the N gene in the region encompassed by nucleotides 1319 and 1474 were analyzed using standard strain PV as a reference.
The topology of the phylogenetic tree (Figure 1) was divided into three clusters: I-Brazilian domestic and wild carnivore-related strains, II- Fixed SAD and CVS strains, and III- Brazilian vampire-bat-related strains. All samples analyzed in this study segregated in cluster III. Sequences from samples 4793, 5446, 5447, 5448, 5449, 5452 and 5453 received the following GenBank accession numbers, respectively: DQ314575, DQ314576, DQ314577, DQ314578, DQ314579, DQ314580 and DQ314574.
Discussion
Suspected cases of human rabies require precise and rapid diagnosis. A reliable diagnosis is important to ensure appropriate patient management and to provide relevant information for public health authorities involved in the control and epidemiological surveillance of rabies, in particular during investigation of fatal encephalitis of unknown etiology.
Most conventional techniques used for postmortem diagnosis of rabies are not suitable for antemortem diagnosis due to the risks of performing a brain biopsy on a living patient [7,12]. The sensitivity of techniques used for antemortem diagnosis of rabies varies greatly, according to the stage of the disease, antibody status, intermittent nature of viral shedding and training of the technical staff [3].
Rabies antigens (nucleocapsid or glycoprotein) may be detected antemortem by using FAT on skin biopsies, which are usually taken from the nuchal area and should include hair follicles, as these contain peripheral nerves [18]. The quality of skin biopsy samples is of paramount importance and is dependent on the ability of the surgeon. This technique may not be practical, as it requires a cryostat in order to prepare frozen tissue sections; in addition, contamination can occur in the cryostat, as various biopsies are processed in the same equipment. Antemortem FAT on corneal impressions is rarely reliable [3], although a small number of laboratories have reported successful use of this technique on corneal impressions [18]. RABV isolation using biological samples obtained by an antemortem procedure can be performed using cell culture or intracranial inoculation of mice (MIT). The biological samples may be saliva or other biological fluids, such as tears or cerebrospinal fluid (the latter is not generally recommended) [3]. There is an important limitation for the detection of rabies antigen by FAT, as there may not be enough cells or viral antigens in the early stages of the disease to yield a positive result. As the disease progresses, viral antigens may also be absent from saliva or biopsies, probably due to neutralizing antibodies [12].
Neutralizing antibodies in the serum and cerebrospinal fluid of nonvaccinated patients can be measured as an antemortem assay with a virus neutralization test [3]. Virus-neutralizing antibodies, however, tend to appear on average eight days after the onset of clinical symptoms, making techniques based on detection of these antibodies less effective for early diagnosis than other antemortem techniques.
Molecular diagnosis has been adopted to detect viral nucleic acid in antemortem or postmortem specimens. RT-PCR has been used successfully for detecting the viral nucleic acid of RABV in antemortem specimens. Molecular detection by RT-PCR technique has the highest sensitivity, but it requires standardization and very stringent quality control in order to avoid false results [9]. The RNA of RABV can be detected in several biological fluids and samples (e.g. saliva, cerebrospinal fluid, tears, urine, skin biopsy and brain samples). Saliva and neck-skin biopsy samples are the most widely-studied specimens for attempting antemortem diagnosis [9,10,12].
We chose the nucleoprotein gene (N) because it is the most conserved RABV gene [19], and analysis of this gene is the most suitable approach for the molecular diagnosis of rabies [20].
We developed and applied an RT-PCR protocol to detect a 249-nucleotide fragment of the N gene. Using this method, we were able to detect viral nucleic acid in 70% of the neck-skin samples, including one of the two samples obtained during antemortem examination. To avoid false positive results, precautions for carrying out RT-PCR [21] were followed strictly, and the sequencing analysis confirmed the RT-PCR results. The addition of the second amplification with the same primers for the PCR allowed us to increase the concentration of the amplified DNA to guarantee the success of the DNA sequencing; this constitutes a new approach for the diagnosis of rabies.
The results for RT-PCR in neck-skin samples obtained postmortem were confirmed by DNA sequencing and phylogenetic analysis in parallel with FAT and MIT of brain specimens from the same patients. The negative results for RT-PCR in neck-skin samples 4795, 5450 and 5451 were probably due to the poor quality of the sample or degradation of viral RNA, as the same samples were positive by MIT. It cannot be argued that sample 5441*, which was negative by FAT, was also negative by RT-PCR due to the small amount of antigen, as the sample was positive by MIT. This is supported by the fact that, in one patient, the brain sample was negative by FAT (5452*) and a neck-skin sample (5452) was positive by RT-PCR.
Though we intended to examine more antemortem samples, the conditions during sample collection at the sites where the deaths occurred, together with the emotional stress that surrounded the cases and the lack of technical conditions, made it difficult to collect antemortem samples. Postmortem collection of neck-skin samples, however, presented fewer difficulties and was of major importance for our study.
In the phylogenetic analysis, the results obtained with the neck-skin samples agreed with the epidemiological field survey, the local ecology where the cases occurred and the antigenic typing carried out at the Evandro Chagas Institute [22]. These results, together with those obtained by antigenic typing and phylogenetic analysis carried out by the Pasteur Institute during the 2004 outbreaks in the municipalities of Viseu and Portel [23], also in the state of Pará, are sufficient to indicate that the vampire bat Desmodus rotundus is the species responsible for RABV transmission in this region.
Also, the experience of the Pasteur Institute with molecular diagnosis, using saliva, cerebrospinal fluid, serum and neck-skin biopsy samples has shown that the latter is the most suitable for use in antemortem diagnosis of rabies, probably due to the late, or less intense, virus neutralization in these samples. All the RT-PCR results for the human samples are registered at the Pasteur Institute.
We conclude that the probability of successful antemortem detection of RABV using neck-skin samples is high and that use of this technique should be encouraged. Given the magnitude of genomic amplification by RT-PCR in postmortem samples, we conclude that the results also reflect the antemortem situation.
This is the first time that RABV has been identified in samples of neck-skin biopsy in Brazil. The successful use of RT-PCR demonstrates that it is a viable, alternative tool for the antemortem diagnosis of rabies, which allows timely use of antivirals and improved patient management.
Acknowledgements
The authors thank all those who were directly or indirectly involved in collecting the samples used in this study and without whom little progress could be made in rabies research, control and treatment.
Received on 22 April 2006; revised 7 August 2006.
- 1. Van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L. et al. Virus Taxonomy: the classification and nomenclature of viruses. The Seventh Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, 2000
-
2World Health Organization - World Survey of Rabies No. 34 for the Year 1998. World Health Organization, Geneva, 2000
-
3WHO Technical Report Series 931, WHO Expert Consultation On Rabies, First Report, 2005
- 5. Instituto Pasteur Secretaria de Estado da Saúde de São Paulo, Informes Técnicos Institucionais. Relato de caso de raiva humana após exumação no Município de São Paulo. Rev Saúde Pública 2004;38:741-2.
- 6. Rupprecht C.E., Cathleen A.H., Hemachudha T. Rabies re-examined. Lancet 2002;2:327-43.
- 7. Trimarchi C.V., Smith J.S. Diagnostic evaluation. In: Jackson A.C., Wunner W.H. eds. Rabies. Academic Press, San Diego, 2002
- 8. Wacharapluesadee S., Hemachudha T. Nucleic-acid sequence based amplification in the rapid diagnosis of rabies. Lancet 2001;358:892-3.
- 9. Crepin P., Audry L., Rotivel Y., et al. Intravitam diagnosis of human rabies by PCR using saliva and cerebrospinal fluid. J. Clin. Microb. 1998;36(4):1117-21.
- 10. David D., Rupprecht C.E., Smith J.S., et al. Antemortem detection and virus characterization of three human rabies fatalities in Israel between 1996-1997. Israel Vet. Med. Assoc. 2005;54(3), 1999 [Online.] http://www.isrvma.org/article/54_3_3.htm Accessed March 14, 2006.
- 11. Willloughby R.E. Jr., Tieves K.S., Hoffman G.M., et al. Survival after treatment of rabies with induction of coma. N Engl J Med 2005;352:2508-14.
- 12. Smith J., Mcelhinney L., Parson G., et al. Case report: rapid ante-mortem diagnosis of a human case of rabies imported into UK from Philippines. J Med Virol 2003;69:150-3.
- 13. Dean D.J., Abelseth M K., Atanasiu P. The fluorescent antibody test. In: Meslin F.X., Kaplan M.M., Koprowski H. eds. Laboratory techniques in rabies, fourth ed. World Health Organization, Geneva, 1996
- 14. Koprowski H. The mouse inoculation test. In: Meslin F.X., Kaplan M.M., Koprowski H. eds. Laboratory techniques in rabies, fourth ed. World Health Organization, Geneva, 1996
- 15. Orciari L.A., Niezgoda M., Hanlon C.A., et al. Rapid clearance of SAG-2 rabies virus from dogs after oral vaccination. Vaccine 2001;19:4511-8.
- 16. Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95-8.
- 17. Kumar S., Tamura K., Jakobsen I.E., et al. MEGA 2: Molecular evolutionary genetic analysis software. Evolutionary Genetics Analysis Software, Arizona State University: Tempe, 2001
- 18. Zaidman G.W., Billingsley A. Corneal impression test for the diagnosis of acute rabies encephalitis. Ophthalmology 1998;105:249-51.
- 19. Wunner W.H. Rabies Virus. In: Jackson A.C., Wunner W.H. eds. Rabies. Academic Press, San Diego, 2002
- 20. Smith J.S. Molecular Epidemiology. In: Jackson A.C., Wunner W.H. eds. Rabies. Academic Press, San Diego, 2002
- 21. Kwok S., Higuchi R. Avoiding false positives with PCR. Nature 1989;339:237-8.
- 22. Travassos da Rosa E.S., Barbosa T.F.S., Pereira A.S., et al. Human rabies outbreak transmitted by hematophagous bats in Augusto Corrêa municipality in Pará State: laboratory findings (p. 186). Abstracts Virus Reviews & Research: XVI National Meeting of Virology (Salvador), Bahia, 2004
- 23. Travassos da Rosa E.S., Brandão P.E., Carrieri M.L., et al. Laboratorial confirmation of human rabies outbreak transmitted by hematophagous bats (Desmodus rotundus) in Pará State (p. 49-50). Abstracts book: XV International Conference Rabies in the Americas (Santo Domingo), Republica Dominicana, 2004
Publication Dates
-
Publication in this collection
31 Jan 2007 -
Date of issue
Oct 2006
History
-
Reviewed
07 Aug 2006 -
Received
22 Apr 2006