Abstracts
Fusarium oxysporum is a common soilborn plant pathogen with a worldwide distribution. Fusarium yellows disease of chickpea (Cicer arientinum) caused by F.oxysporum is one of the most destructive soilborn disease which is a major production constraint in chickpea-growing regions of Iran. Three laboratory methods "amplification of genomic DNA using random primers, enzyme activity staining, and colony size determination" have been used to discriminate between highly virulent (HV) and weakly virulent (WV) isolates of F. oxysporum. On the basis of colony size (a traditional morphological method) and the ability of isolates to produce pectic enzymes, five HV isolates were differentiated from three WV isolates. The HV isolates formed large colony (ranging from 10.1 to 12.6 mm in diameter) and showed the same enzyme pattern,while the WV isolates produced small colony (ranging from 5.8 to 7.8 mm in diameter) and had not detectable enzyme activity in the stained overlaying gel. Twelve arbitrary 10-mer primers were tested on these 8 isolates of F. oxysporum by Polymerase Chain Reaction (PCR). Cluster analysis of the data from the DNA amplification by Random Amplified Polymorphic DNA (RAPD), differentiated HV from WV isolates. The results obtained from RAPD test confirmed the classification of these eight isolates based on pathogenicity test, colony size, and enzyme activity staining into two groups (HV and WV).
Fusarium oxysporum; RAPD; colony size; enzyme activity staining
Fusarium oxysporum é um fitopatógeno amplamente difundido nos solos. A doença amarela do grão-de-bico (Cicer arietinum), causada por ele é destrutiva e um grande problema nas regiões produtoras do Irã. Três métodos de laboratório: amplificação aleatória de DNA genômico (RAPD), coloração de atividade enzimática e determinação das dimensões de colônia, foram usados para diferenciar isolados de Fusarium oxysporum altamente virulentos (HV) e fracamente virulentos (WV). Baseado no tamanho da colônia (método morfológico tradicional) e na capacidade dos isolados produzirem enzimas pécticas, cinco isolados HV foram diferenciados de três WV. Enquanto os isolados HV formaram colônias grandes (entre 10,1 e 12,6 mm de diâmetro) e apresentaram o mesmo padrão enzimático, os isolados WV produziram colônias pequenas (entre 5,8 e 7,8 mm de diâmetro) e não tiveram atividade enzimática detectável no gel de cobertura corado. Doze primers arbitrários foram testados nos 8 isolados de Fusarium oxysporum por reação de polimerase em cadeia (PCR). As análises dos dados da amplificação de DNA pelo método de amplificação aleatória do DNA polimórfico (RAPD) diferenciaram os isolados HV dos WV. Os resultados obtidos pelo teste RAPD confirmaram a classificação dos 8 isolados pelos testes de patogenicidade, tamanho da colônia e atividade enzimática de coloração nos dois grupos, HV e WV.
Fusarium oxysporum; RAPD; tamanho de colônia; coloração de atividade enzimática
Use of RAPD, enzyme activity staining, and colony size to differentiate phytopathogenic Fuzarium Oxysporum isolates from Iran
Uso de RAPD, coloração de atividade enzimática e tamanho de colônia para diferenciação de isolados fitopatogênicos de Fusarium oxysporum obtidos no Irã
Motallebi Mostafa; Zamani Mohammad Reza; Jazayeri Omid; Harighi Mohammad Javad
Department of Biology, Faculty of Science, Razi University, Kermanshah, Iran
Correspondence to Correspondence Department of Biology Faculty of Science Razi University Kermanshah, Iran E-mail: motallebi38@yahoo.com
ABSTRACT
Fusarium oxysporum is a common soilborn plant pathogen with a worldwide distribution. Fusarium yellows disease of chickpea (Cicer arientinum) caused by F.oxysporum is one of the most destructive soilborn disease which is a major production constraint in chickpea-growing regions of Iran. Three laboratory methods "amplification of genomic DNA using random primers, enzyme activity staining, and colony size determination" have been used to discriminate between highly virulent (HV) and weakly virulent (WV) isolates of F. oxysporum. On the basis of colony size (a traditional morphological method) and the ability of isolates to produce pectic enzymes, five HV isolates were differentiated from three WV isolates. The HV isolates formed large colony (ranging from 10.1 to 12.6 mm in diameter) and showed the same enzyme pattern,while the WV isolates produced small colony (ranging from 5.8 to 7.8 mm in diameter) and had not detectable enzyme activity in the stained overlaying gel. Twelve arbitrary 10-mer primers were tested on these 8 isolates of F. oxysporum by Polymerase Chain Reaction (PCR). Cluster analysis of the data from the DNA amplification by Random Amplified Polymorphic DNA (RAPD), differentiated HV from WV isolates. The results obtained from RAPD test confirmed the classification of these eight isolates based on pathogenicity test, colony size, and enzyme activity staining into two groups (HV and WV).
Key words: Fusarium oxysporum- RAPD- colony size- enzyme activity staining
RESUMO
Fusarium oxysporum é um fitopatógeno amplamente difundido nos solos. A doença amarela do grão-de-bico (Cicer arietinum), causada por ele é destrutiva e um grande problema nas regiões produtoras do Irã. Três métodos de laboratório: amplificação aleatória de DNA genômico (RAPD), coloração de atividade enzimática e determinação das dimensões de colônia, foram usados para diferenciar isolados de Fusarium oxysporum altamente virulentos (HV) e fracamente virulentos (WV). Baseado no tamanho da colônia (método morfológico tradicional) e na capacidade dos isolados produzirem enzimas pécticas, cinco isolados HV foram diferenciados de três WV. Enquanto os isolados HV formaram colônias grandes (entre 10,1 e 12,6 mm de diâmetro) e apresentaram o mesmo padrão enzimático, os isolados WV produziram colônias pequenas (entre 5,8 e 7,8 mm de diâmetro) e não tiveram atividade enzimática detectável no gel de cobertura corado. Doze primers arbitrários foram testados nos 8 isolados de Fusarium oxysporum por reação de polimerase em cadeia (PCR). As análises dos dados da amplificação de DNA pelo método de amplificação aleatória do DNA polimórfico (RAPD) diferenciaram os isolados HV dos WV. Os resultados obtidos pelo teste RAPD confirmaram a classificação dos 8 isolados pelos testes de patogenicidade, tamanho da colônia e atividade enzimática de coloração nos dois grupos, HV e WV.
Palavras-chave: Fusarium oxysporum, RAPD, tamanho de colônia, coloração de atividade enzimática
INTRODUCTION
Fusarium yellows disease of chickpea (Cicer arientinum) caused by the vascular wilt pathogen Fusarium oxysporum is a major production constraint in chickpea-growing regions of Iran (1). Isolates of F. oxysporum form a major component of the fungal flora of most cultivated soils and most of these isolates are not perfectly identifiable using phenotypic characters (6). Molecular tools have been used to characterize the diversity among pathogenic isolates of F. oxysporum (2,4,9,12). Molecular markers, such as Random Amplified Polymorphic DNA (RAPD) have been used extensively as genetic markers in different populations (2,4). RAPD markers have some advantages in that they are easy to generate, require only very small amounts of genomic DNA and do not require the use of radioisotopes. RAPD markers can overcome the limitation of other markers because they have the potential to identify a large number of polymorphisms with good coverage of entire genome.
This paper describes three laboratory tests and techniques that confirm the classification of F. oxysporum isolates based on pathogenicity test, as highly virulent (HV) and weakly virulent (WV) isolates. These tests and techniques include colony size, enzyme activity staining and RAPD.
MATERIALS AND METHODS
Fungal isolates and growth conditions
Eight isolates (five highly virulent "HV" and 3 weakly virulent "WV") of F. oxysporum from aerial sections of chickpea from different geographical locations in Iran were collected (17) (
Table 1). Isolates were maintained in wheat seed medium (50 g autoclaved soaked wheat seed in 500 ml flask) at 4ºC. Isolates were grown in shake culture in pectic zymogram (PZ) medium containing 2.64 g, (NH4)2 SO4, 0.34 g KH2PO4, 0.14 g MgSO4.7H2O, 10 g Citrus pectin (Sigma), one litter distilled water, pH adjusted to 4.5 (14). After 6 days growth at 26ºC a liquid culture filtrate was obtained (as crude enzyme) by Whatman filter paper No.1 and stored at -20ºC until using for detecting the enzyme activity. For colony size measurement, the isolates were grown on Modified D-medium contained 0.14g MgSO4 7H2O, 2.6g (NH4)2SO4, 0.5g NaCl, and 20g agar supplemented with 20g L-sorbose in one litter distilled water. To reduce carmelization, the L-sorbose was autoclaved separately from the other ingredients.Enzyme activity staining
For rapid characterization of pectic enzyme activity, 10 µl of culture filtrate was mixed with equal volume of sample buffer containing, a solution of 50 mM Tris-HCl pH 6.8, 2% (w/v) Sodium Dodecyl Sulphate (SDS), 5% (v/v) of 2-b mercaptoethanol, 10% (v/v) glycerol, and 0.05% w/v) bromophenol blue. The proteins were analyzed by Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE). The separated proteins were assayed for polygalacturonase activity with an ultrathin pectate agarose gel by staining with 0.05% ruthenium red for 30 min as described by Ried and Collmer (13).
(a) Preparation of pectate agrose overlay gel
Thin pectate overlay gels (0.75mm) for detection of enzyme activity were cast by using a gel support film for agarose gels (FMC Bio products) on one of the glass plates of vertical apparatus. The overlay contained, in addition of 1% agarose (Sigma), 0.1% polygalacturonic acid in 100 mM potassium acetate buffer, pH 4.5 and 10 mM EDTANa2 (11). The agarose solution was boiled to dissolve the agarose and cooled to 75ºC. The gel mold was heated to 60ºC before casting.
(b) Detection of polygalacturonase activity in SDS-PAGE
After electrophoresis, the gel was washed by three 200 ml changes of 10 mM potassium acetate buffer, pH 4.5 (5). The pectate agarose overlay was laid directly on top of the gel and incubated at 37ºC for 120 min in a moisture lunch box. The overlay was removed and stained with 0.05% (w/v) ruthenium red for 30 min. To enhance recovery of renature enzyme activity, bovine serum albumin (BSA) at 10 µg/ml was incorporated into separating gels as described by Lacks and Springhorn (10).
Colony size
Isolates were grown on PDA plates for 7 days. With a thin platinum needle, 20 stab transfers were made from the PDA to a plate of D-medium. Plates were incubated for 72 hours at 25ºC. The diameters of 20 colonies of each strain were then measured from the underside of the plate using a binocular dissecting microscope equipped with an ocular micrometer. Colony diameters were measured to the nearest 0.1 mm (6). Dendrogram was produced by cluster analysis, using the Unweighted Pair-Group Method Analysis (UPGMA).
DNA extraction
Lyophilized mycelium of different isolates was homogenized and genomic DNA was extracted according to Zamani et al. (16).
Amplification of genomic DNA using RAPD primers
Amplification of DNA fragments was carried out by Polymerase Chain Reaction (PCR) using 10-mer arbitrary primers. Amplification reactions were performed in 50 µl reaction volumes containing one unit of Taq DNA polymerase, 2 µmol/ml each of dATP, dCTP, dGTP and dTTP, 1.5 µmol/ml primer, and 60 ng of genomic DNA.
The reaction mixture was overlaid with sterile mineral oil (50 µl) to prevent evaporation during PCR cycling. The programe comprised 34 cycles of denaturation at 94ºC for 2 min, primer annealing at 32ºC for 2 min, and extension of primer at 72ºC for 2 min. After the cycling steps were completed, the mixture was held at 72ºC for 4 min to allow complete extension of amplified products. A total of 12 Primers were used (
Table 2). Amplified DNA fragments were analyzed by electrophoresis in 2% agarose gel in TBE buffer.RAPD product scoring and Data analysis
Data were compiled as a binary 0/1 matrix by the presence (1) or absence (0) of a band at particular position. Only major RAPD bands were considered for statistical analysis. Dendrogram were produced by cluster analysis using UPGMA.
RESULTS
Colony size
Fifteen isolates of F. oxysporum derived from different geographical regions have been previously studied based on pathogenicity test. From these fifteen isolates five highly virulent isolates (F15, F18, F23, F47 and F59) and three weakly virulent isolates (F2, F21 and F58) were distinguished. (17). These eight isolates were grown on D-medium containing L-sorbose.
The growth of different isolates tested on medium, and each isolate had a characteristic mean colony diameter (
Fig. 1). Isolates F02, F21, and F58 produced small-size colonies, with mean diameters 7.8, 6.4, and 5.8 mm, respectively (Table 1). Isolates F15, F18, F23, F47, and F59 formed large colonies with mean diameters: 11.5, 10.1, 12.6, 11.1, and 10.1 mm, respectively (Table 1). A dendrogram constructed by UPGMA cluster analysis showed a grouping of these isolates into two clusters. One cluster with five isolates (large colony) were highly virulent and second cluster containing three isolates (small colony) were weakly virulent (Fig. 2).RAPD analysis
In order to compare the overall similarity between these isolates at DNA level, 12 decamer oligonucleotide primers were used which generated polymorphic bands. Example of polymorphic bands detected in amplified DNA from different isolates is shown in
Fig. 3.
The data from polymorphic bands were analyzed by UPGMA method. Comparison of the results from pathogenicity test of these isolates with the results of cluster analysis of RAPD poly morphic bands demonstrated that among 12 primers, only the dendrogram obtained from R2 primer (Table 2) was able to differentiate the weakly virulent isolates F2, F21, and F58 as a separate cluster (
Fig. 4).
Enzyme activity staining
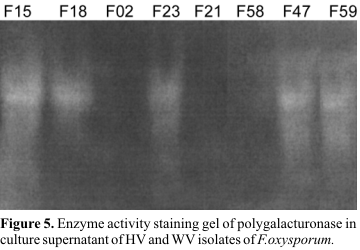
In order to study whether pectic enzymes are present in the culture supernatant, an attempt was made to stain the gel for pectic enzyme activity, using pectate agarose overlay. An equivalent amount of enzyme was loaded onto the gel for all isolates (HV and WV). The obtaining results indicated that all highly virulent isolates show the same enzyme activity pattern (
Fig. 5). Comparison of the banding patterns or enzyme activity for the highly virulent and weakly virulent isolates demonstrated that, a high intensity band is present in the highly virulent isolates which is absent in weakly virulent isolates (Fig. 5).
DISCUSSION
In this study, we have evaluated three laboratory methods for their ability to discriminate between highly virulent and weakly virulent isolates of F. oxysporum: amplification of genomic DNA using random primers, enzyme activity staining, and colony size determination.
In a previous study (17) five highly virulent and three weakly virulent isolates have been identified. In the present investigation, these eight isolates were characterized by other laboratory methods. In the first test, colonies of the different isolates of F. oxysporum were grown on a sorbose-containing medium and their diameters were measured. The second test assessed the ability of the isolates to produce pectic enzymes, as tested by enzyme activity staining on the gel. By cluster analysis of the results from these two tests, these eight isolates have been divided into two distinct clusters. Comparing the arrangement of isolates in Fig. 2 (based on colony size) and Fig. 5 (based on enzyme activity staining) with the classification in pathogenic groups (Table 1), one can see that the classification of the isolates by pathogenicity test is in accordance with the results of these two tests. Further analysis of the results revealed that isolates F15, F18, F23, F47, and F59, which were highly virulent, formed large colonies (ranging from 10.1 to 12.6 mm in diameter) and produced detectable pectic enzyme activity in overlaying gel, while isolates F02, F21, and F58, which were weakly virulent, formed small colonies (ranging from 5.8 to 7.8 mm in diameter) and showed not detectable pectic enzyme activity in overlaying gel. Correll et al. (6) reported that virulent isolates of F. oxysporum have a characteristically large colony size which is in agreement with our finding.
When comparing the RAPD analysis to classification into pathogenicity groups, the results using 12 primers did not present good correlation between amplification patterns and pathotype classification.
However, primer R2 alone allowed differentiation of these isolates, which was strongly supported by pathotype, colony size and overlaying classification. As evidenced in our study and in others as well, RAPD markers were effective for detecting polymorphism in F. oxysporum (3,7,8,15).
Finally, from these results it can be concluded that these three rapid methods (RAPD, colony size, and enzyme activity staining) could be employed to distinguish highly virulent from weakly virulrnt isolates of F. oxysporum. A fast diagnosis of the highly virulent in an early stage of infection may provide advantages for phytosanitary procedures. Nevertheless, standardization of the detection system will be required.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Afshari-Azad from Ministry of Agriculture and Mr. H. Younesi from Agricultural Research Center of Kermanshah for supplying the isolates of F. oxysporum. This work is supported by Ministry of Agriculture of Iran.
Submitted: February 18, 2002; Returned to authors for corrections: July 17, 2002; Approved: December 05, 2002.
- 1. Afshari-Azad, H. Identification of Iranian fungal isolates causing yellow disease in chickpea. 13th Iranian Plant Protection Congress, Karaj, 1998.
- 2. Bentley, S.; Pegg, K.G.; Dale, J.L. Optimizatoin of RAPD-PCR fingerprinting to analyze genetic variation within populations of Fusarium oxysporum f. sp. cubense J. phytopathol, 142: 64-78, 1994.
- 3. Bentley, S.; Pegg, K.G.; Dale, J.L.Genetic variation among a world-wide collection of isolates of Fusarium oxysporum f.sp cubense analyzed by RAPD-PCR fingerprinting. Myco. Res, 99: 1378-1384, 1995.
- 4. Bridge, P.D.; Pearce, D.A.; Rivera, A.; Rutherford, M.A.VNTR derived oligonucleotides as PCR primers for population studies in filamentous fungi. Lett. App. Microb, 24: 426-430, 1997.
- 5. Collmer, A.; Ried, J.L.; Mount, M.S. Assay methods for pectic enzymes. Academic Press, London. 1988, 329-399.
- 6. Correll, J.C.; Puhalla, J.E.; Schneidar, R.W. Identification of Fusarrium oxysporum f.sp apii on the basis of colony size, virulence and vegetative compatibility. Phytopathology, 76: 396-400, 1986.
- 7. Crowhurst, R.N.; King, F.Y.; Hawthorne, B.T.; Sanderson, F.R.; Choi-Pheng, Y. RAPD characterization of Fusarium oxysporum associated with wilt of angsana (Pterocarpus indicus) in Singapore. Mayco. Res, 99: 14-18, 1995.
- 8. Gherbawy, Y.A.M.H. RAPD profile analysis of isolates belonging to different formae speciales of Fusarium oxysporum Cytologia, 64: 269-276, 1999.
- 9. Kim, D.H.; Martyn, R.D.; Magill, C.W. Mitochondrial DNA(mt DNA)-Relatedness among formae speciales of Fusarium oxysporum in the cucurbitaceae. Phytopathololy, 83: 91-97, 1993.
- 10. Lack, S.A.; Springhorn, S.S. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of SDS. J. Biol. Chem., 255: 7467-7473, 1980.
- 11. MacGuire, R.G.; Palenzuela, P.R.; Collmer, A.; Burr, T.J. Polygalacturonase production by Agrobacterium tumefaciens biovar3. Appl. Environ. Microbiol, 57: 660-664, 1991.
- 12. Namiki, F.; Shiomi, T.; Kayamura, T.; Tsuge, T. Characterization of formae speciales of Fusarium oxysporum causing wilts of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Envir. Microb, 60: 2684-2691, 1994.
- 13. Ried, J.L.; Collmer, A. Activity stain for rapid characterization of pectic enzymes in isoelectric focusing and sodium dodecyl sulfate-polyacrilamide gels. Appl. Envir. Microb, 50: 615-622, 1985.
- 14. Sweetingham, M.W.; Cruickshank, R.H.; Wong, D.H. Pectic zymograms and taxonomy and pathogenicity of the ceratobasidiaceae. Trans. Br. Mycol. Sci, 86: 305-311, 1986.
- 15. Wang, P.-H.; Lo, H.S.; Yeh, Y. Identification of F.o.cucumerinum and F. o. luffae by RAPD-generated DNA probes. Lett. Appl. Microb, 33: 397-401, 2001.
- 16. Zamani, M.R.; Motallebi, M.; Hosseinzadeh Colagar, A.Virulence and polymorphic DNA relationships of Ascocyta rabiei to geographic regions. Iranian. J. Biol, 7: 1-17, 1998.
- 17. Zamani, M.R.; Motallebi, M.; Harighi, M.J. Pectic enzyme patterns of Fusarium oxysporum isolates from chickpea in Iran. J. Sci. Isl. Rep. Iran, 12: 17-21, 2001.
Publication Dates
-
Publication in this collection
18 June 2003 -
Date of issue
Dec 2002
History
-
Reviewed
17 July 2002 -
Received
18 Feb 2002 -
Accepted
05 Dec 2002