Abstract
Antibiotic therapy in hematologic patients, often weak and susceptible to a wide range of infections, particularly nosocomial infections derived from long hospitalization periods, is a challenging issue. This paper presents ESBL-producing strains isolated from such hematologic patients treated at the Amazon Hematology and Hemotherapy Foundation (HEMOAM) in the Brazilian Amazon Region to identify the ESBL genes carried by them as well as the susceptibility to 11 antimicrobial agents using the E-test method. A total of 146 clinical samples were obtained from July 2007 to August 2008, when 17 gram-negative strains were isolated in our institution. The most frequent isolates confirmed by biochemical tests and 16S rRNA sequencing were E. coli (8/17), Serratia spp. (3/17) and B.cepacia (2/17). All gram-negative strains were tested for extended-spectrum-beta-lactamases (ESBLs), where: (12/17) strains carried ESBL; among these, (8/12) isolates carried blaTEM, blaCTX-M, blaOXA, blaSHV genes, (1/12) blaTEM gene and (3/12) blaTEM, blaCTX-M, blaOXA genes. Antibiotic resistance was found in (15/17) of the isolates for tetracycline, (12/17) for ciprofloxacin, (1/17) resistance for cefoxitin and chloramphenicol, (1/17) for amikacin and (3/17) cefepime. This research showed the presence of gram-negative ESBL-producing bacteria infecting hematologic patients in HEMOAM. These strains carried the blaTEM, blaSHV, blaCTX-M and blaOXA genes and were resistant to different antibiotics used in the treatment. This finding was based on a period of 13 months, during which clinical samples from specific populations were obtained. Therefore, caution is required when generalizing the results that must be based on posological orientations and new breakpoints for disk diffusion and microdilution published by CLSI 2010.
ESBL; beta-lactams; nosocomial infection; TEM; SHV; OXA; CTX-M
MEDICAL MICROBIOLOGY
Extended-spectrum beta-lactamase-producing bacteria isolated from hematologic patients in Manaus, State of Amazonas, Brazil
Cristina Motta FerreiraI,* * Corresponding Author. Mailing address: Fundação de Hematologia e Hemoterapia do Amazonas (HEMOAM) Avenida Constantino Nery, 4397, Chapada. CEP: 69050-002, Manaus, Amazonas, Brazil.; Tel.: (55)-92-3655-0100.; E-mail: cris_motta_ferr@yahoo.com.br ; William Antunes FerreiraII; Nayanne Cristina Oliveira da Silva AlmeidaIII; Felipe Gomes NavecaIV; Maria das Graças Vale BarbosaV
IUniversidade do Estado do Amazonas, Fundação de Medicina Tropical do Amazonas, Fundação de Hematologia e Hemoterapia do Amazonas, Manaus, AM, Brasil
IIUniversidade do Estado do Amazonas, Fundação de Medicina Tropical do Amazonas, Fundação de Dermatologia Tropical e Venereologia Alfredo da Matta, Manaus, AM, Brasil
IIIUniversidade Federal do Amazonas, Manaus, AM, Brasil
IVInstituto Leônidas e Maria Deane, Fundação Oswaldo Cruz, Manaus, AM, Brasil
VFundação de Medicina Tropical do Amazonas, Universidade do Estado do Amazonas, Manaus, AM, Brasil
ABSTRACT
Antibiotic therapy in hematologic patients, often weak and susceptible to a wide range of infections, particularly nosocomial infections derived from long hospitalization periods, is a challenging issue. This paper presents ESBL-producing strains isolated from such hematologic patients treated at the Amazon Hematology and Hemotherapy Foundation (HEMOAM) in the Brazilian Amazon Region to identify the ESBL genes carried by them as well as the susceptibility to 11 antimicrobial agents using the E-test method. A total of 146 clinical samples were obtained from July 2007 to August 2008, when 17 gram-negative strains were isolated in our institution. The most frequent isolates confirmed by biochemical tests and 16S rRNA sequencing were E. coli (8/17), Serratia spp. (3/17) and B.cepacia (2/17).
All gram-negative strains were tested for extended-spectrum-beta-lactamases (ESBLs), where: (12/17) strains carried ESBL; among these, (8/12) isolates carried blaTEM, blaCTX-M, blaOXA, blaSHV genes, (1/12) blaTEM gene and (3/12) blaTEM, blaCTX-M, blaOXA genes. Antibiotic resistance was found in (15/17) of the isolates for tetracycline, (12/17) for ciprofloxacin, (1/17) resistance for cefoxitin and chloramphenicol, (1/17) for amikacin and (3/17) cefepime. This research showed the presence of gram-negative ESBL-producing bacteria infecting hematologic patients in HEMOAM. These strains carried the blaTEM, blaSHV, blaCTX-M and blaOXA genes and were resistant to different antibiotics used in the treatment. This finding was based on a period of 13 months, during which clinical samples from specific populations were obtained. Therefore, caution is required when generalizing the results that must be based on posological orientations and new breakpoints for disk diffusion and microdilution published by CLSI 2010.
Key words: ESBL, beta-lactams, nosocomial infection, TEM, SHV, OXA, CTX-M
INTRODUCTION
Extended-spectrum beta-lactamases (ESBL) are enzymes produced by gram-negative bacteria such as Klebsiella pneumoniae and Escherichia coli (24) as well as by species from other genera, such as Enterobacter sp., Salmonella sp., Proteus sp., Serratia marcescens, Shigella dysenteriae, Pseudomonas aeruginosa, and Burkholderia cepacia (2, 3, 31, 26).
ESBLs originate from punctual genetic mutations in the active sites of the TEM-1, TEM-2, and SHV-1 enzymes (3, 10, 12, 31, 19, 23, 26, 34), which enable the hydrolysis of extended-spectrum cephalosporins, monobactams, penicillins. (2, 3, 5, 9, 11, 12, 27, 30). However, these plasmid mediated enzymes have their activity inhibited by clavulanic acid, tazobactam and sulbactam. Another important epidemiological characteristic is the possibility of these enzymes being transferred to other species via plasmids or transposons (2, 3, 5, 6, 9, 11, 12, 21, 27, 28, 30, 34).
The emergence of ESBL-producing bacteria, responsible for infectious processes with high levels of morbidity and mortality has complicated the therapeutic options available for elderly, immunocompromised, and debilitated individuals (4, 31). The prevalence of ESBLs among clinical isolates varies according to countries and institutions (31). In the United States, the presence of these enzymes among Enterobacteriaceae has increased from 0% to 25%, with a national average of 3%, whereas the percentage of Klebsiella sp. has demonstrated an increased resistance from 5% to 10% to ceftazidime (6).
In other countries, such as Switzerland, the frequency of ESBL-producing bacteria varies from 3% to 8%, whereas it is 34% in Portugal, 4.8% in Korea, 8.5% in Taiwan, 12% in Hong Kong, and 58% in Turkey. In Latin America, studies have reported frequencies ranging from 30% to 60% (6, 21). The prevalence of ESBL resistance to antibiotics in Brazil has reached 45.4%, 65% of which stand for K. pneumoniae and E. coli, which is considered one of the highest levels in the world. For the city of Manaus, in the State of Amazonas, Brazil, no published data is found regarding the prevalence of such bacteria (3, 5).
ESBL-producing bacteria are responsible for serious infectious processes, commonly associated with outbreaks of nosocomial infections. This situation is relevant as most patients with hematologic diseases undergo chemotherapy that can lead to a deficient immune response. These medical conditions can lead to prolonged hospitalization, elevation of the risks of mortality and morbidity, and increased treatment costs.
This study therefore aimed to detect ESBL-producing strains isolated from such hematologic patients treated at the HEMOAM in the Brazilian Amazon region to identify the ESBL genes carried by them and susceptibility to 11 antimicrobial agents using the E-test method.
MATERIALS AND METHODS
The study was approved by the HEMOAM Ethics Committee (Approval number 170 CAAE-0005.0.112.000-06 - Version 003/06). An informed consent was obtained from each patient before specimens were collected and the results were used for the management of each respective patient.
We conducted a descriptive cross-sectional study including patients of both sexes at any age who had been admitted to the HEMOAM Foundation in the city of Manaus, State of Amazonas, with prior diagnosis of hematologic disease, presenting clinical signs and symptoms suggestive of acute gram-negative bacterial infection, whether feverish or not, or having developed fever during hospital stay in the period from July 2007 to August 2008.
Gram-negative bacteria strains were identified using colonial morphology on blood, MacConkey, Eosin Methylene Blue (EMB), and Mueller-Hinton agar plates (Himedia-Hexasystems, Mumbai, India), followed by biochemical panels like NF II Kit, enterokit C, and enterokit B (Probac, São Paulo, Brazil). Other tests, such as urease, oxidase, glucose, mannose, lactose, and maltose fermentation, were also performed. The 16S rRNA protocol with the bacterial primers 27F (5'-GAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTAC GACTT-3') (Invitrogen, Carlsbad, CA, USA) with a product size around 900 bp, were performed to confirm genus and species (17).
The sequence analysis of the 16S rRNA region was compared with GenBank data using BLAST search with the MegaBLAST algorithm for identifying high-similarity sequences. The sequences with the highest levels of similarity were considered as being consistent for species identified by the biochemical tests.
The antibiotics E-test® strips representing common drugs used in the treatment of suspected gram-negative bacterial infections in our hospital were chosen for antimicrobial testing. Strips included tetracycline, ceftazidime, cefoxitin, chloramphenicol, cefepime, and amikacin (0.016-256.0 µg/mL), and ciprofloxacin and imipenem (0.002-32.00 µg/mL).
Test organisms were suspended in 0.8% saline to 0.5 McFarland standards and then inoculated on Mueller-Hinton agar plates, followed by overnight incubation at 37ºC for 18-24 hours. Interpretation was performed using guidelines laid down in the Clinical and Laboratory Standards (CLSI; Wayne, Pennsylvania, USA) manual, which provides break points corresponding to the diameter of the zone of inhibition.
The isolates were screened for the production of ESBL by the E-test® method with the antibiotics cefotaxime (0.25-16 µg/mL)/cefotaxime (0.016-1 µg/mL) plus 4 µg/mL clavulanic acid, ceftazidime (0.5-32 µg/mL) /ceftazidime (0.064-4 µg/mL) plus 4 µg/mL clavulanic acid, following manufacturer's recommendations (7). Strains of E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality controls for culture media, susceptibility tests, PCR and the ESBL-production E-test®.
The bacteria deemed as positive for ESBL-production were inoculated in 5 ml of Luria-Bertani (LB) broth with ampicillin (50 µg/mL) and incubated at 37ºC for 16 hours for plasmid extraction following the protocol described in the Kit Wizard® Plus SV Minipreps-DNA purification System (Promega®; Madison, WI, USA).
Electrophoresis was performed in 0.8% agarose gel stained with SYBR Safe (Invitrogen, Carlsbad, CA, USA) for the identification of plasmid sizes. The Supercoiled Loading Dye Ladder 2-16 Kb (Promega®) was used as a standard molecular weight. Data were filed by using Excel programs. ESBL isolates were preserved for future molecular analysis. PCR amplification for the detection of chromosomal beta-lactamase resistance AmpC type in Pseudomonas spp. isolates was not included in our study.
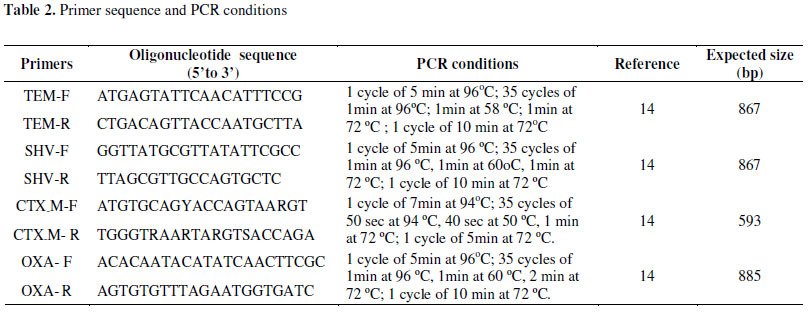
The Polimerase Chain Reaction (PCR) assay to amplification and detection of the blaTEM, blaSHV, blaCTX-M and blaOXA was carried out in a final volume reaction of 25µl using 1X buffer, MgCl2 (1.5µM), dNTP (0.2mM), DNA Taq polimerase (1U) (Invitrogen), 0.5µM of each primer TEM-F, TEM-R, SHV-F, SHV-R, blaCTX-M-F, blaCTX-M-R, blaOXA-F, blaOXA-R and 5µl of plasmidial DNA. Cycling parameters were previously described (Table 2). The amplified products obtained have not been sequenced up to the present moment.
RESULTS
A total of 146 clinical samples were obtained from 69 patients over a period of thirteen months. These samples included the following: urine, pus, blood, marrow aspirate, sputum, feces, abscesses, and oropharyngeal and nasopharyngeal swabs. The strains isolated from different specimen type are presented in Table 5. In relation to gender, (35/69) were male and (34/69) were female. Among the collected samples, we isolated 44 bacterial strains, 17 of which were gram-negative. The most frequent hematological diseases in the patients were acute lymphocytic leukemia (36/69) and acute myeloid leukemia (6/69) (Table 1).
Biochemical tests and 16S rRNA sequencing detected E. coli (8/17), followed by Serratia sp. (2/17), Serratia liquefaciens (1/17) and B. cepacia (2/17) as the most frequent gram-negative strains. All the strains were tested for the production of ESBL and the E-test showed that (12/17) were ESBL producers (Table 3). All of them had their plasmid extracted and the molecular weights were 3.5, 7 and >15 Kb. The PCR assay showed that that (8/12) isolates carried blaTEM, blaCTX-M, blaOXA, blaSHV genes, (1/12) blaTEM gene and (3/12) blaTEM, blaCTX-M, blaOXA genes.
As presented in Table 4, the results from antimicrobial susceptibility tests demonstrated antibiotic resistance in (15/17) of the isolates for tetracycline, (12/17) for ciprofloxacin, (1/17) resistance for cefoxitin/chloramphenicol, and (1/17) for amikacin and (3/17) for cefepime.
Data regarding the molecular epidemiology of ESBL clonality and PCR amplification for the detection of chromosomal beta-lactamase resistance AmpC type in Pseudomonas spp. isolates have not been presented so far, but we believe that this will not expressively impact on this study as it was aimed to detect ESBL-producing strains in hematologic patients only. Our finding was based on a period of 13 months, during which clinical samples were obtained from a specific population. Therefore, caution is required when results are generalized as they must be based on posological orientations and new breakpoints for disk diffusion and microdilution according to the CLSI 2010 manual M100-S20.
DISCUSSION
ESBLs are enzymes that act by inhibiting the action of beta-lactam antibiotics (1, 25). Concerns regarding the clinical and epidemiological aspects of such ESBL-producing pathogens are related to the use of extended-spectrum antimicrobials such as carbapenems for their treatment (12, 22), thus increasing treatment costs as well as patient risks due to longer hospital stay and prolonged therapy (1, 29).
Furthermore, hematologic patients often undergo chemotherapy, which can lead to an immunodeficient status and leave these patients susceptible to a wide range of infections (32, 33). This is particularly important with respect to nosocomial infection in the hospital environment, which can elevate the risks of mortality and morbidity, mainly in the case of infections caused by ESBL-producing bacteria.
In our study, the most frequent ESBLs-producing bacteria were E. coli, B. cepacia, Serratia spp., and Pseudomonas spp. In addition, ESBL-producing strains of E. coli and K. pneumoniae have also been isolated in other studies from patients with outbreaks of bacterial peritonitis (32), bloodstream infections (33) in ICU (1) from different clinical samples from patients in Mwanza (Tanzania) (20), in Enugu (Nigeria) (31), and in France, Portugal (9), Brazil (33), as well as several other countries (2, 13, 16, 19, 37).
These E. coli isolated from samples of the patients point to the possibility of a hospital-acquired infection probably due to inadequate antibiotic therapy, inappropriate prescriptions without culture results and susceptibility testing and cross-infections that contributed to increase the number of these resistant isolates (1, 10, 25, 27, 40).
The detection of ESBL enzymes is very important, because according to CLSI M100-S17 and M100-S18 (25, 27) criteria, these organisms should be reported as resistant to all extended-spectrum beta-lactam antibiotics, regardless of their susceptibility test results. Currently, according to CLSI M100-S20, new interpretive criteria are available for routine ESBL testing (1, 25, 27).
The reduced sensitivity to cephalosporins detected in E. coli strains in this study might be due to the variations in substratum preference, the differing accuracy between tests for enzyme detection, weak inhibition by clavulanic acid, or different combined resistance mechanisms that led either to reduced or no sensitivity (26, 35). This type of phenotype could be demonstrated in other multicenter study developed in Norway in 2003, in which 60% of ESBL-producing isolates showed reduced sensitivity to extended-spectrum cephalosporin, possibly indicating an increased level of resistance (26, 36).
Our findings demonstrated that all the ESBL-producing Enterobacteriaceae were sensitive to imipenem, while 94.1% were sensitive to amikacin, indicating that these agents can still be used in the treatment of infections caused by these pathogens in HEMOAM. Similar results have been observed by Tumbarello et al. (37) in a teaching hospital in Rome (Italy).
The existence of ESBL-producing Pseudomonas spp. is of serious concern as it stands for a pathogen with the capacity to acquire resistance to a wide range of clinically important antimicrobials and it can lead to numerous infectious processes. In this study, we detected 33.4% of the isolates to be resistant to tetracycline and ciprofloxacin (12, 25, 29). Similar results have been observed in India, Iran, Spain, Portugal, Canada, and Brazil. However, in Norway, positive CTX-M isolates showed reduced sensitivity only to ciprofloxacin (12, 16, 24, 36).
In Europe, 20% of hyperproductive ESBL isolates have demonstrated resistance to ceftriaxone and aztreonam, while in Latin America, reports show that the resistance rate is higher in relation to other regions of the world (12, 25). In Manaus (Brazil), our study provides the first data regarding the existence of ESBLs in clinical samples from patients. Thus, it is not possible to compare our results regarding the spectrum of antimicrobial resistance.
The therapeutic alternative for treating infections caused by ESBL-positive pathogens such as Pseudomonas spp. lies in the use of imipenem or meropenem; however, this may stimulate the appearance of multiresistant strains in hospitals, which could lead to additional serious problems for patients as well as difficulties for physicians regarding viable therapeutic options (12, 16, 25, 39).
The control of bacterial resistance is directly related to the detection of this mechanism through laboratory and molecular tests as well as the implementation of rational use of antibiotics in hospitals (26, 39). We documented the presence of blaTEM, blaSHV, blaCTX-M and blaOXA genes in E.coli, Serratia spp., Pseudomonas spp., and B.cepacia strains (Table 3).
Similar results were detected in other studies such as the blaCTX-M gene detected in E.coli isolates in Cambodia, Canada, United Kingdom (28) and in clinical outbreaks of CTX-M-15-type in India, Spain, and Portugal (9), considered as the most prevalent ESBL-encoding gene worldwide, now replacing TEM and SHV as the predominant ESBL isolate in European countries (14).
The prevalence of ESBL production in Enterobacteriaceae in France (18) belonged to CTX-M variants such as CTX-M-I, and CTX-M-9. In Spain and Portugal, they frequently detected TEM-24, CTX-M-15, CTX-M-32 and SHV-12. In Bulgaria, CTX-M-3, CTX-M-15 and SHV-12 in S.marcescens isolates (18). In Amsterdam, CTX-M-1, SHV-1, TEM-116 in P. aeruginosa and CTX-M-1, SHV-1 in S. maltophilia (22). In Brazil, CTX-M-14, TEM-1, CTX-M-15, OXA-1, OXA-4 and OXA-7 (7, 22, 23).
The detection of the blaTEM, blaSHV, blaCTX-M and blaOXA genes in gram-negative isolates in this study allows us to consider the possibility of plasmidial resistance. Both the genes that encode the ESBLs and those that encode resistance to non-beta-lactam antibiotics are frequently located in the same conjugative plasmid, transmitted conjunctively from one isolate to another, showing the ability of being separated by selective pressure due to the use of multiple antibiotics under effective control (16, 26).
The early identification of infectious processes linked to the use of appropriate antimicrobials based on microbiological diagnosis, rational prescription of antibiotics, and effective control can help avoid serious hospital infections. Most significantly, it is important to consider the inclusion of new antibiotics as therapeutic choices. These measures will help the implementation of effective public policies for epidemiological surveillance and control by Brazilian health authorities in the State of Amazonas.
This research showed the presence of gram-negative ESBL-producing bacteria infecting hematologic patients in HEMOAM. These strains carried the blaTEM, blaSHV, blaCTX-M and blaOXA genes and were resistant to different antibiotics used in the treatment. It is necessary to perform screening and confirmatory test for phenotypic detection of these strains in a routine laboratory to avoid health care failure.
ACKNOWLEDGEMENTS
The authors are grateful to acknowledge the support provided by the Amazonas State Research Foundation (Fundação de Amparo a Pesquisa do Estado do Amazonas - FAPEAM). Inputs to carry out this research derived from a project approved by the Research Program of SUS (Brazilian National Health Care System): Shared Health Management - PPSUS-Posting 014/2006-FAPEAM Manaus, State of Amazonas, Brazil. This paper stems from the preliminary findings reported by the author's doctoral thesis at the Amazon State University (UEA) and the Amazon Tropical Medicine Foundation (FMT-AM) in the field of clinical bacteriology.
In addition, the Amazon Hematology and Hemotherapy Foundation (HEMOAM), the Tropical Dermatology and Veneorology Foundation Alfredo da Matta (FUAM), the Graduate Program of Biotechnology (PPGBiotec), the superintendent of the State of the Amazonas (SUFRAMA).
Submitted: September 19, 2010; Returned to authors for corrections: October 21, 2010; Approved: March 14, 2011.
- 1. Ahmed, S.H.; Daef, E.A.; Badary, M.S.; Mahmoud, M.A.; Elsayed, A.A.A. (2009). Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum b-lactamase producing organisms. BMC Research Notes. 2:76.
- 2. Aladag, M.O.; Durak, Y. (2009). Investigating Some Antibiotics, Plasmids Profile and ESBL Characteristic of Klebsiella pneumoniae Isolated from Urinary System Infection. World Appl Sci J 6:630-636 (In Turkish).
- 3. Al-Zarouni, M.; Senok, A.; Rashid, F.; Al-Jesmi, S.M.; Panigrahi, D.M. (2008). Prevalence and antimicrobial susceptibility pattern of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the United Arab Emirates. Medical Principles and Practices 17:32-36.
- 4. Atifah, M.N.; Loo, H.K.C.; Subramaniam, G.; Wong, E.H.; Selvi, P.; Ho, S.; Kamarulzaman, A.; Parasakthi, N. (2005). Faecal prevalence of extended-spectrum β-lactamase (ESBL)-producing coliforms in a geriatric population and among haematology patients. Malaysian J Pathol. 27:75-81.
- 5. Augusti, G.R.; Superti, S.; Zavascki, A.P. (2007). Prevalence of beta-lactamase production of extended-spectrum bacteremia in Klebsiella pneumoniae and Escherichia coli. Scientia Medica 17:192-196 (In Portuguese).
- 6. Bradford, P.A. (2001). Extended-Spectrum-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev. 14:933-951.
- 7. Chmelnitsky, I.; Carmeli, Y.; Leavitt, A.; Schwaber, M.; Venezia, S.N. (2005). CTX-M-2 and new CTX-M-39 enzyme are the major extended-spectrum beta-lacamase in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob Agents Chemother49(11):4745-4750.
- 8. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. Document M100-S20. CLSI, Wayne PA, 2010.
- 9. Cormican, M.G.; Marshall, S.A.; Jones, R.N. (1996). Detection of extended-spectrum β-lactamase (ESBL)-producing strains by the Etest ESBL screen. J Clin Microbiol 34:1880-1884.
- 10. Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Cantón, R.; Nordmann, D.P. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195-200.
- 11. Dalmarco, E.M.; Blatt, S.L.; Córdova, C.M.M. (2006). Laboratory identification of beta-lactamase extended-spectrum (ESBLs) - Review. RBAC. 38:171-177 (In Portuguese).
- 12. Habeeb, K.C.; Surekha, S.; Lakshmi, S.; Narasimha, G. (2007). Multidrug resistance and Beta-lactamase production by Klebsiella pneumoniae. Afr. J. Biotechnol 6:1791-1793.
- 13. Hujer, A.M.; Kslar, K.S.; Dietenberger, N.J.; Bethel, C.R.; Endimiani, A.; Bonomo, R.A. (2009). Detection of SHV β-lactamases in Gram-negative bacilli using fluorescein-labeled antibodies. BMC Microbiology 9:1-4.
- 14. Lim, K.T.; Yasin, R.; Yeo, C.C.; Puthucheary, S.; Thong, K.L. (2009). Characterization of Multidrug Resistant ESBL-Producing Escherichia coli Isolates from Hospitals in Malaysia. J. Biomed. Biotechnol. 1-10.
- 15. Macedo, M.L.A.P.; Cartaxo, R.S.; Almeida, T.C.C.; Souza, L.B.S.; Santana, W.J.; Coutinho, H.D.M. (2005.) Mechanisms of resistance and detection of beta-lactamases. Ciênc Biol Saúde 7:59-63 (In Portuguese).
- 16. Mansouri, M.; Ramazanzadeh, R. (2009). Spread of extended-spectrum beta-lactamase producing Escherichia coli clinical isolates in Sanandaj hospitals. J Biol Sci 9:362-366.
- 17. Miyoshi, T.; Iwatsuki, T.; Naganuma, T. (2005). Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol. 71:1084-1088.
- 18. Mohammad, M.F.; Somayeh, D.; Nafiseh, R.; Araz, M.; Marzieh, A.; Fereshteh, S.; Mahmood, P.; Davud, Y. (2009). Distribution of blaTEM, blaSHV, blaCTX-M Genes Among Clinical Isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. MDR 00(00):1-6.
- 19. Monstein, H.J.; Tärnberg, M.; Nilsson, L.E. (2009). Molecular identification of CTX-M and blaOXY/K1 β-lactamase genes in Enterobacteriaceae by sequencing of universal M13-sequence tagged PCR-amplicons. BMC Infect Dis 9:7. http://www.biomedcentral.com/1471-2334/9/7
- 20. Mshana, S.E.; Kamugisha, E.; Mirambo, M.; Chakraborty, T.; Lyamuya, E.F. (2009). Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzânia. BMC Res Notes. 2:49. Available from: http://www.biomedcentral.com/1756-0500/2/49
- 21. Mulvey, R.M.; Bryce, E.; Boyd, D.; Agostini, M.O.; Christianson, S.; Simor, A.E.; Paton, S. (2004). Ambler Class A extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob Agents Chemother 48:1204-1214.
- 22. Naiemi, N.; Duim, B.; Bart, A. (2006). A CTX-M extended-spectrum beta-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia JMM 55:1607-1608.
- 23. Novella, M.C.C.; Guth, B.E.C.; Castanheira, M.; Carmo, M.S.; Pignatari, A.C.C. (2010). First description of blaCTX-M-14 and blaCTX-M-15-producing Escherichia coli isolates in Brazil. MDR 16(3):177-184 (In Portuguese).
- 24. Pasta, A.A.C.; Fração, F.H.A.; Magalhães, G.L.G.; Quesada, R.M.B. (2008). Prevalence and antimicrobial susceptibility test in strains of extended-spectrum beta-lactamase-producing (ESBL) Klebsiella pneumoniae isolated from patients in University Hospital /UEL. RBAC. 40:137-141 (In Portuguese).
- 25. Perez, F.; Endimiani, A.; Hujer, K.M.; Bonomo, R.A. (2007). The continuing challenge of ESBLs. Curr Opin Pharmacol 7:459-469.
- 26. Picão, R.C.; Gales, A.C. (2007). Extended-spectrum beta-lactamase-producing (ESBL) Pseudomonas aeruginosa: Nightmare or imagination? Prática Hospitalar. 49:79-84 (In Portuguese).
- 27. Romanus, I.I.; Egwu, O.A.; Ngozi, A.T.; Chidiebube, N.A.; Chika, E.P. (2009). Extended-spectrum β-lactamase (ESBL)-mediated resistance to antibiotics among Klebsiella Pneumoniae in Enugu Metropolis. Maced J Med Sci. 2:1-4 (In Nigerian).
- 28. Ruppé, E.; Hem, S.; Lath, S.; Gautier, V.; Ariey, F.; Sarthou, J.L.; Monchy, D.; Arlet, G. (2009). CTX-M β-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis 15:741-748.
- 29. Schmitt, J.; Jacobs, E.; Schmidt, H. (2007). Molecular characterization of extended-spectrum beta-lactamases in Enterobacteriaceae from patients of two hospitals in Saxony, Germany. J Clin Microbiol 56:241-249.
- 30. Seok, H.J.; Kwon, B.; Jung, H.L.; Seung, G.S.; Geun, H.K.; Ghil, J.J.; Young, H.K.; Byeong, C.J.; Sang, H.L. (2004). Molecular characterization of extended-spectrum beta-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean Nationwide Survey. J Clin Microbiol 42:2902-2906.
- 31. Slama, T.G. (2008). Gram-negative antibiotic resistance: there is a price to pay. Review. Critical Care. 12:4. http://ccforum.com/content/12/S4/S4
- 32. Smith, B.A.; Gabasan, A.; Zivin, T.; Sordillo, E.M.; Polsky, B. (2009). Management of an outbreak due to ESBL-positive Klebsiella pneumoniae (ESBL+KP) in an ICU: Focusing on the basics. AJIC 33(5):e121-e122.
- 33. Song, K.H.; Jeonn, J.H.; Park, W.B.; Park, S.W.; Kim, H.B.; Oh, M.D.; Lee, H.S.; Kim, N.J.; Choe, K.W. (2009). Clinical outcomes of spontaneous bacterial peritonitis due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species: A retrospective matched case-control study. BMC Infect Dis 9:41. http://www.biomecentral.com/1471-2334/9/41
- 34. Superti, S.V.; Augusti, G.; Zavascki, A.P. (2009). Risk factors for and mortality of extended-spectrum-b-lactamase-producing Klebsiella pneumoniae and Escherichia coli nosocomial bloodstream infections. Rev Inst Med Trop. S. Paulo 51:211-216 (In Portuguese).
- 35. Thierry, F.; Guillaume, A.; Valerie, G.; Talarmin A.; Raymond, B. (2006). Extended-spectrum-lactamase-producing Enterobacteriaceae, Central African Republic. Emerg Infect Dis 12:863-865.
- 36. Thomson, K.S. (2001). Controversies about extended-spectrum and ampc beta-lactamases. Emerg Infect Dis. 7:333.
- 37. Tofteland, S.; Haldorsen, B.; Dahl, K.H.; Simonsen, G.S.; Steinbakk, M.; Tumbarello, M.; Sali, M.; Trecarichi, E.M.; Leone, F.; Rossi, M.; Fiori, B.; De Pascale, G.; D'Inzeo, T.; Sanguinetti, M.; Fadda, G.; Cauda, R.; Spanu, T. (2008). Bloodstream infections caused by extended-spectrum-lactamase-producing Escherichia coli: Risk factors for inadequate initial antimicrobial therapy. Antimicrob Agents Chemother. 52:3244-3252.
- 38. Venkatachalam, K.V.; Wanzhi, H.; Mark, L.R.; Timothy, P. (1994). Characterization of TEM-1 B-lactamase mutants from position 238 to 241 with increased catalytic efficiency for ceftazidime. J Biol Chem 269:23444-23450.
- 39. Walsh, T.R.; Sundsfjord, A. (2007). Effects of phenotype and genotype on methods for detection of extended-spectrum-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J Clin Microbiol 45:199-205.
- 40. Winn, J.W.C.; Allen, S.D.; Janda, W.M.; Koneman, E.W.; Procop, G.W.; Schreckenberger, P.C.; Woods, G.L. (2008). Microbiological diagnosis. Color Atlas and Text. 6th ed. Guanabara Koogan, Rio de Janeiro.
Publication Dates
-
Publication in this collection
21 Dec 2011 -
Date of issue
Sept 2011
History
-
Accepted
14 Mar 2011 -
Reviewed
21 Oct 2010 -
Received
19 Sept 2010