Abstract
The contamination of ecosystems with heavy metals is an important issue in current world and remediation technologies should be in according to environmental sustainability concept. Bioemulsifier are promising agents to be used in metal removal and could be effective to many applications in environmental industries. The aims of this work was screening the potential production of bioemulsifier by microorganisms isolated from an oil contaminated mangrove, and evaluate cadmium and zinc removal potential of those strains from a hazardous industrial residue. From that, bioemulsifier-producing bacteria were isolated from urban mangrove sediments. Four isolates were identified as Microbacterium sp by 16S rRNA analysis and were able to reduce up to 53.3% of culture medium surface tension (TS) when using glucose as carbon and energy source and 20.2% when sucrose was used. Suspensions containing bioemulsifier produced by Microbacterium sp. strains show to be able to remove cadmium and zinc from contaminated industrial residue, and its ability varied according carbon source. Significant differences in metal removal were observed by all strains depending on the carbon source. When glucose was used, Cd and Zn removal varied from 17 to 41%, and 14 to 68%, respectively. However, when sucrose was used it was observed only 4 to a maximum of 15% of Cd removal, and 4 to 17% of Zn removal. When the same tests were performed after ethanol precipitation, the results were different: the percentages of removal of Zn (7-27%) and Cd (14-32%) were higher from sucrose cultures. This is the first report of heavy metals removal by bioemulsifier from Microbacterium sp.
Biosemulsifier; heavy metals; zinc; cadmium; Microbacterium; Bioremediation
bjm
ENVIRONMENTAL MICROBIOLOGY
Bioemulsifier production by Microbacterium sp. strains isolated from mangrove and their application to remove cadmiun and zinc from hazardous industrial residue
Erick AniszewskiI,II; Raquel Silva PeixotoI; Fábio Faria MotaIII; Selma Gomes Ferreira LeiteII; Alexandre Soares RosadoI,* * Corresponding Author. Mailing address: Instituto de Microbiologia Prof. Paulo de Góes, Universidade Federal do Rio de Janeiro, CCS, Bloco E subsolo, Ilha do Fundão, CEP 21941-590, Rio de Janeiro, RJ, Brasil.; Tel: 55(21) 2562.6740 Fax: 55(21) 2560.8344.; E-mail: asrosado@micro.ufrj.br
IInstituto de Microbiologia Prof. Paulo de Góes, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IIEscola de Química, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
IIILaboratório de Genômica Funcional e Bioinformática e Laboratório de Biologia Computacional e Sistemas, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz, Rio de Janeiro, Brasil
ABSTRACT
The contamination of ecosystems with heavy metals is an important issue in current world and remediation technologies should be in according to environmental sustainability concept. Bioemulsifier are promising agents to be used in metal removal and could be effective to many applications in environmental industries. The aims of this work was screening the potential production of bioemulsifier by microorganisms isolated from an oil contaminated mangrove, and evaluate cadmium and zinc removal potential of those strains from a hazardous industrial residue. From that, bioemulsifier-producing bacteria were isolated from urban mangrove sediments. Four isolates were identified as Microbacterium sp by 16S rRNA analysis and were able to reduce up to 53.3% of culture medium surface tension (TS) when using glucose as carbon and energy source and 20.2% when sucrose was used. Suspensions containing bioemulsifier produced by Microbacterium sp. strains show to be able to remove cadmium and zinc from contaminated industrial residue, and its ability varied according carbon source. Significant differences in metal removal were observed by all strains depending on the carbon source. When glucose was used, Cd and Zn removal varied from 17 to 41%, and 14 to 68%, respectively. However, when sucrose was used it was observed only 4 to a maximum of 15% of Cd removal, and 4 to 17% of Zn removal. When the same tests were performed after ethanol precipitation, the results were different: the percentages of removal of Zn (7-27%) and Cd (14-32%) were higher from sucrose cultures. This is the first report of heavy metals removal by bioemulsifier from Microbacterium sp.
Key words: Biosemulsifier, heavy metals, zinc, cadmium, Microbacterium, Bioremediation
INTRODUCTION
The contamination of ecosystems by heavy metals from human activities is an important issue. Remediation technologies should be in accordance with environmental sustainability concepts. Metals provide several unique challenges for remediation, as they cannot be degraded into innocuous products (18). For example, cadmium and zinc are common contaminants in environmental samples (2). Various bioremediation systems have been proposed to remove or neutralise toxic effects of metal contamination in the environment; however, low bioavailability of those metals in soils may limit the efficiency of removal (14).
For bioremediation approaches such as phytoremediation, synthetic chelators can be used to increase the phytoavailability of heavy metals, but such molecules are too expensive and hazardous to the environment (15). Therefore, utilisation of biosurfactants has been proposed as an alternative (11,15). Different surface active compounds (SACs) are synthesised by many prokaryotic and eukaryotic microorganisms, and due to their properties, microbial SACs have been exploited in environmental remediation techniques (6).
Biosurfactants are produced extracellularly or as part of the cell membrane by bacteria, yeasts and fungi. They comprise a unique class of compounds that have a variety of potential applications in industrial development and in the remediation of organic- and metal-contaminated sites (11). It is estimated that biosurfactants could capture a large part of the surfactant market in the near future (16) and that heavy metal removal is one of the most promising applications (3).
Many types of biosurfactants are synthesised by a number of microorganisms. For example, Pseudomonas aeruginosa produces rhamnolipids, and Bacillus subtilis produces a lipopeptide called surfactin (11). Franzetti and colleagues (6) demonstrated that the crude oil removal by bioemulsans produced by Gordonia sp. BS29 strain was comparable to the rhamnolipid one in the same experimental conditions and the promising use of this bioemulsans as washing agents for remediation of hydrocarbon-contaminated soils.
As far as we know, there is only one report about the production of extracellular polymers (EPS) with surfactant properties by a Microbacterium strain (12). However, utilisation of this bioemulsifier in heavy metals removal has not been evaluated.
This work shows the selection of four Microbacterium bioemulsifier-producing strains and the evaluation of glucose, sucrose and Trypic Soy Broth as carbon sources for bioemulsifier production. In addition, we present the potential utilisation of a suspension containing bioemulsifier produced by the isolated strains in Cd and Zn removal from hazardous industrial waste.
MATERIALS AND METHODS
Bacterial cultures
Bacterial strains were isolated from sediment of an oil-contaminated mangrove in Guanabara Bay (Rio de Janeiro, Brazil) by 1/10 dilution in saline solution. Serial tenfold dilution was prepared from the initial sediment suspensions, and 100-ul aliquots were surface-plated onto Trypic Soy Agar (TSA, Difco, NJ, USA). Plates were incubated at 28ºC. Long-term storage was at -80ºC in 20% (vol/vol) glycerol.
Molecular Identification of the Bioemulsifier producer strains
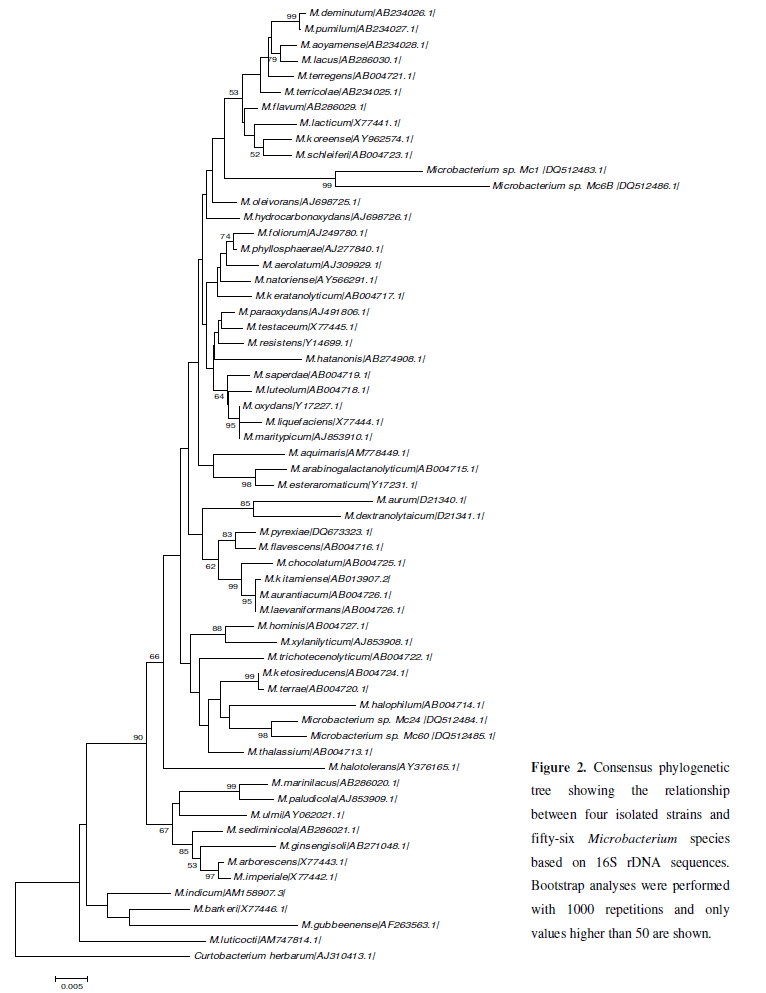
Genomic DNA of bacterial isolates was extracted using the Puregene DNA Isolation Kit (Gentra Systems, QIAGEN Inc.). DNA concentrations were determined by using a Gene Quant spectrophotometer (Amersham Pharmacia Biotech). Amplification of the 16S rDNA region was performed by PCR using universal primers pA and pH as previous described (9). The amplification products were purified with Wizard PCR Preps DNA Purification System and then directly sequenced in both directions by using pA and pH primers and an ABI Prism 3100 automatic sequencer (Applied Biosystems, Foster City). Preliminary identification of sequences was performed by blastn against the Genbank database 16/02/2009 (www.ncbi.nlm.nih.gov/blast) and Seqmatch program against the RDPII database (http://rdp.cme.msu.edu/). For determination of phylogenetic position within Microbacterium genus, 57 related sequences were aligned with ClustalX (17), and then a consensus phylogenetic tree was constructed by MEGA4 software (8) using the Neighbor-Joining method and 1000 replicates bootstrap.
Detection of emulsifying ability and biosurfactantproducing strains
Emulsifying capacity was determined by a technique used by Francy et al. (5). All experiments were performed in triplicate, and the average of the results is presented.
Single colonies with the ability to emulsify diesel oil were inoculated in 250 ml flasks containing 50 ml of three different culture media (mineral medium described previously (5) amended with 2% sucrose or 2% glucose as carbon source and TSB (Difco, NJ, USA). Every 24 hours during incubation, 5 ml of suspension was removed and centrifuged at 5000 x g for 20 minutes at 4º C during ten days. Then, 4.0 ml of the resulting cell free supernatant fluid was overloaded with 10% diesel oil and vortexed for 5 seconds, and the extent of emulsification was recorded after 2 hours of setting time. Surface tension (ST) measurements were taken of the whole culture broth and uninoculated control at room temperature using a Fisher Surface Tensiomat.
Removal of heavy metals from contaminated residue
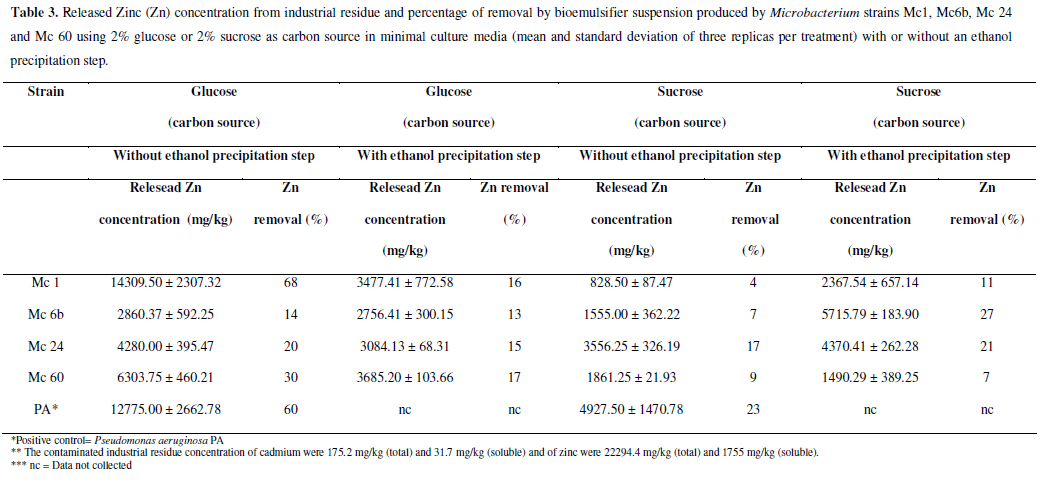
In this work, we utilized the solid residue produced by the Cia Mercantil and Industrial INGÁ, classified as hazardous substance. It is an industrial residue from the extraction process of Zn from calamine ore, situated at the Sepetiba Bay, in Rio de Janeiro, Brazil (43º50'W and 22º56'S). The contaminated industrial residue concentrations of zinc were 22294.4 mg/kg (total) and 1755 mg/kg (soluble) and concentrations of cadmium were 175.2 mg/kg (total) and 31.7 mg/kg (soluble). The heavy metal removal experiments were performed according to Mulligan et al. (10), using cell-free supernatant or suspensions partially purified by ethanol precipitation. The partially purified biomulsans were resuspended in 10 ml distilled water, and this suspension was used to wash the residue of Ingá Industrial Company (v:w/10:1). Summarizing, sediment washing were performed in centrifuge tubes. Pure distillated water was used as negative control. A biosurfactantproducing Pseudomonas aeruginosa PA, obtained from the culture collection, EQ-UFRJ, Brazil, was used as a positive control organism. Samples were taken after 24 h shaking and then centrifuged (5000×g, 10 min). Supernatant metal concentration was analyzed by atomic absorption spectrometry (Varian AA600). The percentage metal removal was determined based on the initial metal content in the sediment and all results are presented as percent metal removal. All experiments were performed in triplicate and the average of the results is presented. The differences between metal removal using sucrose or glucose as carbon source, with or without ethanol precipitation by each replicate of each strain were evaluated by t-test using STATISTICA 7 (StatSoft Inc., OK, USA).
RESULTS AND DISCUSSION
Screening strains for emulsifying ability
A total of 37 isolates were obtained, and 10 of those strains demonstrated the ability to emulsify diesel oil when submitted to an initial screening (data not shown). The 10 isolated strains that exhibited emulsifying ability were tested for the ability to produce cell-free supernatant that retained the emulsifying capacity.
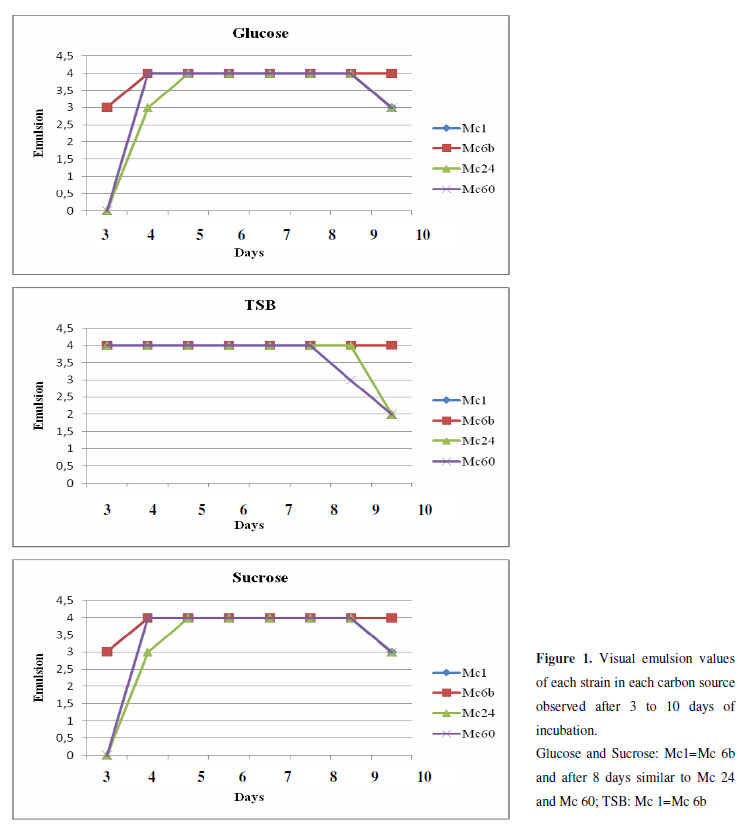
Four strains (Mc 1; Mc 6B; Mc 24 and Mc 60) showed maximal visual emulsification (level 4 of Fancy´s scale) and were able to produce excrete emulsifier to culture medium, as the emulsifying ability remained after cell wash (Figure 1). These strains were selected to further experiments. Krepsky and colleagues. (7) isolated biosurfactant-producing consortia from superficial mangrove sediment contaminated with oil and recommended the selection of biosurfactant producing bacteria from this environment. The authors suggested that the presence of large amounts of organic matter can enhance the biosurfactant production due to the metabolic stimulation. Mangroves are well known to harbour organic matter in abundance; thus, they are good habitats for searching for new bacterial species that produce surface-active agents.
Identification of bacterial strains
The 16S rDNA sequences obtained from isolated strains (Mc 1, Mc 6B, Mc 24 and Mc 60) were deposited in Genbank under accession numbers DQ512483, DQ512484, DQ512485 and DQ512486, respectively. Preliminary identification with blastn and seqmatch showed that all isolated strains belong to the genus Microbacterium. The 16S rDNA sequences from type-strains of all Microbacterium species and the newly isloated sequences (Mc 1, Mc 6B, Mc 24 and Mc 60) were used to construct a consensus phylogenetic tree (Figure 2). The Mc 1 and Mc 6B strains were clustered with M. oleivorans, M. hydrocarbonoxydans, M. schleiferi, M. koreensis, M. lacticum and M. flavum. In addition, blastn analysis showed that the Mc 6B and Mc 1 strains are more closely related to M. oleovarans [AJ698725.1] and M. schleiferi [Y17237.1]. On the other hand, the Mc 24 and Mc 60 strains were closely related to M. halophilum (Figure 2) and M. thalassium (blastn).
Biosurfactants produced by bacteria are capable of lowering surface and/or interfacial tension by partitioning at the water-air and water-oil interfaces improving oil degradation by other bacterial community members, and many studies to evaluate oil bioremediation are being developed. However, there are few studies to evaluate bioemulsifier use to remove metals from contaminated soils and residues, and the rhamnolipids from Pseudomonas aeruginosa has been being certainly the most studied (11).
As far as we know, no report involving heavy metal removal by Microbacterium biosurfactant/emulsifier is available. An important observation in this study is that four isolates from mangrove sediment produced biosurfactants: two Microbacterium strains (Mc1 and Mc6B) closely related to M. oleivorans and two strains (Mc24 and Mc60) related to M. thalassium and M. halophilum. Coincidently, the type-strains of M. thalassium (IFO 16060) and M. halophilum (IFO 16062) were also isolated from mangrove rhizosphere soil. Moreover, Microbacterium species, including M. oleivorans, have the ability to degrade crude oil (13). Altogether, these data could corroborate that the strains isolated in this study may presents bioremediation potential as biosurfactant producers.
Surface tension measurements and evaluation of heavy metal removal by free cell supernatant
Surface tension (ST) measurements were taken in triplicate from Microbacterium strains Mc 1, Mc 6b, Mc 24, and Mc 60 culture broth. When glucose and sucrose were used as carbon sources, all strains were able to reduce surface tension of the culture medium (Table 1). The best result was obtained by the Mc1 strain, which reduced 20% of culture medium ST, using sucrose as the carbon source. In addition, the Mc1 strain showed the best performance using sucrose as the carbon source, reducing 16.5% of culture medium ST. Lower surface tension reduction was observed in strains in TSB medium (approximately 5.8% of ST reduction). Glucose was selected as a carbon source for screening of bioemulsifier producers because glucose supports the production of many types of bioemulsifier, stimulates in situ production of bioemulsifier for remediation applications and is a nontoxic carbon source (1).
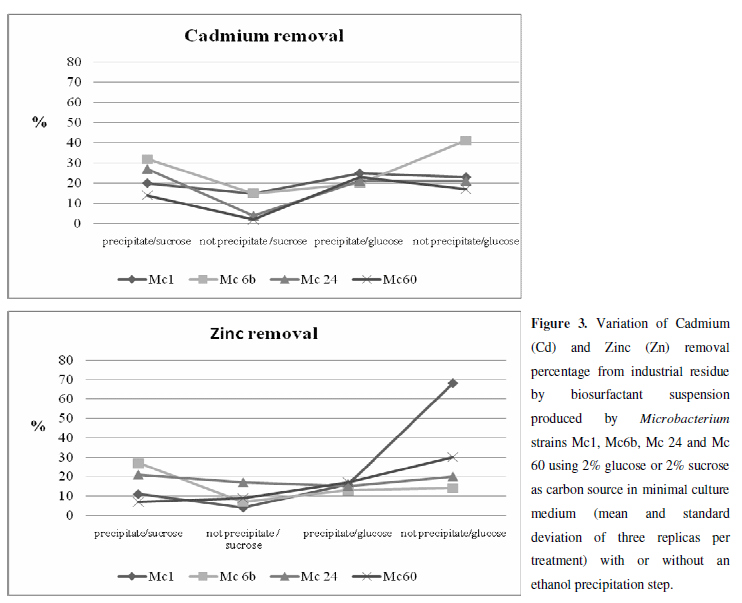
The suspensions containing bioemulsifier produced by selected Microbacterium strains successfully washed heavy metal residue, demonstrating the potential of this methodology of remediation. A residue control washed only with distilled water was used. Significant differences in heavy metals removal were observed in almost all strains according to the carbon source. No significant differences were observed only to strain Mc 1 in cadmium removal and to strain Mc 24 in zinc removal. Apparently, in these strains the carbon source didn´t influence the removal ability of cadmium or zinc, respectively (Tables 2 and 3).
Generally a better efficiency in cadmium removal was obtained by strains that used 2% glucose as the carbon source. In this condition, the strain Mc6b was more efficient for cadmium remediation, removing 41% of the insoluble fraction of cadmium from contaminated residue. Using sucrose as the carbon source, the same strain (Mc 6b) removed only 15% of cadmium, while the positive control strain removed 25% of cadmium (Table 2).
Microbacterium Mc1 strain removed 68% of released zinc from contaminated industrial waste (Table 3) when glucose was used as the carbon source. On the other hand, when sucrose was used, the same strain removed only 4% of Zn. This variance in removal efficiency according to carbon source is because biosurfactant/bioemulsifier production depends on the type of carbon source, as well as the type and amount of other nutrients, in the culture medium. Strains can use (or not as observed in Mc1 and Mc 24 strains to cadimium and zinc removal without ethanol precipitation, respectively (Table 4)) significant different metabolic pathways to form emulsion and to reduce surface tension, and the synthesis of hydrophilic and hydrophobic portions of biosurfactant molecules is substrate-dependent (4). This variation will probably depend to strain, medium conditions, removal target, and other factors.
Despite the better efficiency of metal removal observed when using glucose, sucrose is a valuable carbon source. Sucrose is a low cost carbon source that is an important factor in selecting substrates for producing biotechnology products at industrial levels. It can be obtained from low-cost products such as molasses and sugar cane. Our results show that the Microbacterium strains Mc 1, Mc 6b, Mc 24 and Mc 60 were able to produce bioemulsifier when sucrose was used as the carbon source. Accordingly, sucrose and other cheaper carbon sources should be considered in further applications in biosurfactant production, and we are currently optimising these conditions.
In order to precipitate the emulsifier content from the suspensions free of cells, we performed a partial purification with ethanol. We noticed that partially purified suspensions obtained from strains Mc 1 and Mc 60 showed better efficiency in removing cadmium when grown in glucose, whereas partially purified suspensions from strains Mc6b and Mc24 demonstrated better efficiency when grown in sucrose (Table 2). The removal efficiency of the strains was between 14% and 32% for the insoluble fraction of cadmium residue from sucrose cultures. The strains Mc 1 and Mc 60 demonstrated better efficiency in zinc removal when grown in glucose, whereas the strains Mc6b and Mc24 obtained better efficiency when grown in sucrose (Table 3). The removal efficiencies of the strains were between 7% and 27% of the insoluble fraction of zinc in the residue; higher amounts of zinc were removed in cultures that used sucrose as the carbon source.
The strain Mc 6b demonstrated higher levels of cadmium and zinc removal from contaminated residue when grown in sucrose; however, when grown using the glucose as carbon source, the same strain demonstrated lower levels of cadmium and zinc removal. On the other hand, strains Mc 1 and Mc 60 demonstrated better ability to remove cadmium and zinc when grown in glucose rather than sucrose. Comparing all results together, we observe that, in contrast to the results obtained from cell-free supernatant wash, generally, the metal removal from partially purified biosurfactant was higher when the strains were grown in sucrose (Figure 3). These results suggest that (1) the chemical structures and/or amount of biosurfactants are carbon substrate-dependent, as demonstrated by Das et al. (2008), and (2) different chemical structures can present different specificities, even when produced by the same microorganism.
The high levels of removal of cadmium and zinc by partially purified strains Mc 6b and Mc 24 using sucrose as the carbon source is a promising example of removing heavy metals at lower costs that could stimulate further research with this substrate. The isolation of Microbacterium strains that produce biosurfactants and utilisation of these biosurfactants in heavy metal removal is novel. Searching for new microorganisms and their tenso-active products may be crucial to environmental sustainability, as such work will give more clues to further develop efficient approaches to remediate metal-polluted sites.
ACKNOWLEDGMENTS
This work was supported by grants from the Brazilian National Research Council (CNPq) and Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Submitted: March 24, 2009; Returned to authors for corrections: May 08, 2009; Approved: August 23, 2009.
- 1. Bodour, A.A.; Drees, K.P.; Maier, R.M. (2003). Distribution of Biosurfactant-Producing Bacteria in Undisturbed and Contaminated Soils. Appl. Environ. Microbiol. 69 (6), 3280-3287.
- 2. Buggy, C.J.; Tobin, J.M. (2008). Seasonal and spatial distribution of metals in surface sediment of an urban estuary. Environ. Pollut. 155(2), 308-19.
- 3. Dahrazma, B.; Mulligan, C.N. (2007). Investigation of the removal of heavy metals from sediments using rhamnolipid in a continuous flow configuration. Chemosphere 69(5),705-711.
- 4. Desai, J.D.; Banat, I.M. (1997). Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61(1), 47-64.
- 5. Francy, D.S.; Thomas, J.M.; Raymond, R.L.; Ward, C.H. (1991). Emulsification of hydrocarbons by subsurface bacteria. J. Ind. Microbiol. 8, 237-246.
- 6. Franzetti, A.; Caredda, P.; Ruggeri, C.; La Colla, P.; Tamburini, E.; Papacchini, M.; Bestetti, G. (2009). Potential applications of surface active compounds by Gordonia sp. strain BS29 in soil remediation technologies. Chemosphere 75(6), 801-807.
- 7. Krepsky, N.; Silva, F.S.; Fontana, L.F.; Crapéz, M.A. (2007). Alternative methodology for isolation of biosurfactant-producing bacteria. Braz. J. Biol. 67 (1), 117-124.
- 8. Kumar, S.; Tamura, K.; Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5,150-163.
- 9. Massol-Deya, A.A.; Odelson, D.A.; Hichey, R.P.; Tiedje, J.M. (1995). Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA). In: Akkermans, A.D.L.; van Elsas, J.D.; de Bruijn, F.J. (eds). Molecular Microbial Ecology Manual. Kluwer Academic Dordrecht, The Netherlands, pp 1-8.
- 10. Mulligan, C.N.; Yong, R.N.; Gibbs, B.F. (2001) .Heavy metal removal from sediments by biosurfactants. J. Hazard. Mater. 85, 111-125.
- 11. Mulligan, C. (2005). Environmental applications for biossurfactants. Environ. Pollut. 133, 183-198.
- 12. Ortega-Morales, B.O. ; Santiago-Garcia, M.J. ; Chan-Bacab, X. ; Moppert X.; Miranda-Tello, E.; Fardeau, M.L.; Carrero, J.C.; Bartolo-Pérez, P.; Valadéz-González, A.; Guezennec, J. (2007). Characterization of extracellular polymers synthsized by tropical interdial biofilm bacteria. J. Appl. Microbiol. 102, 254-264.
- 13. Schippers, A.; Bosecker, K.; Sproer, C.; Schumann, P. (2005). Microbacterium oleivorans sp. nov. and Microbacterium hydrocarbonoxydans sp. nov., novel crude-oil-degrading Gram-positive bacteria. Int. J. Syst. Evol. Microbiol 55, 655-60.
- 14. Sheng, X.; Xia, J.J. (2006). Improvement of rape (Brassica napus) plant growth and cadmium uptake by cadmium-resistent bacteria. Chemosphere 64, 1036-1042.
- 15. Sheng, X.; He, L.; Wang, Q.; Ye, H.; Jiang, C. (2008). Effects of inoculation of biosurfactant-producing Bacillus sp. J119 on plant growth and cadmium uptake in a cadmium-amended soil. J. Hazard. Mater. 155(1-2), 17-22.
- 16. Singh, A.; Van Hamme, J.D.; Ward, O.P. (2007). Surfactants in microbiology and biotechnology: Part 2, Application aspects. Biotech Adv 25(1), 99-121.
- 17. Thompson, J.D.; Gibson, T. J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. (1997). The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality tools. Nucleic Acids Res. 24, 4876-4882.
- 18. Wiatrowski, H.; Barkay, T. (2005). Monitoring of microbial metal transformations in the environment. Curr. Opin. Biotech. 16, 261-268.
Publication Dates
-
Publication in this collection
23 Nov 2009 -
Date of issue
Mar 2010
History
-
Accepted
23 Aug 2009 -
Reviewed
08 May 2009 -
Received
24 Mar 2009