Abstract
Early diagnosis plays a vital role in controlling tuberculosis. The conventional methodology is slow, with results taking several weeks, in addition to having low sensitivity, especially in clinical paucibacillary samples. The objective of this study was to evaluate the use of polymerase chain reaction (PCR) on solid medium culture for a rapid diagnosis of tuberculosis, mainly in cases of negative sputum smears. Forty sputum samples were collected from inpatients with tuberculosis treated for less than 2 days. Bacilloscopy, PCR for sputum, culture on Löwestein-Jensen (LJ) solid medium, and daily PCR from culture were performed on each sample. DNA extracted from the BCG vaccine, which contains attenuated bacillus Calmette-Guérin, was used as the positive control. Smear microscopy showed 68.6% sensitivity, 80% specificity, 96% positive predictive value, and 26.7% negative predictive value, with culture on LJ medium as the gold standard. Culture at day 28 showed 74.3% sensitivity and 100% specificity. PCR of DNA extracted from sputum amplified a 1027-bp fragment of the 16s RNA gene, showing 22.9% sensitivity and 60% specificity. PCR performed with DNA extracted from daily culture showed that, from the 17th to the 40th day, the sensitivity (85.7%) and specificity (60%) were constant. We conclude that a 17-day culture is a good choice for rapid diagnosis and to interfere with the transmission chain of tuberculosis.
Tuberculosis; Mycobacteria; PCR
Braz J Med Biol Res, June 2010, Volume 43(6) 543-548
The use of polymerase chain reaction for early diagnosis of tuberculosis in Mycobacterium tuberculosis culture
M. Chagas1,
Correspondence and Footnotes
 R.M. da Silva2, M.L. Bazzo1 and J.I. dos Santos1
R.M. da Silva2, M.L. Bazzo1 and J.I. dos Santos1
1Departamento de Análises Clínicas, Universidade Federal de Santa Catarina, Florianópolis, SC, Brasil
2Programa de Mestrado em Ciências da Saúde, Universidade do Sul de Santa Catarina, Florianópolis, SC, Brasil
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
Early diagnosis plays a vital role in controlling tuberculosis. The conventional methodology is slow, with results taking several weeks, in addition to having low sensitivity, especially in clinical paucibacillary samples. The objective of this study was to evaluate the use of polymerase chain reaction (PCR) on solid medium culture for a rapid diagnosis of tuberculosis, mainly in cases of negative sputum smears. Forty sputum samples were collected from inpatients with tuberculosis treated for less than 2 days. Bacilloscopy, PCR for sputum, culture on Löwenstein-Jensen (LJ) solid medium, and daily PCR from culture were performed on each sample. DNA extracted from the BCG vaccine, which contains attenuated bacillus Calmette-Guérin, was used as the positive control. Smear microscopy showed 68.6% sensitivity, 80% specificity, 96% positive predictive value, and 26.7% negative predictive value, with culture on LJ medium as the gold standard. Culture at day 28 showed 74.3% sensitivity and 100% specificity. PCR of DNA extracted from sputum amplified a 1027-bp fragment of the 16s RNA gene, showing 22.9% sensitivity and 60% specificity. PCR performed with DNA extracted from daily culture showed that, from the 17th to the 40th day, the sensitivity (85.7%) and specificity (60%) were constant. We conclude that a 17-day culture is a good choice for rapid diagnosis and to interfere with the transmission chain of tuberculosis.
Key words: Tuberculosis; Mycobacteria; PCR
Introduction
Since 1998, tuberculosis (TB) has been regarded as a worldwide emergency because it is the leading cause of death by an infectious disease in adults (1-5). The estimate is that approximately two billion people harbor the bacillus (6). The estimated annual global incidence of TB in 2005 was 136 cases per 100,000 inhabitants. In the Americas, Brazil ranked in 15th place, with 112,000 cases (5).
An early diagnosis plays a vital role in controlling TB. However, the conventional methodology is slow, with laboratory results taking several weeks, in addition to having low sensitivity, especially in clinical paucibacillary samples (3,7-9). The reversal of this scenario will require the development of new strategies to increase the quality and speed of TB diagnosis. The estimate for the next 20 years is that the increased detection of cases could reduce the incidence by 41% (5).
Detection of mycobacterial DNA directly from sputum by amplification of the 16S rDNA gene allows rapid species identification (10). However, the amplification of this gene in sputum has been challenging, with sensitivity well below the desired values for the diagnosis (11).
The present study was conducted to evaluate the efficiency of polymerase chain reaction (PCR) directly from sputum and culture on Löwenstein-Jensen (LJ) solid medium in which the colonies are not visible.
Material and Methods
Sputum samples were obtained from inpatients with a clinical diagnosis of TB, with a maximum of 2 days of treatment, admitted to a TB reference hospital, and processed within 2 h after collection.
Each sample was homogenized and divided into three parts, one for slide preparation for bacilloscopy (Ziehl-Nielsen), one for DNA extraction and PCR, and the third for decontamination and distribution in 7 culture tubes containing LJ solid medium. Smear preparation, Ziehl-Nielsen staining and slide reading followed the recommendations outlined in the Manual of Tuberculosis Bacteriology (12).
DNA extraction from sputum was performed by alkaline lysis (13): sputum was resuspended in 400 µL GTE (50 mM glucose, 25 mM Tris-HCl, pH 8.0, and 10 mM EDTA), followed by 400 µL cell lysis solution (1% SDS, 0.2 M NaOH). The solution pH was neutralized with a 3-M solution of potassium acetate, pH 4.8 to 5.0. The sample was then treated with 20 mg/mL proteinase K solution (Invitrogen®, USA). The extraction was performed with phenol/chloroform/isoamyl alcohol (25:24:1) and the DNA was precipitated in ethanol in the presence of salt and resuspended in 20 µL TE (10 mM Tris, pH 7.4, 1 mM EDTA). Quality control of the DNA extracted and verification of the presence of PCR inhibitors were performed by amplifying the 350-bp fragment using the pair of oligonucleotide primers ZR-244 and F-285, which target the 16s RNA gene (14). Oligonucleotide primers were designed from the corresponding nucleotides of the 16S rRNA gene (15,16). Amplification by PCR for the detection of mycobacteria in clinical samples was performed using specific primers, F-285 - 5’-AGAGTTTGATCCTGGCTCAG-3’ and MYC-264 - 3’-TGCACACAGGCCACAAGGGA-5’, which amplify a 1027-bp fragment. The standard sample used as positive control of PCR amplification was DNA extracted from the BCG vaccine, which contains attenuated bacillus Calmette-Guérin.
The samples were cultured on LJ medium by the Petroff technique (12). The tubes with cultures were incubated in an inclined position at temperatures ranging from 35° to 37°C.
To detect the presence of mycobacterial DNA in cultures before visualizing the colonies, material for PCR analysis was collected daily by scraping part of the surface of the medium in the culture tubes. Scraping of the medium was started 24 h after seeding (day 1).
Nine culture tubes were seeded for each sample and numbered 1, 2, 3, 4, 5, 6, 7, 14, and 42. Tube No. 42 was used to check bacterial growth up to 42 days.
A sterile wood stick was used to scrape the culture surface. On day 1, an aliquot was removed from tube 1 by scraping the surface of the LJ medium, and after a further 24 h (day 2) another aliquot was removed from tube 2 and so on, every 24 h, until tube 7. Tubes 1, 2, 3, 4, 5, and 6 were used again to scrape the aliquots of days 8, 9, 10, 11, 12, and 13, respectively. On day 14, tube 14, which was still untouched, was used. The collection of these aliquots was finalized when PCR was positive, or up to 42 days for the negative cultures. All collected aliquots were resuspended in GTE and submitted to the same procedure for PCR and DNA extraction as described previously. The cultures that showed no bacterial growth up to 42 days of culture were considered to be negative.
Sensitivity, specificity, predictive values, and odds ratio were calculated for each assessment with their respective confidence intervals (95%), taking as the gold standard the culture on the 42nd day, using MedCalc for Windows, version 9.6.0.0.
The research project was approved by the Ethics Committee in Human Research (CEPSH) of the Federal University of Santa Catarina under number 227/2006. All patients gave written informed consent to participate in the study.
Results
Bacilloscopy was positive in 25 (62.5%) of the 40 samples evaluated. PCR performed directly on DNA extracted from sputum was positive in 14 (35%) samples and negative in 26 (65%). No relationship was found between the amount of bacilli and the results of PCR.
All samples were cultured on LJ medium. Those which had positive PCR in DNA extracted directly from sputum were cultured and the time of colony growth was recorded. The remaining 26 samples were also cultured, but for these, in addition to the daily checking of colony emergence, a daily scraping of the medium surface was also performed to collect an aliquot for DNA extraction and PCR. Of the 40 cultured samples, five showed no mycobacterial growth by the 42nd day, with two PCR-positive sputum samples. Of the 26 cultured samples, which showed no positive PCR in sputum, 23 (88.5%) were positive by PCR on different days and 3 (11.5%) remained negative after the 40th PCR and culture day.
Of the total 40 samples cultured, 5 (12.5%) were negative, 5 (12.5%) were positive by 7 days, 5 (12.5%) were positive by 14 days, 6 (15%) were positive by 21 days, 10 (25%) by 28 days, 1 (2.5%) by 40 days, and 8 (20%) by 42 days of culture.
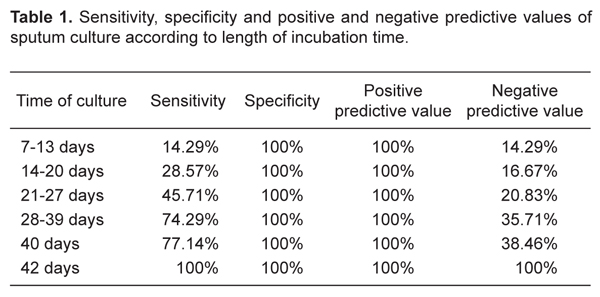
Bacilloscopy showed 68.6% sensitivity, 80% specificity, 96% positive predictive value, and 26.7% negative predictive value, considering the culture as the gold standard. Sensitivity, specificity, positive and negative predictive values for different incubation periods are listed in Table 1.
Figure 1A illustrates the receiver operating characteristic (ROC) curve for culture at day 21 of incubation. The area under the curve was 0.729 and the P value 0.0311; sensitivity was 45.7% and specificity 100%. Figure 1B presents the ROC curve for culture at day 28 of incubation. The area under the curve was 0.871 and the P value 0.0001; sensitivity was 74.3% and specificity 100%.
Table 2 shows the sensitivity, specificity, positive and negative predictive values grouped into different incubation periods for PCR in samples of solid culture medium.
The analysis of all values defines the cut-off point of PCR at day 17 of culture. Statistical significance (P < 0.05) was observed only from that day on. All values remained constant and did not vary.
Figure 2 shows the ROC curve for PCR on solid culture medium after 17 days of incubation. The area under the curve was 0.729, the P value 0.0311; sensitivity was 85.7% and specificity 60%.
Receiver operating characteristic curve for PCR at day 17 of culture. The area under the curve was 0.729, the P value 0.0311. Sensitivity was 85.7% and specificity 60.0%.
Sensitivity, specificity and positive and negative predictive values of sputum culture according to length of incubation time.
Sensitivity, specificity, positive predictive value and negative predictive value of polymerase chain reaction (PCR) for sputum samples and for samples at different days of cultivation.
Discussion
An increased detection of pulmonary TB cases was considered to be an additional global strategy for the 2006-2009 period (17).
The sampling of this study consisted of 40 respiratory specimens. On average, each sample was tested 10 times by PCR and, therefore, about 415 PCR assays were performed, a very significant number, considering that about 5000 samples are tested each year in the State of Santa Catarina, 1500 of which are positive (18).
This study was carried out to identify a method capable of improving the sensitivity of PCR culture. This procedure seems to improve the ability of PCR detection of the target gene, since it extracts pure DNA from mycobacteria in contrast to what occurs with DNA extracted directly from sputum. In the tests performed here, we observed that PCR from colonies grown on LJ presented 100% sensitivity in the amplification of 16S rDNA, while sensitivity values in sputum ranged from 22 to 72.4%, with these values being always linked to sputum quality (11,19). Lucchini et al. (20) used a liquid medium to assess PCR in cultures and obtained increased sensitivity.
Most studies that evaluate molecular biology techniques for TB diagnosis have been conducted in industrialized countries, with few series evaluating the clinical usefulness of such techniques under routine conditions. In developing countries, the use of molecular biology techniques for diagnosis is not yet a priority due to the high cost, technical difficulties and operational implementation, even in reference laboratories. Furthermore, there is a lack of information about the clinical pertinence and/or cost-effectiveness in different clinical scenarios (17).
In the present study, bacilloscopy showed 68.6% sensitivity and 80% specificity. The positive predictive value was 96%. Bacilloscopy presents sensitivity that can range from 20 to 80% (11,21-26). This method is recommended by the Stop TB program (6) because, although its sensitivity varies, it has already been shown that if three samples are used there is improvement in sensitivity to values between 80 and 90%. The other bacteriological and molecular methods shall be reserved for cases in which bacilloscopy is negative. In coming years there should be changes in this conduct because of the challenges encountered by the frequent isolation, in humans, of new species of mycobacteria considered nonpathogenic or even mycobacteria of the TB complex, that show a different sensitivity profile to tuberculostatic agents and to tuberculinic tests. In this case, the use of techniques for isolation, species identification and determination of susceptibility profile is necessary (27,28).
The intensity of amplification is different for each sample and appears to be related to the number of bacilli found by bacilloscopy. Ieven and Goossens (29) stated that some investigators ascribe the different results obtained by different methodologies when using the same sample to the unequal distribution of mycobacteria present in sputum and the difficulty in obtaining perfect sample homogeneity.
According to Cheng et al. (30), early studies on nucleic acid amplification tests were carried out in laboratories that did not use clinical information in the decision-making process. However, clinical suspicion of tuberculosis is very important in determining the utility of these tests.
Data show that there is no correlation between the results of bacilloscopy and PCR, probably, as already stated (29), because of the difficulty in obtaining perfect homogenization of bacilli in sputum. Ratnam et al. (31) showed that the decontamination procedures used to facilitate mycobacteria isolation also killed them, and that the percentage of these dead organisms varies depending on the method used and the species of mycobacteria present in the sample (31).
Culture on solid medium, because of the time of Mycobacterium tuberculosis (MTB) growth, can become positive in up to 42 days. This growth time may vary if the sample contains only MTB or is mixed, containing rapid growth mycobacteria, which seems to have been the case in the present study, since growth was observed in 10 samples between 7 and 14 days of cultivation.
In a review article, Wattal (32) analyzed 7 studies that compared different culture media with regard to the time of mycobacterial growth. For the LJ solid medium, the median growth was 26.5 days, in agreement with the results presented here.
We believe that the rapid growth in up to 14 days can be due to mycobacteria other than MTB of rapid growth or simply to the fact that the samples were processed within a maximum of 2 h after collection, resulting in a lower loss of mycobacteria by the action of enzymes present in sputum, which may occur in stored samples.
Although the odds ratio for the 21st day of culture is 9.308, the P value only shows statistical significance from the 28th day of culture on and, therefore, the cut-off point for culture in this study was 28 days. Negi et al. (9), in a comparative study of solid and liquid culture and PCR, found 48.9% sensitivity and 100% specificity on the 24th day of culture, values which were detected on the 21st day in the present study. This can be seen in Figure 1, which compares the ROC curves for days 21 and 28 of culture. Sensitivity was 45.7 and 74.3% on the 21st and 28th days, respectively. The area under the curve for day 28 was 0.871, a higher value than found for day 21, which was 0.729. According to Buijtels and Petit (33), mycobacterial growth is highly affected by the process of sample decontamination; however, this procedure is very important because sputum contains a variety of microorganisms that can grow much faster than MTB. Other studies have shown that this procedure kills 70 to 90% of viable bacilli, changing the low viability of the sample both for culture and for PCR (29,33).
The sensitivity and specificity of PCR performed from DNA extraction of MTB culture indicate a cut-off point at day 17 of cultivation followed by PCR for the detection of mycobacterial DNA. Regarding the culture cut-off point, the present study expedited diagnosis within 11 days. Lucchini et al. (20) conducted a study for the early detection of M. tuberculosis in liquid culture medium followed by PCR and obtained a small positive sample. They concluded that PCR performed on samples cultured in liquid medium after 5 days of inoculation can detect M. tuberculosis earlier than culture alone, reinforcing the importance of culture associated with a molecular technique.
From the 14th day of cultivation followed by PCR the odds ratio (6.0) increased considerably, but the P value showed significance only from the 17th day (P = 0.03), which remained constant until the 39th day.
The ROC curve for PCR after 17 days of cultivation showed 85.7% sensitivity and 60% specificity. Moreover, the area of the ROC curve with a value of 0.729 and a P value of 0.03 indicated statistical significance in relation to the gold standard and increased the value of PCR sensitivity. In this case, PCR sensitivity was much higher than the 22% value obtained when PCR was performed directly from sputum. The comparative ROC curve of the methods used indicated that PCR performed with DNA extracted from 17-day cultivation was the best alternative to achieve a more rapid TB diagnosis, and recommended its combination with techniques traditionally employed in public health clinical laboratories.
The present results show that the use of PCR combined with culture can reduce the time of TB diagnosis to 17 days and can effectively intervene in the epidemiological chain of TB.
References
1. Lopez-Moreno S, Garrido-Latorre F, Hernandez-Avila M. [Historical development of epidemiology: is growth as scientific discipline]. Salud Publica Mex 2000; 42: 133-143.
2. Hong SW. Preventing nosocomial Mycobacterium tuberculosis transmission in international settings. Emerg Infect Dis 2001; 7: 245-248.
3. Neonakis IK, Gitti Z, Krambovitis E, Spandidos DA. Molecular diagnostic tools in mycobacteriology. J Microbiol Methods 2008; 75: 1-11.
4. Rosseti MLR, Sperhacke RD. Tuberculose. In: Rosseti ML, da Silca CD, Rodrigues J (Editors), Doenças infecciosas: Diagnóstico molecular. Vol. 1. Rio de Janeiro: Guanabara Koogan; 2005. p 47-59.
5. World Health Organization. Implementing the Stop TB strategy. A handbook for national tuberculosis control programmes. Geneva: WHO/HTM/TB/2008; 2008.
6. World Health Organization. The global plan to stop TB. Geneva: WHO; 2006.
7. World Health Organization. Global tuberculosis control. Geneva: WHO. WHO Report 2001; 2001.
8. Soini H, Musser JM. Molecular diagnosis of mycobacteria. Clin Chem 2001; 47: 809-814.
9. Negi SS, Khan SF, Gupta S, Pasha ST, Khare S, Lal S. Comparison of the conventional diagnostic modalities, Bactec culture and polymerase chain reaction test for diagnosis of tuberculosis. Indian J Med Microbiol 2005; 23: 29-33.
10. Waleria-Aleixo A, Kroon EG, Campos MA, Margutti-Pinto ME, Bonjardim CA, Ferreira PC. Heteroduplex mobility assay for rapid, sensitive and specific detection of mycobacteria. Diagn Microbiol Infect Dis 2000; 36: 225-235.
11. Bazzo ML. Método de identificação e caracterização de micobactérias para uso em diagnóstico de rotina nos laboratórios de saúde e determinação da resistência. [Doctoral thesis]. Belo Horizonte: Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais; 2006.
12. Ministério da Saúde. Fundação Nacional de Saúde. Coordenação Nacional de Pneumologia Sanitária. Centro de Referência Prof. Hélio Fraga. Manual de bacteriologia da tuberculose. 2ª edn. Rio de Janeiro: Fundação Nacional de Saúde; 1994.
13. Sambrook J, Russel DW. Molecular cloning. A laboratory manual. 3rd edn. Vol. 3. New York: Cold Spring Harbor Laboratory Press; 2001.
14. Rogall T, Flohr T, Bottger EC. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol 1990; 136: 1915-1920.
15. Hughes MS, Skuce RA, Beck LA, Neill SD. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol 1993; 31: 3216-3222.
16. Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger EC. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol 1990; 28: 1751-1759.
17. Kritski AL. Evaluating the efficiency of polymerase chain reaction in diagnosing pulmonary tuberculosis in indigenous and non-indigenous patients. J Bras Pneumol 2006; 32: 13-14.
18. SPS/DVE/SES/SC. Secretaria de Políticas de Saúde - Diretoria de Vigilância Epidemiológica. Florianópolis: Secretaria de Estado da Saúde de Santa Catarina; 2008.
19. Bazzo ML, Ferreira LAP, Silva RM, Scheffer M, Chagas M, Severino JL, et al. Relação entre a qualidade de amostras de escarro e o diagnóstico de micobacterioses por PCR. Arq Cat Med 2004; 33: 23-27.
20. Lucchini GM, Wegmann K, Sauer M, Altwegg M. Early detection of Mycobacterium tuberculosis by culture on BBL MGIT medium followed by PCR. J Microbiol Met 1996; 27: 97-107.
21. Chakravorty S, Dudeja M, Hanif M, Tyagi JS. Utility of universal sample processing methodology, combining smear microscopy, culture, and PCR, for diagnosis of pulmonary tuberculosis. J Clin Microbiol 2005; 43: 2703-2708.
22. Bruchfeld J, Aderaye G, Palme IB, Bjorvatn B, Kallenius G, Lindquist L. Sputum concentration improves diagnosis of tuberculosis in a setting with a high prevalence of HIV. Trans R Soc Trop Med Hyg 2000; 94: 677-680.
23. Garay JE. Analysis of a simplified concentration sputum smear technique for pulmonary tuberculosis diagnosis in rural hospitals. Trop Doct 2000; 30: 70-72.
24. Perera J, Arachchi DM. The optimum relative centrifugal force and centrifugation time for improved sensitivity of smear and culture for detection of Mycobacterium tuberculosis from sputum. Trans R Soc Trop Med Hyg 1999; 93: 405-409.
25. Aslanzadeh J, de la Viuda M, Fille M, Smith WB, Namdari H. Comparison of culture and acid-fast bacilli stain to PCR for detection of Mycobacterium tuberculosis in clinical samples. Mol Cell Probes 1998; 12: 207-211.
26. Habeenzu C, Lubasi D, Fleming AF. Improved sensitivity of direct microscopy for detection of acid-fast bacilli in sputum in developing countries. Trans R Soc Trop Med Hyg 1998; 92: 415-416.
27. Somoskovi A, Dormandy J, Mayrer AR, Carter M, Hooper N, Salfinger M. "Mycobacterium canettii" isolated from a human immunodeficiency virus-positive patient: first case recognized in the United States. J Clin Microbiol 2009; 47: 255-257.
28. Viana-Niero C, Lima KV, Lopes ML, Rabello MC, Marsola LR, Brilhante VC, et al. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol 2008; 46: 850-855.
29. Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev 1997; 10: 242-256.
30. Cheng VC, Yew WW, Yuen KY. Molecular diagnostics in tuberculosis. Eur J Clin Microbiol Infect Dis 2005; 24: 711-720.
31. Ratnam S, Stead FA, Howes M. Simplified acetylcysteine-alkali digestion-decontamination procedure for isolation of mycobacteria from clinical specimens. J Clin Microbiol 1987; 25: 1428-1432.
32. Wattal C. Improving bacteriological diagnosis of tuberculosis. Indian J Pediatr 2002; 69 (Suppl 1): S11-S19.
33. Buijtels PC, Petit PL. Comparison of NaOH-N-acetyl cysteine and sulfuric acid decontamination methods for recovery of mycobacteria from clinical specimens. J Microbiol Methods 2005; 62: 83-88.
Address for correspondence: R.M. da Silva, Rua Moçambique, 852, 88060-415 Florianópolis, SC, Brasil. Fax: +48-3260-7008. E-mail: rosemaurici@hotmail.com
Received November 7, 2009. Accepted April 9, 2010. Available online April 22, 2010. Published June 11, 2010.
The Brazilian Journal of Medical and Biological Research is partially financed by
- 1. Lopez-Moreno S, Garrido-Latorre F, Hernandez-Avila M. [Historical development of epidemiology: is growth as scientific discipline]. Salud Publica Mex 2000; 42: 133-143.
- 2. Hong SW. Preventing nosocomial Mycobacterium tuberculosis transmission in international settings. Emerg Infect Dis 2001; 7: 245-248.
- 3. Neonakis IK, Gitti Z, Krambovitis E, Spandidos DA. Molecular diagnostic tools in mycobacteriology. J Microbiol Methods 2008; 75: 1-11.
- 4. Rosseti MLR, Sperhacke RD. Tuberculose. In: Rosseti ML, da Silca CD, Rodrigues J (Editors), Doenças infecciosas: Diagnóstico molecular. Vol. 1 Rio de Janeiro: Guanabara Koogan; 2005. p 47-59.
-
5World Health Organization. Implementing the Stop TB strategy. A handbook for national tuberculosis control programmes Geneva: WHO/HTM/TB/2008; 2008.
- 6. World Health Organization. The global plan to stop TB Geneva: WHO; 2006.
- 7. World Health Organization. Global tuberculosis control. Geneva: WHO. WHO Report 2001; 2001.
- 8. Soini H, Musser JM. Molecular diagnosis of mycobacteria. Clin Chem 2001; 47: 809-814.
- 9. Negi SS, Khan SF, Gupta S, Pasha ST, Khare S, Lal S. Comparison of the conventional diagnostic modalities, Bactec culture and polymerase chain reaction test for diagnosis of tuberculosis. Indian J Med Microbiol 2005; 23: 29-33.
- 10. Waleria-Aleixo A, Kroon EG, Campos MA, Margutti-Pinto ME, Bonjardim CA, Ferreira PC. Heteroduplex mobility assay for rapid, sensitive and specific detection of mycobacteria. Diagn Microbiol Infect Dis 2000; 36: 225-235.
- 11. Bazzo ML. Método de identificação e caracterização de micobactérias para uso em diagnóstico de rotina nos laboratórios de saúde e determinação da resistência. [Doctoral thesis]. Belo Horizonte: Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais; 2006.
- 12. Ministério da Saúde. Fundação Nacional de Saúde. Coordenação Nacional de Pneumologia Sanitária. Centro de Referência Prof. Hélio Fraga. Manual de bacteriologia da tuberculose 2ª edn. Rio de Janeiro: Fundação Nacional de Saúde; 1994.
- 13. Sambrook J, Russel DW. Molecular cloning. A laboratory manual 3rd edn. Vol. 3. New York: Cold Spring Harbor Laboratory Press; 2001.
- 14. Rogall T, Flohr T, Bottger EC. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol 1990; 136: 1915-1920.
- 15. Hughes MS, Skuce RA, Beck LA, Neill SD. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol 1993; 31: 3216-3222.
- 16. Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger EC. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol 1990; 28: 1751-1759.
- 17. Kritski AL. Evaluating the efficiency of polymerase chain reaction in diagnosing pulmonary tuberculosis in indigenous and non-indigenous patients. J Bras Pneumol 2006; 32: 13-14.
-
18SPS/DVE/SES/SC. Secretaria de Políticas de Saúde - Diretoria de Vigilância Epidemiológica Florianópolis: Secretaria de Estado da Saúde de Santa Catarina; 2008.
- 19. Bazzo ML, Ferreira LAP, Silva RM, Scheffer M, Chagas M, Severino JL, et al. Relação entre a qualidade de amostras de escarro e o diagnóstico de micobacterioses por PCR. Arq Cat Med 2004; 33: 23-27.
- 20. Lucchini GM, Wegmann K, Sauer M, Altwegg M. Early detection of Mycobacterium tuberculosis by culture on BBL MGIT medium followed by PCR. J Microbiol Met 1996; 27: 97-107.
- 21. Chakravorty S, Dudeja M, Hanif M, Tyagi JS. Utility of universal sample processing methodology, combining smear microscopy, culture, and PCR, for diagnosis of pulmonary tuberculosis. J Clin Microbiol 2005; 43: 2703-2708.
- 22. Bruchfeld J, Aderaye G, Palme IB, Bjorvatn B, Kallenius G, Lindquist L. Sputum concentration improves diagnosis of tuberculosis in a setting with a high prevalence of HIV. Trans R Soc Trop Med Hyg 2000; 94: 677-680.
- 23. Garay JE. Analysis of a simplified concentration sputum smear technique for pulmonary tuberculosis diagnosis in rural hospitals. Trop Doct 2000; 30: 70-72.
- 24. Perera J, Arachchi DM. The optimum relative centrifugal force and centrifugation time for improved sensitivity of smear and culture for detection of Mycobacterium tuberculosis from sputum. Trans R Soc Trop Med Hyg 1999; 93: 405-409.
- 25. Aslanzadeh J, de la Viuda M, Fille M, Smith WB, Namdari H. Comparison of culture and acid-fast bacilli stain to PCR for detection of Mycobacterium tuberculosis in clinical samples. Mol Cell Probes 1998; 12: 207-211.
- 26. Habeenzu C, Lubasi D, Fleming AF. Improved sensitivity of direct microscopy for detection of acid-fast bacilli in sputum in developing countries. Trans R Soc Trop Med Hyg 1998; 92: 415-416.
- 27. Somoskovi A, Dormandy J, Mayrer AR, Carter M, Hooper N, Salfinger M. "Mycobacterium canettii" isolated from a human immunodeficiency virus-positive patient: first case recognized in the United States. J Clin Microbiol 2009; 47: 255-257.
- 28. Viana-Niero C, Lima KV, Lopes ML, Rabello MC, Marsola LR, Brilhante VC, et al. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol 2008; 46: 850-855.
- 29. Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev 1997; 10: 242-256.
- 30. Cheng VC, Yew WW, Yuen KY. Molecular diagnostics in tuberculosis. Eur J Clin Microbiol Infect Dis 2005; 24: 711-720.
- 31. Ratnam S, Stead FA, Howes M. Simplified acetylcysteine-alkali digestion-decontamination procedure for isolation of mycobacteria from clinical specimens. J Clin Microbiol 1987; 25: 1428-1432.
- 32. Wattal C. Improving bacteriological diagnosis of tuberculosis. Indian J Pediatr 2002; 69 (Suppl 1): S11-S19.
- 33. Buijtels PC, Petit PL. Comparison of NaOH-N-acetyl cysteine and sulfuric acid decontamination methods for recovery of mycobacteria from clinical specimens. J Microbiol Methods 2005; 62: 83-88.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
07 June 2010 -
Date of issue
June 2010
History
-
Received
07 Nov 2009 -
Accepted
09 Apr 2010