Abstract
Vertebrates have a central clock and also several peripheral clocks. Light responses might result from the integration of light signals by these clocks. The dermal melanophores of Xenopus laevis have a photoreceptor molecule denominated melanopsin (OPN4x). The mechanisms of the circadian clock involve positive and negative feedback. We hypothesize that these dermal melanophores also present peripheral clock characteristics. Using quantitative PCR, we analyzed the pattern of temporal expression of Opn4x and the clock genes Per1, Per2, Bmal1, and Clock in these cells, subjected to a 14-h light:10-h dark (14L:10D) regime or constant darkness (DD). Also, in view of the physiological role of melatonin in the dermal melanophores of X. laevis, we determined whether melatonin modulates the expression of these clock genes. These genes show a time-dependent expression pattern when these cells are exposed to 14L:10D, which differs from the pattern observed under DD. Cells kept in DD for 5 days exhibited overall increased mRNA expression for Opn4x and Clock, and a lower expression for Per1, Per2, and Bmal1. When the cells were kept in DD for 5 days and treated with melatonin for 1 h, 24 h before extraction, the mRNA levels tended to decrease for Opn4x and Clock, did not change for Bmal1, and increased for Per1 and Per2 at different Zeitgeber times (ZT). Although these data are limited to one-day data collection, and therefore preliminary, we suggest that the dermal melanophores of X. laevis might have some characteristics of a peripheral clock, and that melatonin modulates, to a certain extent, melanopsin and clock gene expression.
Xenopus laevis; Dermal melanophores; Melatonin; Melanopsin; Per1; Per2; Bmal1; Clock
Abstract
Introduction
Material and Methods
Results
Discussion
References
Acknowledgments
Braz J Med Biol Res, August 2012, Volume 45(8) 730-736
The expression of melanopsin and clock genes in Xenopus laevis melanophores and their modulation by melatonin
A.P.C. Bluhm, N.N. Obeid, A.M.L. Castrucci and  Correspondence and Footnotes
Correspondence and Footnotes
 M.A. Visconti
M.A. Visconti
Departamento de Fisiologia, Instituto de Biociências, Universidade de São Paulo, São Paulo, SP, Brasil
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Vertebrates have a central clock and also several peripheral clocks. Light responses might result from the integration of light signals by these clocks. The dermal melanophores of Xenopus laevis have a photoreceptor molecule denominated melanopsin (OPN4x). The mechanisms of the circadian clock involve positive and negative feedback. We hypothesize that these dermal melanophores also present peripheral clock characteristics. Using quantitative PCR, we analyzed the pattern of temporal expression of Opn4x and the clock genes Per1, Per2, Bmal1, and Clock in these cells subjected to a 14-h light:10-h dark (14L:10D) regime or constant darkness (DD). Also, in view of the physiological role of melatonin in the dermal melanophores of X. laevis, we determined whether melatonin modulates the expression of these clock genes. These genes show a time-dependent expression pattern when these cells are exposed to 14L:10D, which differs from the pattern observed under DD. Cells kept in DD for 5 days exhibited overall increased mRNA expression for Opn4x and Clock, and a lower expression for Per1, Per2, and Bmal1. When the cells were kept in DD for 5 days and treated with melatonin for 1 h, 24 h before extraction, the mRNA levels tended to decrease for Opn4x and Clock, did not change for Bmal1, and increased for Per1 and Per2 at different Zeitgeber times (ZT). Although these data are limited to one-day data collection, and therefore preliminary, we suggest that the dermal melanophores of X. laevis might have some characteristics of a peripheral clock, and that melatonin modulates, to a certain extent, melanopsin and clock gene expression.
Key words: Xenopus laevis; Dermal melanophores; Melatonin; Melanopsin; Per1; Per2; Bmal1; Clock
There are three structures in the core of the circadian system that regulate and control several forms of rhythmic expression in vertebrates, namely the retinas, the pineal complex (pineal body and the eye/parietal organ) and the suprachiasmatic nucleus (SCN) (1). The circadian system also includes hierarchically organized peripheral oscillators located outside the main oscillator itself (1). These peripheral oscillators can be coupled or not, even though they are linked to the SCN. Thus, their expression is connected and depends on the temporal structure of the organism, even though they are self-sustained and therefore independently capable of generating oscillation for the cell or tissue where they are located (2).
SCN and peripheral clocks work through a transcriptional circuit of clock genes, which generate the rhythmic patterns of protein expression during metabolism, and participate in several other functions (3-5). In mammals, the transcription factors CLOCK and BMAL1 act as positive regulators, whereas the three PER proteins (PER1, PER2 and PER3) and the two CRY proteins (CRY1 and CRY2) operate as negative agents (3,6). The molecular mechanism of the circadian clock has also been confirmed in other organisms such as mice and humans (7).
Clock gene circadian oscillation controls the expression of genes involved in multiple cell functions, during a period of approximately 24 h by at least two mechanisms: 1) through direct interaction with E-boxes in the promoters of these genes, and 2) by the regulation of other clock-controlled genes, which also act as transcription factors (3,5).
The molecular mechanism involved in maintaining circadian rhythmicity is defined by transcription/translation in feedback loops, which are based on mRNA and protein rhythmic expression of circadian clock components. SCN synchronization by light induces expression of a group of clock genes, including Per2 and Per1 (8). Even though Per genes are critical elements for clock operation, their specific functions have not been well defined. For example, in Drosophila, a single Per gene is sufficient as a negative loop, whereas in the mammalian system there are three homologous genes (Per1, Per2 and Per3), two of which (Per1 and Per2) are essential for SCN operation. Apparently these three homologous genes possess distinct functions (9).
Peripheral clocks are widely distributed from fish to mammals (2,10-13). These peripheral clocks are reset by the central oscillator through neurohormonal signals, or by metabolic signals and food (13). The independence of these oscillators from the central oscillator has been demonstrated (2,10-13), suggesting the presence of their own synchronizing elements (13). Circadian expression observed in peripheral tissues can be directly synchronized by exposure to light/dark cycles, both in whole organisms and in various cell types in culture (14-17).
In the amphibian Xenopus laevis, an opsin was characterized in the dermal melanophores, and denominated melanopsin (OPN4) (14). Subsequently a melanopsin orthologous to that of X. laevis was identified in a small population of intrinsically photoreceptive cells (ipRGCs) in the retinas of mice (15).
OPN4 is expressed in all classes of vertebrates examined to date, from teleosts to mammals (2,18,19). Furthermore, melanopsin is also found in other encephalic regions, such as the pineal gland and the deeper brain regions of non-mammalian vertebrates in which circadian photoreception is manifest (2,18,20-23).
Melanopsin is also expressed in pigment cells and mediates the photo-response of melanophore proliferation (24) and melanosome dispersion (22,25,26). To date, little is known about the regulatory mechanisms of melanopsin or other opsins, such as rhodopsin, in vertebrate pigment cells. Data from previous studies from our laboratory demonstrated rhodopsin modulation by alpha-melanocyte-stimulating hormone (α-MSH) in pigment cells of Carassius auratus (GEM-81 erythrophoroma cells) (24). More recently, Farhat et al. (27) demonstrated the biphasic action of endothelin-1 on Opn4x expression in Danio rerio ZEM-2S cells, in which low concentrations of this hormone stimulated Opn4x expression and high concentrations inhibited it.
The pineal gland of non-mammalian vertebrates contains photoreceptors, which permit nocturnal production and release of melatonin, thereby synchronizing light/dark cycles. In amphibians, reptiles and birds, the pineal gland, together with the lateral eyes that are structures similar to retinal photoreceptors, efficiently assumes this function (28). In vertebrates, melatonin is released by the pineal gland during darkness (29). In unicellular animals, bacteria and invertebrates, it also marks this period (30). In vertebrates, melatonin is further synthesized by the retina, thereby participating in the adaptive processes of night vision (31).
It has been shown that the exogenous administration of melatonin is capable of synchronizing circadian rhythms (32). The daily rhythms of N-acetyltransferase activity and melatonin secretion can be observed in pineal cell cultures. These rhythms can be initiated by light/dark cycles of 24 h in fish, lizards and birds (32).
In amphibian dermal melanocytes, melatonin induces the aggregation of melanosomes and is a potent skin-lightening agent (29). Embryonic dermal melanophores of X. laevis express Opn4x and Opn4m. Furthermore, when these melanophores are exposed to light, there is a dispersion of pigment granules throughout the cytoplasm (33). Considering the existence of Opn4x and clock genes in several types of cells in X. laevis, our hypothesis is that the dermal melanophores of this amphibian also present peripheral clock characteristics. As a preliminary approach to this hypothesis, we analyzed 24-h data of temporal expression of Opn4x, and the clock genes Per1, Per2, Bmal1 and Clock in these cells, when subjected to a 14-h light:10-h dark (14L:10D) regime or constant darkness (DD). Also, in view of the physiological role of melatonin in the dermal melanophores of X. laevis, we determined whether expression of Opn4x and the clock genes in these cells would be modulated by melatonin under the experimental conditions utilized.
Cell culture
X. laevis melanophores were kept in 60% L-15 medium supplemented with 10% fetal calf serum, 480 mg/L galactose, 5 mg/L insulin/transferrin/selenium, 4 mg/L uridine, 87.6 mg/L L-glutamine, 25 mg/L L-asparagine, 152 mg/L CaCl2, 49.6 mg/L MgCl2, 51.7 mg/L MgSO4, 0.4X MEM non-essential amino acids, 0.2X MEM essential amino acids, 0.3X MEM vitamin solution, 2X hypoxanthine-thymidine supplement, 0.5% penicillin/streptomycin (all from Gibco, USA), and 0.1 nM α-MSH (Sigma Aldrich, USA), pH 7.5, at 25°C. The culture medium was replaced once a week and the cells were subcultured at confluence. In general, six to ten culture flasks were used in the experiments.
Assays monitoring Opn4x gene expression were carried out in the absence and presence of α-MSH, and with the reduction of bovine fetal serum from 10 to 2% for 1 to 8 days. The results of these tests (data not shown) demonstrated that removal of the hormone from the culture medium and/or reduction in serum led to an increase in Opn4x expression in the first days of DD. Nevertheless, 4 to 5 days after the removal of both components, mRNA levels stabilized. Based on these data, we chose to withdraw α-MSH from the culture medium prior to gene analysis.
Cell culture for total RNA extraction
In order to carry out experimental assays, the concentration of fetal bovine solution from cells maintained as described above was reduced to 2%, and the α-MSH was withdrawn from the culture medium, which was then supplemented with 50 nM retinaldehyde. The cells were seeded onto 6-well dishes in duplicate (2 x 106/well), and incubated for 9 days, either in a regime of DD or a photoperiod of 14L:10D (light at 650 lux). On the 10th day, RNA was extracted every 3 h (0, 3, 6, 9, 12, 15, 18, and 21 Zeitgeber time, ZT) in such a way that 0 h of each experiment corresponded to the time the lights were turned on (9 am). All experiments were set up at the same time (3 pm). The same procedure was used for the analysis of all genes. During the periods when the cells were in darkness, medium changes and RNA extraction were performed under red light (Kodak GBX-2S Safelight filter).
Hormone assays
For hormone assays, the concentration of bovine fetal serum was reduced to 2% and α-MSH was withdrawn from the culture medium. The cells were seeded in duplicate onto 6-well dishes, and then incubated for 8 days in DD. The culture medium was aspirated on the 9th day, and the cells were washed twice with phosphate-buffered saline. The experimental culture medium containing 1.0 nM melatonin (Calbiochem, USA) and 1.0 mM stock solution in ethanol (Merck, USA) was added under red light. On the 10th day, RNA was extracted every 3 h as outlined above. All experiments were set up at the same time (3 pm). The same procedure was used for all gene analyses.
Extraction of total RNA, RT-PCR, and PCR
Total RNA was extracted with Trizol reagent (Invitrogen, USA) according to manufacturer instructions. The RNA pellet was resuspended in water containing diethylpyrocarbonate (DEPC; Ambion Inc., USA) and treated with DNase according to manufacturer instructions (turbo-DNA-free™, Ambion Inc.). RNA concentration was determined with a spectrophotometer (GeneQuant Pro, Biochrom Ltd., UK), and 1 µg was used in the reverse transcriptase reaction (SuperScript III RT, Invitrogen).
Quantitative PCR was carried out with primer pairs specific for the genes (Table 1), designed with the Primer Express software (ABI, USA; synthesis by IDT, USA). Separate solutions containing the primers (300 nM for Opn4x, Clock, Bmal1, Per1, and Per2; 50 nM for RNA 18S), 2X iQ Sybr Green Supermix (100 mM KCl, 40 mM Tris-HCl, 1.6 mM dNTPS, 50 U/mL iTaq DNA polymerase, and 6 mM MgCl2, BioRad Laboratories, USA) and autoclaved MilliQ water were prepared to a final volume of 23 µL/well. Each solution was divided into Eppendorf tubes, each aliquot being sufficient for 2 wells and the cDNA of each sample being added to the corresponding tube. The solutions, already with cDNA, were then distributed onto 96-well slides in duplicate (23 µL/well). Negative controls containing autoclaved MilliQ water instead of cDNA were included in the assays. All assays were carried out in an iCycler (BioRad Laboratories) thermal cycler, under the following conditions: 1X 95°C for 7 min, 50X 95°C for 10 s, 60°C for 1 min, followed by the melting curve.
Data analysis
Data analysis was carried out using the ΔΔCT method. The values were obtained by tracing the so-called threshold line, which intercepts the log phase of the amplification curves. By knowing the number of cycles where this line passes (CT), ΔCT was calculated as the difference between CT for each gene of interest and for 18S RNA. Next, the values for each well were subtracted from the average of wells where expression was lower (calibrator) in DD, thereby obtaining ΔΔCT. This value was placed as the negative power in base 2 (2-ΔΔCT), thus obtaining the number of times that the gene was expressed at the various time points, as compared to the minimal expression. The log values were averaged and expressed as percentage of the minimal value in DD.
Means ± SEM of 6 to 10 cultures were calculated. Eventual temporal differences in each experimental condition (DD, 14L:10D, or DD in the presence of melatonin) were analyzed by ANOVA followed by the Tukey test. The various treatments were compared by two-way ANOVA followed by the Bonferroni test. Differences were considered significant when P ≤ 0.05.
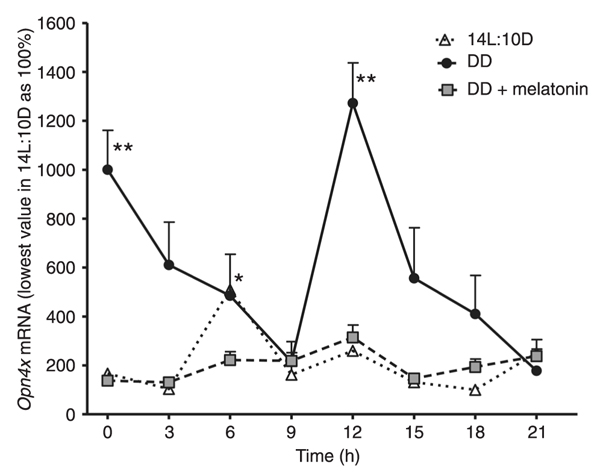
Under DD, significantly high values of the expression of Opn4x were observed at 0 and 12 h, with a gradual and significant reduction between 3-9 and 15-21 h. In 14L:10D, there was a statistically significant increase in Opn4 mRNA values at 6 h. Compared to DD, mRNA levels were remarkably lower under 14L:10D conditions (Figure 1). When cells were subjected to DD and treated with 1.0 nM melatonin for 1 h, 24 h before starting mRNA extraction, significantly lower values in mRNA relative expression were noted at 12 h. Compared to DD, there was a noticeable flattening of the expression with hormone treatment (Figure 1).
In DD, significantly high values of Clock expression were observed at 0 and 12-15 h. During the 14L:10D photoperiod, there was a statistically significant increase in mRNA values at 12 h. Compared to DD, it could be noted that there was an increase in mRNA values at 12 h under 14L:10D conditions (Figure 2). When cells were subjected to DD and treated with 1.0 nM melatonin for 1 h, 24 h before starting mRNA extraction, the mRNA values were reduced compared to DD (Figure 2).
In DD, Bmal1 expression was low and constant, except at 9 and 18 h, where a small but significant increase was observed. Under 14L:10D conditions, there was a statistically significant decrease in Bmal1 values between 3-9 h, as compared to the other times. Comparing 14L:10D to DD, it can be noted that, as a rule, the values of Bmal1 mRNA were elevated during the dark phase of the L:D cycle (Figure 3). When the cells were subjected to DD, and treated with 1.0 nM melatonin for 1 h, 24 h prior to starting mRNA extraction, no change in the time course of mRNA relative values could be observed. When compared to DD, one can note that hormone treatment did not change these levels (Figure 3).
In DD, similar to Bmal1, Per1 expression was also low and constant. Under the 14L:10D photoperiod, there was a statistically significant increase in the time course of mRNA values during the light phase of the cycle. When the DD condition was compared to the 14L:10D cycle, mRNA values were significantly higher between 3 and 12 h (Figure 4). When the cells remained in DD and were treated with 1.0 nM melatonin for 1 h, 24 h before starting mRNA extraction, there was a significant increase in Per1 mRNA values between 0 and 3 h in hormone-treated cells (Figure 4).
In DD, significantly high values of Per2 expression were observed at 3 and 12 h. Under the 14L:10D cycle, Per2 mRNA was low and constant between 6 and 12 h, with a gradual increase towards 12 h. When DD was compared to the 14L:10D photoperiod, significantly lower mRNA values were observed at 3 and 21 h in the L:D cycle (Figure 5). When the cells in DD were treated with 1.0 nM melatonin for 1 h, 24 h before starting mRNA extraction, significantly high values of mRNA were observed at 12 and 21 h. Compared to DD, melatonin did not affect Per2 levels, with the exception of the 3-h point, where the values were lower in the presence of the hormone (Figure 5).
Relative Opn4x expression under a 14-h light:10-h dark (14L:10D) regime, constant darkness (DD) or DD plus 1.0 nM melatonin. Each point is the mean ± SEM for 6-10 cultures. * and **P < 0.05 for time course differences in 14L:10D and DD, respectively (ANOVA followed by the Tukey test).
Relative Clock expression under a 14-h light:10-h dark (14L:10D) regime, constant darkness (DD) or DD plus 1.0 nM melatonin. Each point is the mean ± SEM for 6-10 cultures. * and **P < 0.05 for time course differences in 14L:10D and DD, respectively (ANOVA followed by the Tukey test).
Relative Bmal1 expression under a 14-h light:10-h dark (14L:10D) regime, constant darkness (DD) or DD plus 1.0 nM melatonin. Each point is the mean ± SEM for 6-10 cultures. * and **P < 0.05 for time course differences in 14L:10D and DD, respectively (ANOVA followed by the Tukey test).
Relative Per1 expression under a 14-h light:10-h dark (14L:10D) regime, constant darkness (DD) or DD plus 1.0 nM melatonin. Each point is the mean ± SEM for 6-10 cultures. * and **P < 0.05 for time course differences in 14L:10D and DD + melatonin, respectively (ANOVA followed by the Tukey test).
Relative Per2 expression under a 14-h light:10-h dark (14L:10D) regime, constant darkness (DD) or DD plus 1.0 nM melatonin. Each point is the mean ± SEM for 6-10 cultures. *, ** and ***P < 0.05 for time course differences in 14L:10D, DD and DD + melatonin, respectively (ANOVA followed by the Tukey test).
Biological clocks have been identified in various tissues in the most diverse species. However, the clocks in the various types of cells may present different operational characteristics, and constitute cell and/or self-sufficient molecular oscillators adjusted by environmental cycles, especially light-dark. In Drosophila, for example, in which circadian clocks were encountered in all the tissues studied, the main pacemaker maintains oscillations for a long time even under conditions of constant darkness, with little or no rhythm damping. Nevertheless, the differences between the central pacemaker and the machinery of the clock itself, and in the remaining vertebrate and invertebrate systems, have not yet been entirely clarified (34).
The dermal melanophores of X. laevis respond to light by dispersing melanosomes, a characteristic of embryonic melanophores of this amphibian (33). The ability of amphibian melanophores to react directly to light, although frequent, is not always present. Two primary chromatic responses are perceived, the most common being the dispersion of pigment granules throughout the cytoplasm in light-exposed melanophores (33).
In previous studies, it was observed that Opn4, the already cited photopigment discovered in X. laevis dermal melanophores, is expressed in the retina of all classes of vertebrates examined to date from teleosts to mammals, including humans (14,18). In the present study, we were able to confirm the function of photopigments, whose presence was confirmed by the Western blot technique (data not shown), in a cell-line that had been preserved in our laboratory for about 5 years. It was also possible to suggest that the melanophores of this amphibian respond to light in a way similar to that described for other cell types (17).
Besides Opn4, the embryonic dermal melanophores of X. laevis also express the clock genes Clock, Per1, Per2, and Bmal1. According to data from the present study on this cell line, when the cells were exposed to a light:dark cycle, Per1, Per2, and Bmal1 genes showed an expression pattern apparently dependent on time of day. However, in constant darkness there is no variation of time-dependent expression, except maybe for Bmal1. Carr and Whitmore (17) observed that in DD each cell of a population continues to oscillate, but with fluctuations in free-running period, and these oscillations are not in phase. As we collect data of cell populations, the expression of clock genes may also be attenuated due to anti-phase oscillations of individual cells.
In our experiments, ZT 0 (0 h) corresponded to the time when the light was switched on (9 am). Furthermore, all experiments were set up at the same time of day (3 pm), the culture medium being changed simultaneously when necessary. This procedure was required since Yoshikawa et al. (35) showed that the rhythmicity of cell cultures, obtained from organs containing central or peripheral circadian oscillators, are influenced by the time when the cultures were obtained and the medium was changed.
In the mammalian circadian system, Per1 and Per2 gene expression is light-entrained (9); in Xenopus retina, under a 12L:12D regime, Per2 rhythm is light-entrained with lower expression just before the beginning of the light phase, and peaking during the day. On the other hand, in this amphibian, Per1 retina expression is rhythmic under light-dark and DD conditions (36). In X. laevis melanophores under a 14L:10D photoperiod regime, there is an increase in Per2 expression during the light period, similar to what occurs in the retina. Furthermore, Per1 expression is higher in the light phase, thereby implying that its expression is dependent on the time of day, similar to what is observed in mammals.
As to the Bmal1 gene, results indicate that, in a 14L:10D photoperiod regime, there is a decrease during the 3- to 9-h period, contrary to what is observed for Per1 and, to a lesser extent, for Per2, in the light phase. Furthermore, there is a visible increase in expression during the 12- to 15-h period (end of the light and beginning of the dark phases), which is maintained at a high level until 18 h. Overall, these results are the opposite of those observed in Per1 and Per2 expression.
Similar results were observed by Durgan et al. (37) in a mammalian model (rat cardiomyocytes) in which they analyzed oscillation patterns over a period of 24 h, with sample collection every 3 h, using the same protocol as ours. Under a 12L:12D photoperiod, the authors reported a decrease in Bmal1 expression in the light phase and an increase in the dark, the opposite occurring with Per2 expression. Farhat et al. (27) showed that under the 12L:12D condition, there was robust daily expression of Per1 and Cry1b clock genes, whereas Opn4x and Clock expression seemed to vary in an ultradian pattern. For both Per1 and Cry1b genes, expression was higher during the light phase, whereas for the Clock gene there was an increase in expression coincident with the dark phase during the subjective night.
Generally speaking, a circadian cycle begins in the early morning hours with activation of Per and Cry transcription by CLOCK/BMAL1. Furthermore, according to Albrecht and Eichele (38), transcription levels reach their apex around midday and, in fact, our data indicate an increase in expression over the 12-15 ZT period for both Per genes, this corresponding to the end of the light phase and the beginning of the dark one. Bmal1 expression remains high throughout the dark phase. According to the proposed model for mammals, this high expression would facilitate synthesis of the BMAL1 protein, and the consequent dimerization with CLOCK, thereby leading to an increase in Per expression. Our data with these transcripts corroborate this model (6).
Melatonin, the hormone secreted by the pineal gland, is directly related to biological clock machinery. In all species, it is secreted at night and is associated with sleep, the maintenance of body temperature, immunological response and other nocturnal physiological events. There is evidence that the exogenous administration of melatonin can adjust these rhythms to both endogenous cortisol secretion and to that of melatonin itself (39). Under certain circumstances, melatonin can synchronize the circadian rhythm (24 h) in free-running individuals, i.e., it is able to rescue the endogenous period of their internal biological clocks (40). In X. laevis dermal melanophores, our data indicate that melatonin, after a pulse of 1 h, practically does not affect melanopsin or clock gene expression, but, in general, causes a flattening of the temporal curve.
The results obtained here for X. laevis dermal melanophores, although less complex than the mammalian system, may contribute to a better understanding of the inherent molecular mechanisms present in most organisms, which could be confirmed by investigation from a phylogenetic perspective.
1. Moore-Ede MC, Sulzman FM, Fuller CA. Characteristics of a circadian clock. In: Moore-Ede MC, Sulzman FM, Fuller CA (Editors), The clocks that time us: physiology of the circadian timing system. Cambridge: Harvard University Press; 1982.
2. Davidson AJ, London B, Block GD, Menaker M. Cardiovascular tissues contain independent circadian clocks. Clin Exp Hypertens 2005; 27: 307-311.
3. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002; 418: 935-941.
4. Okamura H, Yamaguchi S, Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res 2002; 309: 47-56.
5. Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 2007; 47: 593-628.
6. Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci 2004; 21: 359-368.
7. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 2006; 15 (Spec No. 2): R271-R277.
8. Cajochen C, Jud C, Munch M, Kobialka S, Wirz-Justice A, Albrecht U. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci 2006; 23: 1082-1086.
9. Besharse JC, Zhuang M, Freeman K, Fogerty J. Regulation of photoreceptor Per1 and Per2 by light, dopamine and a circadian clock. Eur J Neurosci 2004; 20: 167-174.
10. Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, et al. Circadian rhythms in isolated brain regions. J Neurosci 2002; 22: 350-356.
11. Bartell PA, Miranda-Anaya M, Menaker M. Period and phase control in a multioscillatory circadian system (Iguana iguana). J Biol Rhythms 2004; 19: 47-57.
12. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. Period: Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 2004; 101: 5339-5346.
13. Costa A, Castanon-Cervantes O, Menaker M, Piccione G, Caola G. Daily rhythm of lactate dehydrogenase in rat (Rattus norvegicus) carrying a Per1-luciferase transgene: assessment on serum and liver. Vet Res Commun 2005; 29 (Suppl 2): 183-186.
14. Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: An opsin in melanophores, brain, and eye. Proc Natl Acad Sci U S A 1998; 95: 340-345.
15. Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci 2000; 20: 600-605.
16. Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 2000; 404: 87-91.
17. Carr AJ, Whitmore D. Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol 2005; 7: 319-321.
18. Rollag MD, Berson DM, Provencio I. Melanopsin, ganglion-cell photoreceptors, and mammalian photoentrainment. J Biol Rhythms 2003; 18: 227-234.
19. Terakita A. The opsins. Genome Biol 2005; 6: 213.
20. Provencio I, Rollag MD, Castrucci AM. Photoreceptive net in the mammalian retina. This mesh of cells may explain how some blind mice can still tell day from night. Nature 2002; 415: 493.
21. Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, et al. Melanopsin is required for non-image-forming photic responses in blind mice. Science 2003; 301: 525-527.
22. Isoldi MC, Rollag MD, Castrucci AM, Provencio I. Rhabdomeric phototransduction initiated by the vertebrate photopigment melanopsin. Proc Natl Acad Sci U S A 2005; 102: 1217-1221.
23. Nayak SK, Jegla T, Panda S. Role of a novel photopigment, melanopsin, in behavioral adaptation to light. Cell Mol Life Sci 2007; 64: 144-154.
24. Im LH, Isoldi MC, Scarparo AC, Visconti MA, Castrucci AM. Rhythmic expression, light entrainment and alpha-MSH modulation of rhodopsin mRNA in a teleost pigment cell line. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 691-696.
25. Moriya T, Miyashita Y, Arai J, Kusunoki S, Abe M, Asami K. Light-sensitive response in melanophores of Xenopus laevis: I. Spectral characteristics of melanophore response in isolated tail fin of Xenopus tadpole. J Exp Zool 1996; 276: 11-18.
26. Rollag MD. Amphibian melanophores become photosensitive when treated with retinal. J Exp Zool 1996; 275: 20-26.
27. Farhat FP, Martins CB, De Lima LH, Isoldi MC, Castrucci AM. Melanopsin and clock genes: regulation by light and endothelin in the zebrafish ZEM-2S cell line. Chronobiol Int 2009; 26: 1090-1119.
28. Golombek DA, Roblero RA. Mecanismos de temporização nos vertebrados. In: Marques N (Editor), Cronobiologia: princípios e aplicações. São Paulo: EDUSP; 2003. p 163-190.
29. Filadelfi AM, Castrucci AM. Comparative aspects of the pineal/melatonin system of poikilothermic vertebrates. J Pineal Res 1996; 20: 175-186.
30. Hardeland R, Poeggeler B. Non-vertebrate melatonin. J Pineal Res 2003; 34: 233-241.
31. Tosini G, Fukuhara C. Photic and circadian regulation of retinal melatonin in mammals. J Neuroendocrinol 2003; 15: 364-369.
32. Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 2005; 6: 544-556.
33. Rollag MD, Provencio I, Sugden D, Green CB. Cultured amphibian melanophores: a model system to study melanopsin photobiology. Methods Enzymol 2000; 316: 291-309.
34. Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol 2003; 1: E13.
35. Yoshikawa T, Yamazaki S, Menaker M. Effects of preparation time on phase of cultured tissues reveals complexity of circadian organization. J Biol Rhythms 2005; 20: 500-512.
36. Zhuang M, Wang Y, Steenhard BM, Besharse JC. Differential regulation of two period genes in the Xenopus eye. Brain Res Mol Brain Res 2000; 82: 52-64.
37. Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem 2006; 281: 24254-24269.
38. Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev 2003; 13: 271-277.
39. Markus RP, Ferreira ZS, Fernandes PA, Cecon E. The immune-pineal axis: a shuttle between endocrine and paracrine melatonin sources. Neuroimmunomodulation 2007; 14: 126-133.
40. Zawilska JB, Skene DJ, Arendt J. Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol Rep 2009; 61: 383-410.
Xenopus laevis melanophores were a gift from Dr. Mark Rollag, Uniformed Services University of Health Sciences, Bethesda, MD, USA. We are thankful to Márcio P. Lima and Telma Pazini for technical assistance. Research partially supported by FAPESP. A.P.C. Bluhm and N.N. Obeid were recipients of fellowships from CAPES and CNPq, respectively.
 Address for correspondence: M.A. Visconti, Departamento de Fisiologia, Instituto de Biociências, USP, Rua do Matão, travessa 14, 101, 05508-900 São Paulo, SP, Brasil. Fax +55-11-3091-7568. E-mail: maviscon@usp.br
Address for correspondence: M.A. Visconti, Departamento de Fisiologia, Instituto de Biociências, USP, Rua do Matão, travessa 14, 101, 05508-900 São Paulo, SP, Brasil. Fax +55-11-3091-7568. E-mail: maviscon@usp.br
Received December 11, 2011. Accepted May 11, 2012. Available online May 25, 2012. Published August 3, 2012.
The Brazilian Journal of Medical and Biological Research is partially financed by
All the contents of this journal, except where otherwise noted, is licensed under a Creative Commons Attribution License
- 1. Moore-Ede MC, Sulzman FM, Fuller CA. Characteristics of a circadian clock. In: Moore-Ede MC, Sulzman FM, Fuller CA (Editors), The clocks that time us: physiology of the circadian timing system Cambridge: Harvard University Press; 1982.
- 2. Davidson AJ, London B, Block GD, Menaker M. Cardiovascular tissues contain independent circadian clocks. Clin Exp Hypertens 2005; 27: 307-311.
- 3. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002; 418: 935-941.
- 4. Okamura H, Yamaguchi S, Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res 2002; 309: 47-56.
- 5. Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 2007; 47: 593-628.
- 6. Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci 2004; 21: 359-368.
- 7. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 2006; 15 (Spec No. 2): R271-R277.
- 8. Cajochen C, Jud C, Munch M, Kobialka S, Wirz-Justice A, Albrecht U. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci 2006; 23: 1082-1086.
- 9. Besharse JC, Zhuang M, Freeman K, Fogerty J. Regulation of photoreceptor Per1 and Per2 by light, dopamine and a circadian clock. Eur J Neurosci 2004; 20: 167-174.
- 10. Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, et al. Circadian rhythms in isolated brain regions. J Neurosci 2002; 22: 350-356.
- 11. Bartell PA, Miranda-Anaya M, Menaker M. Period and phase control in a multioscillatory circadian system (Iguana iguana). J Biol Rhythms 2004; 19: 47-57.
- 12. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. Period: Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 2004; 101: 5339-5346.
- 13. Costa A, Castanon-Cervantes O, Menaker M, Piccione G, Caola G. Daily rhythm of lactate dehydrogenase in rat (Rattus norvegicus) carrying a Per1-luciferase transgene: assessment on serum and liver. Vet Res Commun 2005; 29 (Suppl 2): 183-186.
- 14. Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: An opsin in melanophores, brain, and eye. Proc Natl Acad Sci U S A 1998; 95: 340-345.
- 15. Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci 2000; 20: 600-605.
- 16. Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 2000; 404: 87-91.
- 17. Carr AJ, Whitmore D. Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol 2005; 7: 319-321.
- 18. Rollag MD, Berson DM, Provencio I. Melanopsin, ganglion-cell photoreceptors, and mammalian photoentrainment. J Biol Rhythms 2003; 18: 227-234.
- 19. Terakita A. The opsins. Genome Biol 2005; 6: 213.
- 20. Provencio I, Rollag MD, Castrucci AM. Photoreceptive net in the mammalian retina. This mesh of cells may explain how some blind mice can still tell day from night. Nature 2002; 415: 493.
- 21. Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, et al. Melanopsin is required for non-image-forming photic responses in blind mice. Science 2003; 301: 525-527.
- 22. Isoldi MC, Rollag MD, Castrucci AM, Provencio I. Rhabdomeric phototransduction initiated by the vertebrate photopigment melanopsin. Proc Natl Acad Sci U S A 2005; 102: 1217-1221.
- 23. Nayak SK, Jegla T, Panda S. Role of a novel photopigment, melanopsin, in behavioral adaptation to light. Cell Mol Life Sci 2007; 64: 144-154.
- 24. Im LH, Isoldi MC, Scarparo AC, Visconti MA, Castrucci AM. Rhythmic expression, light entrainment and alpha-MSH modulation of rhodopsin mRNA in a teleost pigment cell line. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 691-696.
- 25. Moriya T, Miyashita Y, Arai J, Kusunoki S, Abe M, Asami K. Light-sensitive response in melanophores of Xenopus laevis: I. Spectral characteristics of melanophore response in isolated tail fin of Xenopus tadpole J Exp Zool 1996; 276: 11-18.
- 26. Rollag MD. Amphibian melanophores become photosensitive when treated with retinal. J Exp Zool 1996; 275: 20-26.
- 27. Farhat FP, Martins CB, De Lima LH, Isoldi MC, Castrucci AM. Melanopsin and clock genes: regulation by light and endothelin in the zebrafish ZEM-2S cell line. Chronobiol Int 2009; 26: 1090-1119.
- 28. Golombek DA, Roblero RA. Mecanismos de temporização nos vertebrados. In: Marques N (Editor), Cronobiologia: princípios e aplicações São Paulo: EDUSP; 2003. p 163-190.
- 29. Filadelfi AM, Castrucci AM. Comparative aspects of the pineal/melatonin system of poikilothermic vertebrates. J Pineal Res 1996; 20: 175-186.
- 30. Hardeland R, Poeggeler B. Non-vertebrate melatonin. J Pineal Res 2003; 34: 233-241.
- 31. Tosini G, Fukuhara C. Photic and circadian regulation of retinal melatonin in mammals. J Neuroendocrinol 2003; 15: 364-369.
- 32. Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 2005; 6: 544-556.
- 33. Rollag MD, Provencio I, Sugden D, Green CB. Cultured amphibian melanophores: a model system to study melanopsin photobiology. Methods Enzymol 2000; 316: 291-309.
- 34. Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol 2003; 1: E13.
- 35. Yoshikawa T, Yamazaki S, Menaker M. Effects of preparation time on phase of cultured tissues reveals complexity of circadian organization. J Biol Rhythms 2005; 20: 500-512.
- 36. Zhuang M, Wang Y, Steenhard BM, Besharse JC. Differential regulation of two period genes in the Xenopus eye Brain Res Mol Brain Res 2000; 82: 52-64.
- 37. Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem 2006; 281: 24254-24269.
- 38. Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev 2003; 13: 271-277.
- 39. Markus RP, Ferreira ZS, Fernandes PA, Cecon E. The immune-pineal axis: a shuttle between endocrine and paracrine melatonin sources. Neuroimmunomodulation 2007; 14: 126-133.
- 40. Zawilska JB, Skene DJ, Arendt J. Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol Rep 2009; 61: 383-410.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
31 July 2012 -
Date of issue
Aug 2012
History
-
Received
11 Dec 2011 -
Accepted
11 May 2012