Abstracts
The aim of the present work was to characterize changes in the protein profile throughout seed development in O. catharinensis, a recalcitrant species, by two-dimensional gel electrophoresis. Protein extraction was undertaken by using a thiourea/urea buffer, followed by a precipitation step with 10% TCA. Comparative analysis during seed development showed that a large number of proteins were exclusively detected in each developmental stage. The cotyledonary stage, which represents the transition phase between embryogenesis and the beginning of metabolism related to maturation, presents the highest number of stage-specific spots. Protein identification, through MS/MS analysis, resulted in the identification of proteins mainly related to oxidative metabolism and storage synthesis. These findings contribute to a better understanding of protein metabolism during seed development in recalcitrant seeds, besides providing information on established markers that could be useful in defining and improving somatic embryogenesis protocols, besides monitoring the development of somatic embryos in this species.
2-DE; comparative proteomics; embryo development; storage protein; Lauraceae
O objetivo deste trabalho foi a caracterização dos perfis protéicos expressos durante o desenvolvimento da semente de O. catharinensis, uma espécie recalcitrante, via eletroforese bidimensional. A extração de proteínas foi realizada utilizando tampão uréia/tiouréia seguido de uma etapa de precipitação com TCA 10%. A análise comparativa durante o desenvolvimento da semente mostrou um grande número de proteínas exclusivas em cada estádio do desenvolvimento. O estádio cotiledonar, que representa a fase de transição entre a embriogênese e o começo do metabolismo relacionado a maturação, apresentou o maior número de proteínas estádio-específicas. Proteínas relacionadas ao metabolismo oxidativo e a síntese de reservas foram identificadas via MS/MS. Estes resultados contribuem para o melhor entendimento do metabolismo de proteínas durante o desenvolvimento de sementes recalcitrantes, além de prover informações no estabelecimento de parâmetros para protocolos de embriogênese somática.
2-DE; desenvolvimento embrionário; Lauraceae; proteômica comparativa; proteínas de reserva
Two-dimensional gel electrophoretic protein profile analysis during seed development of Ocotea catharinensis: a recalcitrant seed species
Leonardo L.C. DiasI* * Corresponding author: carnevalli45@yahoo.com.br , Tiago S. BalbuenaI,Vanildo SilveiraII, Claudete Santa-CatarinaIII, Andrej SchevchenkoIV And Eny I. S. FlohI
IPlant Cell Biology Laboratory, Department of Botany, IB-University of São Paulo, POBox 11461, CEP: 05422-970. São Paulo-SP, Brazil.
IIBiotechnology Laboratory, CBB-North Fluminense State University (UENF). Campos do Goytacazes-RJ, Brazil.
IIITissue and Cellular Biology Laboratory, CBB- North Fluminense State University (UENF). Campos dos Goytacazes-RJ, Brazil.
IVMax Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
Abstract
The aim of the present work was to characterize changes in the protein profile throughout seed development in O. catharinensis, a recalcitrant species, by two-dimensional gel electrophoresis. Protein extraction was undertaken by using a thiourea/urea buffer, followed by a precipitation step with 10% TCA. Comparative analysis during seed development showed that a large number of proteins were exclusively detected in each developmental stage. The cotyledonary stage, which represents the transition phase between embryogenesis and the beginning of metabolism related to maturation, presents the highest number of stage-specific spots. Protein identification, through MS/MS analysis, resulted in the identification of proteins mainly related to oxidative metabolism and storage synthesis. These findings contribute to a better understanding of protein metabolism during seed development in recalcitrant seeds, besides providing information on established markers that could be useful in defining and improving somatic embryogenesis protocols, besides monitoring the development of somatic embryos in this species.
Key words: 2-DE; comparative proteomics; embryo development; storage protein; Lauraceae
Resumo
O objetivo deste trabalho foi a caracterização dos perfis protéicos expressos durante o desenvolvimento da semente de O. catharinensis, uma espécie recalcitrante, via eletroforese bidimensional. A extração de proteínas foi realizada utilizando tampão uréia/tiouréia seguido de uma etapa de precipitação com TCA 10%. A análise comparativa durante o desenvolvimento da semente mostrou um grande número de proteínas exclusivas em cada estádio do desenvolvimento. O estádio cotiledonar, que representa a fase de transição entre a embriogênese e o começo do metabolismo relacionado a maturação, apresentou o maior número de proteínas estádio-específicas. Proteínas relacionadas ao metabolismo oxidativo e a síntese de reservas foram identificadas via MS/MS. Estes resultados contribuem para o melhor entendimento do metabolismo de proteínas durante o desenvolvimento de sementes recalcitrantes, além de prover informações no estabelecimento de parâmetros para protocolos de embriogênese somática.
Palavras-chave: 2-DE; desenvolvimento embrionário; Lauraceae; proteômica comparativa; proteínas de reserva
INTRODUCTION
Ocotea catharinensis Mez. (Lauraceae) is an endangered native forest tree species of the southern Brazilian Atlantic Rain Forest, and catalogued in the IUCN (International Union for Conservation of Nature and Natural Resources) Red List of Threatened Species as vulnerable - VU A1cd (www.iucnredlist.org). Natural propagation of O. catharinensis through seeds is hindered by its recalcitrancy, which means that seeds cannot be stored over long periods without a serious loss of viability (Silva et al.1998; Viana and Mantell 1999; Santa-Catarina et al. 2006).
In the life cycle of higher plants, seed development is the key process connecting two distinct sporophytic generations (Baud et al. 2002). Much related work has been undertaken on metabolic changes and genetic expression (Gallardo et al. 2003; Finnie et al. 2004; Fait et al. 2006; Gutierrez et al. 2007), although most referring to orthodox seed species. For tropical species, in which 52% present seeds with recalcitrant characteristics, studies related to physiological alterations throughout seed development are quite scarce (Tweddle et al. 2003).
Orthodox seeds present higher dehydration tolerance, whereas recalcitrant seeds are sensitive to this process, as much throughout development as after shedding from the parent plant (Berjak and Pammenter 2007). The mechanisms of dehydration tolerance in recalcitrant seeds can be absent or ineffective. Nevertheless, information on the biochemical and cellular alterations produced by dehydration in recalcitrant seeds is rare, thus making a better understanding of the events related to this process obscure (Berjak and Pammenter 2003; Flores 2003). Although several hypotheses have been put forward to explain the physiological basis of seed recalcitrancy (Connor and Bonner 2001), dehydration tolerance is usually correlated with the presence of considerable amounts of oligosaccharides and specific proteins, such as LEA (late embryogenesis abundant), stored during the maturation process (Hoekstra et al. 2001).
Over latter years, several studies have focused on characterizing protein dynamics during plant development, as well as the associated genomes and transcriptomes involved (Roberts 2002; Heazlewood and Millar 2003; Chen and Harmon 2006; Gallardo et al. 2007; Dam et al. 2009). It has been demonstrated that proteomic analysis can generate relevant information towards elucidating the identity of those molecular mechanisms involved in developmental processes. Furthermore, the search is under way to trace biochemical and molecular markers in specific stages of embryogenesis in tree species (Silveira et al. 2004; Rossignol et al. 2006; Santa-Catarina et al. 2006; Silveira et al. 2008). The characterization of profiles and changes in protein content during seed development allows for comparing in vitro and in vivo embryo development. However, although several embryogenically expressed proteins have been identified so far, there have been few reports focused on woody plants (Marsoni et al. 2008), even less so regarding proteomic characterization throughout recalcitrant seed development (Pawlowski et al. 2007; Silveira et al. 2008; Balbuena et al. 2009).
In the present study, our approach was the proteomic characterization of changes in protein profiles throughout the different stages of development in O.catharinensis seeds. The results allow for a better understanding of protein metabolism during developmental stages, as well as the establishment of possibly useful markers for improving and monitoring the development of both recalcitrant and orthodox somatic embryos, as many orthodox species present recalcitrant behavior when cultured in vitro (Moon and Hildebrand 2003; Griga et al. 2007).
MATERIAL AND METHODS
Plant material: Seeds of O. catharinensis were harvested at the Cantareira State Park in São Paulo - Brazil (23º 24' S / 46º 35' W) at the following times after flowering: 170, 230 and 300 days, representing the early-cotyledonary, cotyledonary and mature embryonic stages, respectively (Figure 1), as described by Santa-Catarina et al. (2006). The seeds were stored at -80 ºC, prior to analysis.
Protein extraction: Protein extracts were prepared in biological triplicates for each developmental stage. Due to changes in seed tissue fresh weight throughout seed development, each biological sample was prepared from a bulk of six seeds and ground to a fine powder under liquid nitrogen. For protein extraction, 500 mg of each were transferred into clear 2 mL microtubes containing 1.5 mL of extraction thiourea/urea. The buffer consisted of 7 M urea, 2 M thiourea, 1% dithiothreitol (DTT), 2% Triton X-100, 0.5% Pharmalyte® (GE Healthcare®, Buckinghamshire, UK), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 5 µM pepstatin. The material was vortexed for 15 min and centrifuged for another 5 min at 12.000 g at 4 ºC. Supernatants containing the protein fraction were collected and stored at -20 ºC until 2-DE analysis.
Interfering elements were removed from protein extracts by precipitation with 10% TCA (v/v). Samples were incubated in 10% TCA (v/v) for 30 min at 4 °C. After incubation, samples were centrifuged at 12.000 g for 10 min at 4 ºC. Supernatants were removed and pellets rinsed three times with ice-cold acetone (0.2% DTT) followed by centrifugation (5 min at 12.000 g, 4 ºC) between each rising step. Pellets were air-dried, solubilized in 1 mL of thiourea/urea buffer, and then centrifuged for 5 min at 12.000 g at 4 ºC. Supernatants containing the protein fractions were collected and stored at -20 ºC until 2-DE analysis. Two gels were prepared for each biological sample. Protein concentration was determined using the 2D-Quanti Kit (GE-Healthcare®), with bovine serum albumin (BSA) as standard.
Two-dimensional electrophoresis (2-DE): Aliquots of sample extracts containing 180 µg of protein were used to reach a final volume of 375 µL in a rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG Buffer, 1% DTT, 0.002% bromophenol blue). Prior to this, protein extracts were separated across a broad-range pH 3-10 IPG strip, which resulted in low resolution gels containing spots preferentially distributed in the pH 4-7 range. IPG with a linear separation range of pH 4-7 was chosen for better 2-DE resolution through comparative analysis, and spot gel excision for mass spectrometer. Samples in a rehydration buffer were applied to 18 cm IPG gel strips with a linear separation range of pH 4-7 (GE Healthcare®) and kept in rehydration for 12 h at 20 ºC. First-dimension IEF was carried out on the IPGphor II (GE Healthcare®) at 20 ºC with current limit at 50 µA/strip. Prior to second dimension analysis, individual strips were equilibrated for 20 min in 3 mL of an equilibration buffer (50 mM Tris, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue) containing DTT (125 mM), followed by 20 min in a 3 mL equilibration buffer containing iodoacetamide (125 mM).
The second dimension was performed on 12% SDS-PAGE gels (1 mm x 18 cm x 20 cm) using a Protean II system (Bio-Rad®) coupled to a MultiTemp III Thermostatic Circulator (GE-Healthcare®) cooling bath. Running was set at 12.5 mA/gel for 50 min and 25 mA/gel until the dye reached the bottom of the gel. Low molecular weight protein markers (14 - 97 kDa) (Bio-Rad®) were used as standard. Proteins were stained by mass spectrometry compatible silver staining, according to Shevchenko et al. (1996).
Gel image analyses: Stained gels were scanned with an ImageScanner and analyzed with Image Master Platinum software v.6 (GE-Healthcare®). The authenticity of each spot of protein was validated and edited by visual inspection. A synthetic gel, containing only spots present in all the six gels of each embryo developing stage (three biological samples with two gels each), was obtained and used to validate the presence of spots in all repetitions. The number, percentage volume criterion and distribution of spots were evaluated. Synthetic gels were used to quantify the number of spots per stage, differential expression of proteins using the percentage volume criterion, and differential distribution of proteins in accordance with molecular weight and isoelectric point.
After analysis gels were dehydrated in a solution containing 30% methanol and 3% glycerol, and then stored at room temperature until mass spectrometry identification of differentially expressed polypeptides.
In gel digestion: In gel digestion was performed as described by Shevchenko et al. (2007). Protein spots were excised from the gels with a clean scalpel and cut into approximately 1 mm3 pieces and placed into 600 µL microtubes. Gel pieces were dehydrated in 500 µL acetonitrile for 10 min and then reduced in 50µL of a DTT solution (10mM DTT in 100 mM ammonium bicarbonate) for 30 min at 56 oC, followed by a dehydration step in acetonitrile for 10 min. For alkylation, 50 µL of 55 mM iodoacetamide in 100 mM ammonium bicarbonate was added and gel pieces incubated at room temperature and in the dark for 30 min. Prior to digestion, gel pieces were completely dehydrated by acetonitrile, followed by the increment of grade porcine trypsine (Promega®, Madison, USA) at 16 ng. µL-1. After 120 min, samples were placed into an air circulation thermostat and incubated overnight at 37oC. Following digestion, the gel pieces were saturated with 100 µL of extraction buffer 5% formic acid (FA): acetonitrile (1:2, v/v) and incubated for 15 min at 37 oC in shaker. Supernatants were then collected and dried down in a vacuum centrifuge. For mass spectrometer analysis, 0.05% trifluoroacetic acid (TFA) (10µL) was added into dried tubes, incubated for 2 min and vortexed for a further 10 min at 7 g.
Mass spectrometry: Tryptic digests were injected into a nanoLC-MS/MS Ultimate system (Dionex®, California, USA) interfaced on-line to a linear ion trap LTQ (ThermoElectron®, Massachusetts, USA). Pepetides were first loaded onto a 1 mm x 300 µm i.d. trapping microcolumn packed with C18 PepMap100 5 µm particles in 0.05% TFA at the flow rate of 20 µL/min. After 4 min of washing, they were back-flushed and separated onto a 15 cm x 75 µm i.d. nanocolumn packed with C18 PepMAP100 3 µm particles at the flow rate of 200 nL/min using the following mobile phase gradient: from 5 to 20% of solvent B in 20 min, 20-50% B in 16 min, 50-100% B in 5 min, 100% B during 10 min and then back to 5% B in 5 min. Solvent A was 95:5 water/acetonitrile (v/v) with 0.1% formic acid and solvent B was 20:80 water/acetonitrile (v/v) with 0.1% formic acid. Peptides were eluted into the mass spectrometer via a dynamic nanospray probe (Thermo Electron®) by using an uncoated silicatip needle with a spray voltage of 4.5 kV, with the transfer capillary temperature set to 200 oC. The data dependent acquisition cycle consisted of a survey scan covering the range of m/z 350-1500 followed by MS/MS fragmentation of the three most intense precursor ions under relative collision energy of 35%. The m/z of fragmented precursor ions was excluded for a further 60 s. Spectra were exported as dta files using BioWorks 3.1 software (Thermo Electron®).
Database searches: All dta files from each acquisition cycle were merged into a single mgf file using BioWorks 3.1 software, and prior to database research, mgf files were against 12009 typical background spectra using EagleEye software (Junqueira et al. 2008a, b). Database research was then carried out against the NCBI database by MASCOT v.2.2 (Matrix Science®, Boston, USA). Mass tolerance for precursor and fragmented ions was 2.0 and 0.5 Da, respectively, and carbamidomethyl was set as fixed modification and oxidation (methionine) as variable modification.
Complementary protein identification was done through de novo mass spectra interpretation via PepNovo software (Frank and Pevzner 2005). For each interpretation, seven partially redundant candidate sequences were produced, each containing a quality score which stood for the expected number of confidently determined amino acid residues in the most accurate sequence proposal. Candidate sequences with scores higher than six were merged into a single search string in arbitrary order and MS BLAST searches were performed against the NCBInr database at http://genetics.bwh.harvard.edu/msblast/ under the settings proposed by the engine for LC-MS/MS analysis. Protein identification was carried out according to the MS BLAST identification scheme proposed by Habermann et al. (2004). In cases when different protein accessions were retrieved by MASCOT and MS BLAST searches, bl2seq BLAST was executed between sequences and homology reported.
RESULTS AND DISCUSSION
2-DE analysis of the different stages in O. catharinensis seed development permitted characterizing protein dynamics involved in seed development, disclosing differential metabolism with specific protein, differential expression at each stage, and those in common during the developmental process (Figure 2).
2-DE comparative analysis during seed development revealed that several proteins were observed exclusively at each developmental stage, with 70, 187 and 91 spots related to the early-cotyledonary, cotyledonary and mature stages, respectively (Figure 3). The cotyledonary stage is characterized by highest metabolism indicating the end of embryo development and the beginning of metabolism related to maturation and germination (Gallardo et al. 2003). It was observed that the cotyledonary stage presents the highest number of stage-specific spots, about 45% of the total, suggesting the importance of protein metabolism during transition from embryo development to maturation in the seeds of O. catharinensis. While there was a decrease in the number of spots during the last stages of seed development, this was counterbalanced by an increase in protein concentration (Figure 4), probably due to increased expression by specific proteins at the mature stage, such as LEA and those involved in storage, as well as those related to protection against insects (Sheoran et al. 2005).
During seed development in O. catharinensis, insoluble proteins were detected at increasing concentrations only in the last stages of zygotic embryogenesis (Santa-Catarina et al. 2006). In more tolerant species, dehydration insoluble reserves accumulated in seeds, thereby providing mechanical support (Greggains et al. 2001). Nevertheless, soluble protein concentration increased continuously, reaching a maximum amount in the mature embryo (Santa-Catarina et al. 2006). This was also observed herein, with a significant increase in protein content at the mature stage (Figure 5). Probably, this increase is due to a system of active absorption of amino acids from cotyledons during the final stages of embryo development, when large amounts of storage proteins are synthesized in the seed (Weber et al. 1997).
Among the spots observed during seed development in O. catharinensis, 103 were common to all stages, suggesting few changes in protein metabolism throughout the process. It has been suggested that recalcitrant species present continuous cellular metabolism throughout seed development, without maturation and dehydration phases (Farrant et al. 1997; Brown et al. 2001). Comparison between stages indicated 62 spots in common between early-cotyledonary and cotyledonary, 64 between cotyledonary and mature stages and only 6 between early-cotyledonary and mature stages (Figure 3). Interestingly, certain proteins are frequently identified in proteomic studies due to their abundance, such as enolases and ATP synthase beta subunits (Petrak et al. 2008).
Through 2-DE analysis, alterations in the number of spots in the different ranges of molecular weight indicated the diverse protein profiles during seed development (Figure 5). A decrease in the number of proteins with low molecular weight and an increase in those with high molecular weight, during seed development were evident. An increase in high molecular weight protein expression is fundamental for embryo development, as they are related to maintenance activities and storage, whereas low-weight proteins are mainly related to synthesis (Vensel et al. 2005).
Several studies have shown the synthesis and accumulation of storage proteins in different stages of seed development in tree species (Chatthai and Misra 1998; Silveira et al. 2004; Panza et al. 2007). The accumulation of these proteins during the last phase of development is fundamental, as these proteins will be degraded and used during embryo germination, thereby supplying free amino acids for the beginning of seedling development (Shewry et al. 1995).
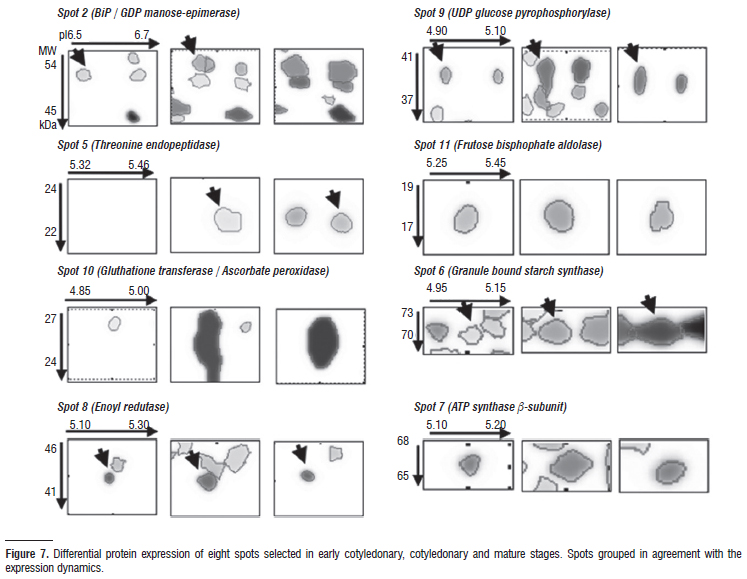
Aiming at a better understanding of the process of seed development in recalcitrant species, protein identification in O. catharinensis seeds was carried out in eight different spots with varied expression: spots exclusive in the final stages (spots 2, 5 e 10), spots with an increase throughout development (spots 8, 9 e 11), and spots with constant expression (spots 6 e 7) (Figs. 6 and 7).
Mass spectrometry analysis and database research resulted in the identification of thirteen proteins from the eight spots analyzed (Table 1). One of the difficulties related to proteomic analysis in most tree species refers to the absence of genomic sequences and expressed sequence databases (ESTs) (Dias et al. 2010). From the thirteen proteins identified, three are directly related to the synthesis of reserves whose expression was increased at the mature stage. The maturation phase is characterized by the storage of reserves, the three main types being: proteins, lipids and polysaccharides (Bewley and Black 1994).
Spot 2 was identified as a BiP protein (Luminal binding protein), a member of the heat shock 70 protein (HSP70) family found in the endoplasmic reticulum (ER) and involved in translocation and protein processing (Hatano et al. 1997). High levels of BiP expression are associated with the accumulation of protein intermediates and folding in storage proteins (Hatano et al. 1997; Forward et al. 2002). The protein identified in spot 6 is Granule-bound starch synthase I, that responsible for forming the extra-long chains in amylopectins, the major type αglucans in starch (Hanashiro et al. 2008; Stensballe et al. 2008). Silva et al. (1998) have already verified that O. catharinensis seeds present a similar protein and carbohydrate concentration. During seed development, enoyl-ACP redutase (spot 8) is synthesized throughout the period of lipid biosynthesis and deposition (Poghosyan et al. 2005). The identification of these proteins reveals storage synthesis during the mature stage, especially since BiP proteins are extremely abundant throughout seed development. As with O. catharinensis seeds, Brown et al. (2001) observed that seeds of Durio zibethinus do not achieve desiccation tolerance but store reserves which they begin to mobilize prior to maturation, suggesting the addition of a fourth category to the proposal by Farrant et al. (1997).
A further two spots were identified as UDP-glucose-pyrophosphorylase (UDPase) (spot 9) and frutose-biphosphate aldolase (spot 11), and are described as key enzymes in carbohydrate metabolism and cell wall biosynthesis (Chen et al. 2007). UDPase catalyzes the production of glucose-1-phosphate and uridine triphosphate from UDP-glucose and pyrophosphate, or the reverse, depending on the metabolic status of the tissues (Chen et al. 2007). In cereal endosperms, it was demonstrated that UDPase can be transformed into AGPase for starch synthesis as an alternative route for starch storage synthesis during the final stages (Kleczkowski 1996).
Metabolic activity and the mitochondrial respiration rate are high during seed development, thereby implying that a significant amount of reactivated oxygen species (ROS) can be generated (Pukacka and Ratajczak 2007). The ascorbate system appears as the main route in oxidative metabolism during embryogenesis and seed filling (De Tulio and Arrigoni 2003). Proteins involved in oxidative metabolism, such as ascorbate peroxidase and glutathione transferase (spot 10), presented increased expression during seed development in O. catharinensis. Moreover, high expression of GDP-mannose3´,5´-epimerase was observed (spot 2). This could be correlated with high concentrations of ascorbic acid (Bulley et al. 2009). Ascorbic acid is involved in the redox system, the redox state can be important in primary metabolism, especially during stress response (Noctor 2006). During seed development, ascorbic acid can be of aid in modulating phytohormones synthesis, as ethylene, giberellin and abscisic acid, thereby affecting growth (Pastori et al. 2003). Increased expression in proteins related to the oxidative system is in agreement with observations by Pukacka and Ratajczak (2007). These authors demonstrated that recalcitrant seeds, the case of O. catharinensis, are characterized by high ascorbate content and intensely active ascorbate system enzymes, when compared with orthodox seeds, in which ascorbic acid contents and ascorbate peroxidase activity decrease significantly or totally disappear during maturation and dehydration phases.
Spot 5, identified as a protein like proteasome, was not expressive in the early stages, although highly so in mature stage. Proteasomes are multicatalytic complexes involved in protein degradation, required for protein activation in the processing of inactive precursors (Etienne et al. 2000). In some cases, the proteinases are synthesized during seed maturation and remain inactive until germination (Callis 1995). Proteins associated to the later stages of embryogenesis may not only be regulated during the maturation phase, but also during the last phases of germination (Finnie et al. 2002; DellAquila 2004). It is important to point out that expression in this group of proteins undergoes expressive increment during mature stage, keeping higher levels throughout the germinative process (data not shown).
The study of differential protein expression furnishes important information towards understanding metabolism and the various roles in seed development. The present study permitted characterizing 2D protein profiles and identifying various proteins during seed development in O. catharinensis. It could be suggested that protein expression related to oxidative metabolism and the BiP proteins observed hereby appear as good makers in the final phase of development. Further studies are fundamental in storage protein form evaluation and those protease activities which mediate in the mobilization of these proteins.
Finally, these findings contribute to our understanding of protein expression dynamics during the development of recalcitrant seeds. Furthermore, identified proteins could be useful as markers for improving and monitoring somatic embryo development, seeing that zygotic and somatic embryogenesis events are very much alike.
Acknowledgments: This research was carried out with financial support from the State of São Paulo Research Foundation (FAPESP) and the National Council for Scientific and Technological Development (CNPq).
Received: 25 February 2010; Accepted: 05 April 2010
- Balbuena TS, Silveira V, Junqueira M, Dias LLC, Santa-Catarina C, Shevchenko A, Floh EIS (2009) Changes in the 2-DE protein profile during zygotic embryogenesis in the Brazilian Pine (Araucaria angustifolia). J.Proteome Res. 72 : 337-352.
- Baud S, Boutin J, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Bioch. 40: 151-160.
- Berjak P, Pammenter NM (2003) Orthodox and recalcitrant seeds. In: Vozzo JA (Ed.) Tropical tree seed manual. USDA Forest Service, Washington DC, Agricultural, 899p.
- Berjak P, Pammenter NW (2007) From Avicennia to Zizania: Seed recalcitrance perspective. Ann. Botany 21: 1-16.
- Bewley JD, Black M (1994) Seeds: Physiology of development and germination. 2nd ed. New York: Plenum Publishing.
- Brown MJ, Hor YL, Grenwood JS (2001) Reserve accumulation and protein storage vacuole formation during development of recalcitrant seed of Durio zibethinus L. Seed Sci. Res. 11: 293-303.
- Bulley SM, Rassam M, Hoser D, Otto W, Schunemann N, Wright M, Macrae E, Gleave A, Laing W (2009) Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C byosynthesis. J. Exp. Botany 72: 1-14.
- Callis J (1995) Regulation of protein degradation. Plant Cell 7: 845-857.
- Chatthai M, Misra S (1998) Sequence and expression of embryogenesis-specific cDNAs encoding 2S seed storage proteins in Pseudotsuga menziesii (Mirb). Franco. Planta 206: 138145.
- Chen S, Harmon AC (2006) Advances in plant proteomics. Proteomics 6: 55045516.
- Chen R, Zhao X, Shao Z, Zhu L, He G (2007) Multiple isoforms of UDP-glucose pyrophosphorylase in rice. Physiol. Plant. 129: 725-736.
- Connor KF, Bonner FT (2001) The effects of dessication on seeds of Acer saccharinum and Aesculus pavia: recalcitrance in temperate tree seeds. Tree 15: 131-136.
- Dam S, Laursen BS, Ornfelt JH, Jochimsen B, Staerfeldt HH, Friis C, Nielsen K, Goffard N, Besenbacher S, Krusell L, Sato S, Tabata S, Thogersen IB, Enghild JJ, Stougaard J (2009) The proteome of seed development in the model legume Lotus japonicus Plant Physiol. 149: 1325-1340.
- De Tulio MC, Arrigoni O (2003) The ascorbic acid system in seeds: to protect and to serve. Seed Sci. Res. 13: 249-260.
- DellAquila A (2004) Protein patterns, characterized by computer image analysis, of lentil embryo axes germinating under salt stress. Biol. Plant. 28: 237242.
- Dias LLC, Santa-Catarina C, Floh EIS, Balbuena TS, Santos ALW, Guerra MP, Steiner N, Silveira V (2010) Proteômica aplicada ŕ biotecnologia de plantas. In: Iacomini V (Ed.) Biotecnologia e novos direitos. Ithala, Curitiba, Brasil. (In press)
- Etienne P, Petitot AS, Houot V, Blein JP, Suty L (2000) Induction of tcl7, a gene encoding a ß-subunit of proteasome in tobacco plants treated with elicitins, salicylic acid or hydrogen peroxide. FEBS Lett. 466: 213-218.
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochiniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 142: 839-854.
- Farrant JM, Pammenter NM, Berjak P, Walters CW (1997) Subcellular organization and metabolic activity during the development of seeds that attain different levels of desiccation tolerance. Seed Sci. Res. 7: 135-144.
- Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis or grain filling and seed maturation in barley. Plant Physiol. 129: 13081319.
- Finnie C, Maeda K, Ostegaard O, Bak-Jensen KS, Larsen J, Svensson B (2004) Aspects of the barley seed proteome during development and germination. Bioch. Soc. Trans. 32: 517-519.
- Flores EM (2003) Seed biology. In: Vozzo, J.A. (Ed.) Tropical tree seed manual. Washington DC, USDA Forest Service. 899p.
- Forward BS, Osusky M, Misra S (2002) The Douglas-fir BiP promoter is functional in Arabidopsis and responds to wouding. Planta 215: 569-576.
- Frank A, Pevzner PP (2005) PepNovo: De novo peptide sequencing via probabilistic network modeling. Ann. Chem. 77: 964-973.
- Gallardo K, Signor C, Vandekerckhove J I, Thompson RD, Burstin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic process related to reserve accumulation. Plant Physiol. 133: 664682.
- Gallardo K, Firnhaber C, Zuber H, Hericher D, Belghazi M, Henry C, Kuster H, Thompson RD (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds. Mol.Cell Proteomics 6: 2165-2179.
- Greggains V, Finch-Savage E, Atherton NM, Berjak P (2001) Viability loss and free radical processes during desiccation of recalcitrant Avicennia marina seeds. Seed Sci. Res. 11: 235-242.
- Griga M, Horacek J, Klenoticova H (2007) Protein patterns associated with Pisum sativum somatic embryogenesis. Biol. Plant. 51: 201-211.
- Gutierrez L, Wuytswinkel OV, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci. 12: 294-300.
- Habermann B, Oegema J, Sunyaev S, Shevchenko A (2004) The power and the limitations of cross-species protein identification by mass spectrometry driven sequence similarity searches. Mol. Cell Proteomics 3: 238-249.
- Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, Matsugasako JI, Takeda Y (2008) Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amilopectin in rice. Plant Cell Physiol. 49: 925-933.
- Hatano K, Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I (1997) A rapid increase in the level of Binding Protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 38: 344-351.
- Heazlewood JL, Millar AH (2003) Integrated plant proteomics putting green genomes to work. Funct. Plant Biol. 30: 471482.
- Hoekstra FA, Golovina EA, Buitinik J (2001). Mechanisms of plant dessication tolerance. Trends Plant Sci. 6: 431-438.
- Junqueira M, Spirin V, Balbuena TS, Thomas H, Adzhubei I, Sunyaev S, Shevchenko A (2008a) Protein identification pipeline for the homology-driven proteomics. J. Bioch. Biophys. Meth. 71: 346-356.
- Junqueira M, Spirin V, Balbuena TS, Waridel P, Surendranath V, Kryukov G, Adzhubei I, Thomas H, Sunyaev S, Shevchenko A (2008b) Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 7: 3382-3395.
- Kleczowski LA (1996) Back to the drawing board: redefining starch synthesis in cereals. Trends Plant Sci. 1: 363-364.
- Marsoni M, Bracale M, Espen L, Prinsi B, Negri AS, Vannini C (2008) Proteomic analysis of somatic embryogenesis in Vitis vinifera Plant Cell Rep. 27: 347-356.
- Moon H, Hildebrand DF (2003) Effects of proliferation, maturation, and desiccation methods on conversion of soybean somatic embryos. In Vitro Cell. Dev. Biol.-Plant 39: 623-628.
- Noctor G (2006) Metabolic signaling in defense and stress: the central roles of soluble redox couples. Plant Cell Environm. 29: 409-425.
- Panza V, Distéfano AJ, Carjuzaa P, Láinez V, del Vas M, Maldonado S (2007) Detection of dehydrin-like proteins in embryos and endosperm of mature Euterpe edulis seeds. Protoplasma 231: 1-5.
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanic S, Verrier PJ, Noctor G, Foyer CH (2003) Leaf vitamin C contents modulate plant defensive transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939-951.
- Pawlowski TA (2007) Proteomics of European beech (Fagus sylvatica L.) seed dormancy breaking: influence of abscisic and giberellic acids. Proteomics 7: 2246-2257.
- Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD (2008) Déjŕ vu in proteomics. A hit parade pf repeatedly identified differentially expressed proteins. Proteomics 8: 1744-1749.
- Poghosyan ZP, Giannoulia K, Katinakis P, Murphy DJ, Hatzopoulos P (2005) Temporal and transient expression of olive enoyl-ACP reductase gene during flower and fruit development. Plant Physiol. Bioch. 43: 37-44.
- Pukacka S, Ratajczak E (2007) Ascorbate and glutathione metabolism during development and desiccation of orthodoz and recalcitrant seeds of the genus Acer. Funct.Plant Biology 34: 601-613.
- Roberts JKM (2002) Proteomics and a future generation of plant molecular biologists. Plant Mol Biol. 48: 143154.
- Rossignol M, Peltier JB, Mock HP, Matros A, Maldonado AM, Jorrín JV (2006) Plant proteom analysis: a 2004 2006 update. Proteomics 6: 55295548.
- Santa-Catarina C, Silveira V, Balbuena TS, Viana AM, Estelita MEM, Handro W, Floh EIS (2006) IAA, ABA, polyamines and free amino acids associated with zygotic embryo development of Ocotea catharinensis Plant Growth Reg. 49: 237247.
- Sheoran IS, Olson DJH, Ross ARS, Sawhney VK (2005) Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics 5: 37523764.
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometry sequencing of proteins from silver-stained polyacrylamide gels. Ann. Chem. 68: 850-858.
- Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 1: 2856-2860.
- Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945956.
- Silva A, Aguiar IB, Damiăo-Filho CF, Durigan JF (1998) Caracterizaçăo morfológica e química de frutos e sementes de canela-preta (Ocotea catharinensis Mez. Lauraceae). Rev.Inst. Flor. 10: 217-228.
- Silveira V, Balbuena TS, Santa-Catarina C, Floh EIS, Guerra MP, Handro W (2004) Biochemical changes during seed development in Pinus taeda L. Plant Growth Reg. 44: 147156.
- Silveira V, Santa-Catarina C, Balbuena TS, Moraes FMS, Ricart CAO, Sousa MV, Guerra MP, Handro W, Floh EIS (2008) Endogenous abscisic acid and protein contents during seed development of Araucaria angustifolia Biol. Plant. 52: 101-104.
- Stensballe A, Hald S, Bauwm G, Blennow A, Welinder KG (2008) The amyloplast proteome of potato tuber. FEBS J. 275: 1723-1741.
- Tweddle JC, Dickie JB, Baskin CC, Baskin JM (2003) Ecological aspects of seed desiccation sensitivity. J. Ecol. 91: 294-304.
- Vensel WH, Tanaka CK, Cai N, Wong JH, Buchanan BB, Hurkman WJ (2005) Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 5: 15941611.
- Viana AM, Mantell H (1999) Somatic embryogenesis of Ocotea catharinensis an endangered tree of the Mata Atlântica (S. Brasil). In: Jain, S., Gupta, P., Newton, R. (Eds.) Somatic embryogenesis in woody plants. Kluwer Publishers, Dordrecht, pp. 330.
- Weber H, Borisjuk L, Wobus W (1997) Sugar import and metabolism during seed development. Trends Plant Sci. 2: 169174.
Publication Dates
-
Publication in this collection
23 July 2010 -
Date of issue
2010
History
-
Received
25 Feb 2010 -
Accepted
05 Apr 2010