Abstract
The aim of data mining is to find useful knowledge inout of databases. In order to extract such knowledge, several methods can be used, among them machine learning (ML) algorithms. In this work we focus on ML algorithms that express the extracted knowledge in a symbolic form, such as rules. This representation may allow us to ''explain'' the data. Rule learning algorithms are mainly designed to induce classification rules that can predict new cases with high accuracy. However, these sorts of rules generally express common sense knowledge, resulting in many interesting and useful rules not being discovered. Furthermore, the domain independent biases, especially those related to the language used to express the induced knowledge, could induce rules that are difficult to understand. Exceptions might be used in order to overcome these drawbacks. Exceptions are defined as rules that contradict common believebeliefs. This kind of rules can play an important role in the process of understanding the underlying data as well as in making critical decisions. By contradicting the user's common beliefves, exceptions are bound to be interesting. This work proposes a method to find exceptions. In order to illustrate the potential of our approach, we apply the method in a real world data set to discover rules and exceptions in the HIV virus protein cleavage process. A good understanding of the process that generates this data plays an important role oin the research of cleavage inhibitors. We consider believe that the proposed approach may help the domain expert to further understand this process.
data mining; exceptions; HIV; protease
BIOINFORMATICS
RESEARCH ARTICLE
Looking for exceptions on knowledge rules induced from HIV cleavage data set
Ronaldo Cristiano Prati; Maria Carolina Monard; André C. P. L. F. de Carvalho
Universidade de São Paulo, Instituto de Ciências Matemáticas e de Computação, São Carlos, SP, Brazil
Correspondence Correspondence to Ronaldo Cristiano Prati Universidade de São Paulo, Instituto de Ciências Matemáticas e de Computação Caixa Postal 668, 13560-970 São Carlos, SP, Brazil E-mail: prati@icmc.usp.br
ABSTRACT
The aim of data mining is to find useful knowledge inout of databases. In order to extract such knowledge, several methods can be used, among them machine learning (ML) algorithms. In this work we focus on ML algorithms that express the extracted knowledge in a symbolic form, such as rules. This representation may allow us to ''explain'' the data. Rule learning algorithms are mainly designed to induce classification rules that can predict new cases with high accuracy. However, these sorts of rules generally express common sense knowledge, resulting in many interesting and useful rules not being discovered. Furthermore, the domain independent biases, especially those related to the language used to express the induced knowledge, could induce rules that are difficult to understand. Exceptions might be used in order to overcome these drawbacks. Exceptions are defined as rules that contradict common believebeliefs. This kind of rules can play an important role in the process of understanding the underlying data as well as in making critical decisions. By contradicting the user's common beliefves, exceptions are bound to be interesting. This work proposes a method to find exceptions. In order to illustrate the potential of our approach, we apply the method in a real world data set to discover rules and exceptions in the HIV virus protein cleavage process. A good understanding of the process that generates this data plays an important role oin the research of cleavage inhibitors. We consider believe that the proposed approach may help the domain expert to further understand this process.
Key words: data mining, exceptions, HIV, protease.
Introduction
The convergence of computing and communication has changed scientific research almost beyond recognition. For instance, in the study of biological processes, especially genetics and molecular biology, high volumes of data have been produced. The sequencing of the human genome and that of other organisms is just one element of an emerging trend in the life sciences. While the knowledge, experience, and insight of researchers remains indispensable, the understanding of life processes is increasingly a data-driven enterprise. Thus, it is necessary to develop sophisticated methods to analyse such data.
Several methods can be used to analyse the collected data, among them, methods based on mathematical and statistical modelling and Machine Learning (ML). Common to all these methods is a frequent that they mainly focus on prediction, i.e., forecasting what will happen in new situations from data that describe what happened in the past. However, we are often interested in methods that, in addition to forecasting, can be used for data explanation and understanding.
Symbolic machine learning can be regarded as the acquisition of structural descriptions from examples. These descriptions should support explanation and understanding as well as prediction. In our view, insights that might be gained by the user are of major interest in the majority of practical machine learning applications. Indeed, this is one of the symbolic machine learning's major advantages over other methods.
In spite of successful machine learning applications reported in the machine learning and data mining literature, some of them highly correlated to specific biological fields, such as medicine (Lavrac et al., 1997) and molecular biology (Cootes et al., 2003), the discovered knowledge is seldom communicable on the application's domain (Dzeroski and Langley 2001). This is mainly due to the fact that machine learning and data mining framework employ formalisms developed by artificial intelligence researchers, such as decision trees and sets of rules. Although such methods can produce highly accurate predictive models, their output representation is not necessarily presented in terms that are familiar to domain experts.
Furthermore, since life is an evolutionary process, the underline underlying processes that generate the data are often a moving target. In this scenario, the currently accepted models are often based in on general rules to create a big picture and exceptions to explain uncharacterised uncharacterisedtic situations. Moreover, in life sciences in general and in genetics and molecular biology in particular, rare objects are of the most interest. This is to say that, although general theories form the core of knowledge in such areas, researchers are often interested in identifying truly unknown exceptions. As a matter of fact, it is almost impossible to come up with models that explain the data without taking into consideration its their possible exceptions.
All things considered, in order to popularise the employment of machine learning methods in such domains, it would be interesting not only to employ machine learning methods in the yet unexplained data but also to adapt these methods in order to make them as close as possible to the current representation of accepted theories. In this work, we present an approach that aims to frame the acquired knowledge in such a way that to increases its acceptance and utilization. Its main characteristic is to incorporate exceptions into the representation used by machine learning algorithms.
This work is organized as follows: Material and Methods presents our proposed approach to discover knowledge, highlighting its main component: exceptions. Case Study presents the HIV protease cleavage case study and results obtained using the standard as well as the proposed approach to extract knowledge from this data using machine learning algorithms. Although this is an intensively studied problem, often employing sophisticatedmodels, we believe that our approach might contribute to the understanding of viral protease function in general, thereby leading to a better understanding of protease families and their substrate characteristics. Finally, Discussion presents some concluding remarks and discussion.
Materials and Methods
Exceptions
In this work, we are interested in symbolic ML algorithms that induce rules1 1 Some symbolic ML algorithms also induce Decision Trees. As we can always rewrite a decision tree as a set of rules, from now on the term rule represents either a rule directly induced by aML algorithm or the one obtained by rewriting a branch of a decision tree as a rule. from a data set D, which consists of a set of n instances described by m distinctive attributes X1, X2, ..., Xm. The rules induced from such a data set are targeted at a specific attribute, often named class attribute, which can assume one of k possible distinct values, named labels or classes.
The aim of the set of rules induced by the learning algorithm is to classify any new instance that has an unknown class value in one of the k possible classes. We focus on symbolic algorithms because the user normally wants to know both the unknown class of an instance and how the other attributes are related to the target attribute. Thus, we do not consider neural networks or other kinds of ''black box'' classifiers, as they are not capable of explicitly expressing knowledge in a symbolic manner. The set of rules Rj, j = 1, ..., p, induced by symbolic algorithms are generally in the format
R: if <condition> then <class = Ci>.
The if <condition> is also called Body or B and <class = Ci> is also called Head or H. Hereafter, to refer to a rule, we will use the notation Body ® Head or, in brief, B ® H. For rule learning algorithms that have the same representational power as propositional logic, each condition is a disjoint of restrictions among the attributes, such as Xi op value, where op can be any operator from the set {=, <, >, < , > , Î} and Ci is one of the k possible classes.
Classical rule learning algorithms are mainly developed to induce sets of rules for classification or prediction tasks, whose aim is to predict or classify new instances with as high an accuracy as possible. In other words, these algorithms try to induce rules with high accuracy and support, so that these rules are gathered in a final set of rules, called classifiers, able to predict the class of new instances with high accuracy. Although this approach produces consistent classifiers, some of the induced rules may be either trivial or difficult to understand by humans.
The straightforward way to discover novel knowledge is to individually evaluate the rules that constitute the classifier, filtering the whole set of rules in order to select those most interesting, according to some objective or subjective criteria (Freitas, 1999). Since these rules are mainly induced by focusing on the classification accuracy bias, they generally express common sense knowledge (i.e., they are common sense or general rules). Even though general rules are consistent with the experts' expectations, in some activities is interesting to find out other kinds of rules outside besides the general ones.
Another important issue is how we interpret and understand the induced rules. ML algorithms can induce both disjointed and overlaid rules. Furthermore, overlaid rules can be either ordered (decision lists) or unordered (independent rules). From a knowledge discovery point of view, rules in a decision list are difficult to understand by the domain expert, since they are meaningful only in the context of all the preceding rules. Alternatively, disjointed and unordered rules can be individually interpreted. Nevertheless, the rules presented in these sets of rules are uncorrelated with each other. In many Data Mining (DM) applications, establishing a type of relationship among those rules can play an important role in obtaining a good overall understanding of the underlying relationships in the domain.
In our view, the construction of classifiers where the main emphasis lies on the classifier's accuracy fails to reflect the way humans construct and express hypothesis. Although the classifier's accuracy is an important issue, it is important to note that at the knowledge discovery level, and in some practical applications, it is almost worthless the direct application of learning algorithms strongly based on accuracy bias is almost worthless, since they it fails on searching in the search for novelty patterns and/or expressing the discovered knowledge closer than humans do (Dzeroski and Langley, 2001).
From a knowledge representation point of view, one of the main features of rules is that they tend to have exceptions (Kivinen et al., 1994). If we could represent the induced rules in this manner, they would be more intuitive for humans, since humans generally talk about knowledge in terms of general patterns and special cases. For instance, in medical applications, physicians generally say that people with certain characteristics tend to have a particular disease; however, in some special situations, they may not develop the disease. Thus, more realistic rules are of the form 'if P then u unless Q'. To represent such a rule we can refine common sense rules by adding exceptions.
Intuitively, exceptions contradict a general or common sense rule. A common sense rule represents a common phenomenon that comes with high support and confidence in a particular domain. Therefore, exceptions to the rules are weak in terms of support, but have confidence similar to the common sense rules (Hussain et al., 2000). Support is a measure related to the relative frequency of the instances covered by a rule and confidence is related to its accuracy.
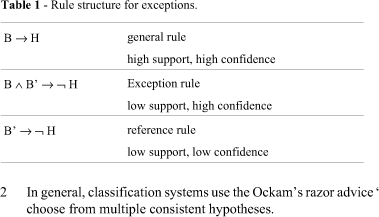
In this work, we use the exception concept given in (Hussain et al., 2000), which structurally defines exception as shown in Table 1, where the term B' also represents a non-empty set of conjunctions of restrictions among the attributes. For instance, if we have the common sense rule ''if a gene X and gene Y are expressed, then the cellular cycle halts'', we could have an exception such as ''if a gene X and gene Y are expressed, but a gene Z is also expressed, then the cellular cycle does not halt''. In this case, the rule ''a gene Z is expressed'' represents the reference rule, which explains the exception. Reference rules should have low support and low confidence and they are difficult do discover.
Exceptions help to solve the understandability problem. A set of isolated rules is not intuitive to the domain expert, since these rules fragment the knowledge; sets of rules expressed in this manner are generally difficult to read and understand because the domain expert cannot see any relationship among the rules. The induced rules have the property that most of the examples are covered by the high-level (general) rules. Lower-level (reference) rules represent exceptions. Once the knowledge is represented in this way, the domain expert can either get a good feeling for what the rules mean by ignoring all the deeper structures and looking only at the first levels or focus his/her attention on some specific points that are unexpected and interesting. It is worth noticing noting that exceptions have the same representation power and logical constraints as production rules. Thus, it is possible not only to represent the concept in an easy-to-understand way but also apply all the available formalisms, such as evaluation metrics.
A related problem is the discovery of interesting or useful rules. The quest for a simple set of rules of existing classification systems2 2 In general, classification systems use the Ockams razor advice " prefer the simplest hypothesis consistent with the data", in order to choose from multiple consistent hypotheses. results in many interesting and useful rules not being discovered. By contradicting the common sense rules, exceptions are generally more interesting and useful to the users. For instance, an exception can play an important role in a cell process regulation. If the biologist recognizes this role as an exception, he or she could better understand the related process.
Another problem is the one concerned towith the static models generated by machine learning algorithms. As some processes are not static (i.e., they may change with time) or new data may be acquired, it is interesting to have a model that could evolve without reengineering. Aside from that, people generally cope with the acquisition and maintenance of complex knowledge structures by making incremental changes to them within a well-defined context such that the effect of changes is locally contained in a well-defined manner (Gaines and Compton, 1995). Standard production rule systems do not have this property. The modularity of the rules themselves is not reflected in a the modularity of the consequences of changes in these rules. Small changes can lead, through complex interactions, to major effects, making the development and maintenance of rule-based systems far more difficult than it appears at first.
Proposed approach
This section describes a new method to find exceptions out from general classification rules. This method is mainly based in the following three key principles (Prati et al., 2003):
1. A reasonable rule induction algorithm can summarize data and learn rules;
2. This algorithm has biases that favour the induction of rules with high support;
3. Exceptions should have low support in the whole data set; otherwise, they would be a common sense rule.
These three principles make difficult the direct induction of exceptions by traditional ML algorithms difficult. The direct extraction of exceptions from a data set is not a trivial task since ML algorithms biases favour the induction of general rules. Although we may relax these biases in order to induce rules with lower support, there is a high chance that the knowledge would be fragmented among the induced rules. All things considered, our proposed approach is divided in the following two steps:
Step 1 induction of common sense rules
In this step we use a traditional rule learning algorithm in order to induce general classification rules. As we are mainly interested in the induction of general rules, its main objective is to avoid the induction of highly specialized rules. This can be achieved by properly configuring the parameters of the learning algorithm.
Normally, a user can stop here for his/her preliminary data mining probing. The user can also apply a filtering step in order to select the most interesting rules. In our approach we also apply a filtering stage, but with the intent of finding and focusing on some rules to be further treated on in the next step.
Step 2 looking for exceptions
In this step we focus on rules that are general (cover several examples) but also have high misclassification rates in order to search for possible exceptions. After identifying such rules, the search for reference rules that might be exceptions is initiated. For each of these rules, and only using the subset of instances that are misclassified by that rule, we look for associations with attribute instances and the negative(s) class(es)3 1 Some symbolic ML algorithms also induce Decision Trees. As we can always rewrite a decision tree as a set of rules, from now on the term rule represents either a rule directly induced by aML algorithm or the one obtained by rewriting a branch of a decision tree as a rule. foreseen by the rule. If those associations have a minimum support and confidence values in the subset of instances, they represent a reference rule and the pair of rules (general rule, reference rule) represents representing one exception. It is worth noticing noting that, although reference rules have high support in the subset of instances used to look for exceptions, they probably have small support in the whole data set.
A major advantage regarding our approach is that besides extracting exceptions out of general classification rules, it also preserves the locality concept of exceptions. This offers a powerful mechanism of expressing knowledge. Another point is that the model is not static, since incremental modifications can be made to a set of rules by adding exceptions to existing rules rather than by reengineering the entire set (Gaines and Compton, 1995).
Case Study
The biological problem
Intact HIV (Human Immunodeficiency Virus) virions are endocytosed (inserted into a cell) via specific cellular receptors on human cells. For 'retroviruses', a single stranded RNA sequence (typically between 8-12 kilobases and containing at least 9 genes, including genes for producing core protein precursors (gag), envelope proteins (env) and pol (reverse transcriptase, integrase and protease)) is then transcribed by one of the enzymes accompanying the RNA sequence into double stranded DNA (by the reverse transcriptase enzyme) and integrated with the host genome (by the integrase enzyme). The DNA provirus (originally reverse transcribed from RNA or single stranded DNA, or simply the original double stranded inserted viral DNA), when expressed, is transcribed into messenger RNA (mRNA) and translated into a protein chain (viral polyproteins), giving rise to new viral molecules which then reassemble to form complete virions that are then released for the infection of further cells.
Viral protease is the third enzyme typically accompanying viral DNA or RNA into the cell, although protease can also self-cleave itself naturally from the viral polyprotein if it is not introduced through endocytosis. It cleaves the precursor viral polyproteins (the substrate) at specific cleavage-recognition sites when they emerge from the ribosome of the host cell as one long sequence. This cleavage step is essential in the final maturation step of HIV. That is, protease is responsible for the post-translation processing of the viral gag and gag-pol polyproteins to yield the structural proteins and enzymes of the virus for further infection (Figure 1). If viral protease action can be inhibited by drugs so that such cleavage-recognition sites cannot be identified, viral replication can be stopped.
Results
In order to illustrate the potential of our approach, we chose a real world data set4 4 Available at http://www.dcs.ex.ac.uk/~anarayan/ismbdatasets/ related to where a viral protease cleaves HIV viral polyprotein amino acid residues. This data set is also used by (Narayanan et al., 2002, Cai and Chou, 1998). Table 2 summarizes this data set.
Each instance of the HIV data set consists of eight attributes that represent a recognition sequence followed by its class, related to its cleavage-ability. In its turn, each attribute on the recognition sequence represents one amino acid. The attributes on the recognition sequence are sequentially ordered, i.e., the first attribute refers to position one in the sequence, the second attribute to position two, and so forth. The size of the recognition sequence is the same as the size of the viral protease. When one of the recognition sequences matches its counterpart in the viral protease, the cleavage occurs between positions 4 and 5 of the recognition site. Whenever it does not match, the cleavage does not occur.
To avoid a possibility of exceptions over-fitting the data, we repeated the experiment three times using different training and testing samples; the data set was divided into three different disjoint subsets, and in each experiment two subsets were used for training and the other one for testing (3-fold cross-validation).
To apply the first step of the proposed methodology, we used the See5 program, which induces symbolic decision trees. To generate smaller trees, the option of generating nodes having subsets of possible values for conditions rather than individual values was set up. The overall error rate achieved by See5 is 14.63% with an standard deviation of 5.95%. Using our proposed approach, the error rate is estimated in at 13.53% with a standard deviation of 6.24%. This result is significantly better with a 95% of confidence level, using a paired t-test to assess significance. Regarding sensitivity (true positive rate) our approach also outperforms See 5 with a 95% of confidence level. The sensitivity achieved by See5 is 85.09% with a 7.60% of standard deviation while our approach achieved 93.86% of sensitivity with a 4.02% of standard deviation.
As the transcription of the decision trees into rules produced a small number of rules, we selected decided to apply the second step to all the rules. On In the second step, each rule selected in the first step defines a subset of instances where we need to look for exceptions. As stated before, this subset contains all the instances that are covered by a rule but do not have the same class as the one foreseen by the rule. In order to look for possible reference rules, we applied the association rule-mining algorithm APRIORI (Agrawal et al., 1993). We only select as possible reference rules the ones that present at least a support value of at least 0.5. Furthermore, if the possible reference rule appears at least twice in all the conducted experiments, we assume it is a reference rule and the pair (general rule, reference rule) as a truely exception.
Table 3 shows the final hypothesis found by our proposed approach. In this hypothesis we have an exception to the rule R1. Without the exception, this rule covered 229 instances in the whole data set, where 17 of them were mistakenly covered. When we added the exception, 10 of these 17 examples were correctly covered.
The induced rules confirm the importance on onf the cleavage process of ofn the amino acids in positions 4 and 5 of the substrate. This point is the one where the linkage of the catalytic process occurs in order to cleave the substrate (scissile linkage). The generated exception also shows the importance of the position 6 in this process, which does not appear oin the rules directly induced by the ML algorithm 4. The importance of position 6 in this process is also related in (Narayanan et al., 2002).
In order to compare our proposed approach to a traditional ML algorithm, we also apply the See5 (rules) program, with default parameters, to the whole data set. Table 4 shows the rule set induced by See5 (rules) on the data set. The numbers in parentheses at the end of each rule represent, respectively, the number of instances correctly and incorrectly covered by the rule. The overall misclassification rate, assessed using the 3-fold cross validation technique, is 14.08% with a standard deviation of 6.43%. In this case, although the nominal rate is slightly better, our approach does not outperform See5 with a 95% of confidence level. As can be observed, a domain expert is barely able to have an overall understanding of the related data.
We also tried to find out the exception using only a traditional ML algorithm (in this case, we used again the See5 algorithm, without setting up the option that generates subsets of values in the nodes). To this end, we stepwise relaxed the pruning confidence factor in by 5% until the attribute position 6 appeared in the induced tree (we stepwise relaxed this value from 25% (default) until to 60%). The result is an induced decision tree with 55 leave nodes that can be translated into 55 rules. In two of these 55 rules, the disjoint if pos6 = E appears. However, in these cases, we cannot see the relationship between the generated rules and the exception.
Discussion
HIV protease is one of the most studied processes aiming in an effort to develop drugs against AIDS (Acquired Immune Deficiency Syndrome) infection (Wlodawer and Vondrasek, 1998). Although an understanding of the HIV lifecycle indicates that inhibition of HIV protease could lead to a treatment of HIV infections, the creation of HIV protease inhibitors requires a detailed understanding of the molecules involved and their interactions.
However, studies on protease inhibitors are mainly carried out ''ad-hoc'', generally focusing only on some HIV variations and developing complex computer models or analysing crystallized forms of proteases using X-rays. These processes are both costly and time consuming. Besides, it is also known that proteases seem to be able to evolve in response to virus mutations (Narayanan et al., 2002). Some HIV protease inhibitors (e.g., Saquinivir, Ritonavir, and Indinavir) have been produced and are currently on the market. These drugs have been proved a successful treatment against proven successful in treating HIV infection (Wlodawer and Vondrasek, 1998), although serious side effects (the virus life cycle is intertwined with the cell life cycle) and the development of resistance are still major unsolved problems (Wlodawer and Vondrasek, 2002). In this sense, due to the high degree of viral mutation, it is generally accepted that protease inhibitors may have to ''co-evolve'' with their protease targets.
However, even though there are more than 150 cleavage proteins deposited at the Protein Data Bank (PDB), there is still little understanding of how viral polyproteins are cut into their functional units. Moreover, it is clear that the structures provided only a small fragment of information necessary for drug design (Wlodawer and Vondrasek, 2002). Thus, the challenge is to investigate whether it is possible to generalise from known cleavage sites to unknown ones. A general understanding of viral protease specificity may help the development of future anti-viral drugs involving protease inhibitors by identifying specific features of protease activity for further experimental investigation.
We believe that the approach proposed in this work to find exceptions out of general rules is especially suitable for such analysis. It allows a more compact and easy to understand model description, helping the domain expert to understand the underline underlying process. While the general induced rules are related to the biological process, as is already known that phenylalanine(F), tryptophan(W) or tyrosine(Y) are generally present in on either side of the cleavage point in the substrate (Pettit et al., 1991) and R2 capture this pattern, the exception found also, it is well know that carboxylic acids, such as Glutamic (E) preset in the found exception, have a fundamental role in the formation of peptide bonds which link amino acids together to form the backbone of the protein (Landis et al., 2004) can provide some insights in order to help the domain expert to understand the underlying data.
A natural extension of this work is the analysis and validation of the generated rules by domain experts. To this end, the results reported here could be used as a pointer compass needle towards for future laboratorial experiments, which would take into account patterns of residuals instead of focusing only on some HIV variations. Finally, it is worth noticing noting that the model built in this work is somehow somewhat simplistic, since it neither explores possible relations between two (or more) positions in the substrate nor takes into account the biochemistry and physical properties of the amino acid residues. It would be interesting to further refine the model by incorporating such properties as background knowledge and using learning algorithms that can directly incorporate such background knowledge and learning first order rules that can represent relationships. However, due to the exponential search associated with such algorithms, the domain expert's help and guidance is of fundamental importance in order to constraint the search.
We conclude by claiming stating that in this task, like others related to molecular biology and bioinformatics, the use of machine learning approaches which are able to provide relevant insights to the domain experts are often more useful than approaches that only look at classification accuracy.
Acknowledgments
This work is partly supported was supported in part by the Brazilian research councils CAPES and FAPESP.
Received: August 15, 2003; Accepted: August 20, 2004
- Agrawal R, Imielinski T and Swami AN (1993) Mining association rules between sets of items in large databases. ACM SIGMOD International Conference on Management of Data, Washington, D.C., pp 207216.
- Cai Y-D and Chou K-C (1998) Artificial neural network model for predicting HIV protease cleavage sites in protein. Advances in Engineering Software 29:119128.
- Cootes A, Muggleton S and Sternberg M (2003) The automatic discovery of structural principles describing protein fold space. Journal of Molecular Biology 330:839-850.
- Dzeroski S and Langley P (2001) Computational discovery of communicable knowledge: Symposium report. Proceedings of the Fourth International Conference on Discovery Science, v 2226 of Lecture Notes in Computer Science, Whasington, DC, pp 4549.
- Freitas AA (1999) On rule interestingness measures. Knowledge-Based Systems 12:309315.
- Gaines BR and Compton P (1995) Induction of ripple-down rules applied to modeling large databases. Journal of Intelligent Information Systems 5:211228.
- Hussain F, Liu H, Suzuki E and Lu H (2000) Exception rule mining with a relative interesting measure. PAKDD-2000, v 1805 of Lecture Notes on Artificial Intelligence, Kyoto, Japan, pp 8697.
- Kivinen J, Mannila H and Ukknonen E (1994) Learning rules with local exceptions. Computational Learning Theory: EuroCOLT '93, Oxford, Clarendon Press, pp 3546.
- Landis C, Cleveland T, Cloninger M and Pollock D (2004) Inhibitors of hiv protease: An introduction to carbonyl chemistry. http://www.chem.wisc.edu/~newtrad/CurrRef/AIDStopic/AIDStext/AIDStoc.html, acessedaccessed in July, 22.
- Lavrac N, Keravnou E and Zupan B (eds) (1997) Intelligent Data Analysis in Medicine and Pharmacology, v 414 of The Kluwer International Series in Engineering and Computer Science. Kluwer Academic Publishers, Boston.
- Narayanan A, Wu X and Yang ZR (2002) Mining viral protease data to extract cleavage knowledge. Bioinformatics 18(Suppl. 1): S5S13.
- Pettit SC, Simsic J, Loeb DD, Everitt L, Hutchinsin III CA and Swanstrom R (1991) Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements os of the p1 amino acid. J Biol Chem 266:1453914547.
- Prati RC, Monard MC and Carvalho ACPLF (2003) A method for refining knowledge rules using exceptions. In: Acosta G (ed), Proceedings of the V Argentine Symposium on Artificial Intelligence, 11 pp.
- Vondrasek, J. and Wlodawer A (2002) HIVdb: A database of the structures of human immunodeficiency virus protease. PROTEINS: Structure, Function, and Genetics 49:429431.
- Wlodawer, A. and Vondrasek J (1998) Inhibitors of HIV-1 protease: A major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct 27:249284.
Publication Dates
-
Publication in this collection
14 Jan 2005 -
Date of issue
2004
History
-
Accepted
20 Aug 2004 -
Received
15 Aug 2003