Abstracts
The American continent was colonized in the 16th century by Europeans who first introduced cattle of Bos taurus origin. Accounts register introduction of Bos indicus cattle into South America in the 19th and continuing through the 20th century, and most reported imports were males derived from the Indian subcontinent. In the present study we show, by using mitochondrial DNA (mtDNA) polymorphism, major participation of matrilineages of taurus origin in the American Zebu purebred origin, i.e., 79, 73 and 100% for the Nellore, Gyr and Brahman breeds, respectively. Moreover, we have created a restriction map identifying polymorphism among B. taurus and B. indicus mtDNA using three restriction enzymes. Results are discussed concerning American Zebu origins and potential use of this information for investigating the contribution of cytoplasmic genes in cattle production traits.
O continente americano foi colonizado no século XVI por europeus que fizeram as primeiras introduções de bovinos, de origem taurina. Os registros das primeiras importações de Zebus para a América do Sul datam do século XIX e continuam até o século XX, constituídos na maioria por machos do sub-continente indiano. Neste artigo, demonstramos, através dos estudos de polimorfismos no DNA mitocondrial (mtDNA), uma participação majoritária de matriarcas de origem taurina na formação do Zebu PO americano (79% dos animais analisados da raça Nelore, 73% na Gir e 100% na Brahman). Ainda, criamos um mapa de restrição com os polimorfismos descritos de três enzimas de restrição. Os resultados estão discutidos em termos da origem do Zebu americano e da aplicação deste conhecimento no estudo dos efeitos do genoma citoplasmático, nas características produtivas dos bovinos.
IS THE AMERICAN ZEBU REALLY Bos indicus?
Flávio V. Meirelles 1,3,4, Artur J.M. Rosa1, Raysildo B. Lôbo1, Joaquim M. Garcia2, Lawrence C. Smith3 and Francisco A.M. Duarte1
1Departamento de Genética, FMRP, USP, Ribeirão Preto, SP, Brasil.
2Departamento de Reprodução Animal, FCAVJ, UNESP, Jaboticabal, SP, Brasil.
3Centre de Recherche en Reproduction Animale Univ. De Montréal PQ Canada.
4Departamento de Ciências Básicas, FZEA, USP, Pirassununga, SP, Brasil. Send correspondence to F.V.M. Av. Duque de Caxias Norte, 225, Caixa Postal 23, 13630-970 Pirassununga, SP, Brasil. E-mail: meirellf@usp.br
ABSTRACT
The American continent was colonized in the 16th century by Europeans who first introduced cattle of Bos taurus origin. Accounts register introduction of Bos indicus cattle into South America in the 19th and continuing through the 20th century, and most reported imports were males derived from the Indian subcontinent. In the present study we show, by using mitochondrial DNA (mtDNA) polymorphism, major participation of matrilineages of taurus origin in the American Zebu purebred origin, i.e., 79, 73 and 100% for the Nellore, Gyr and Brahman breeds, respectively. Moreover, we have created a restriction map identifying polymorphism among B. taurus and B. indicus mtDNA using three restriction enzymes. Results are discussed concerning American Zebu origins and potential use of this information for investigating the contribution of cytoplasmic genes in cattle production traits.
INTRODUCTION
Domestic cattle breeds can be separated into two major groups, no hump taurine (Bos taurus) and humped Zebu (Bos indicus). Although classical Linnean nomenclature distinguishes these as separate species, due to the clinal nature of the allozyme variability and complete interfertility, many authors consider both as subspecies (Manwell and Baker, 1980). Moreover, although fossil evidence has suggested a single origin for both groups within the 10,000-year history of animal husbandry (Epstein, 1971; Epstein and Mason, 1984), molecular clock evidence based on mitochondrial DNA sequence divergence in the displacement loop (D-loop) has more recently suggested two independent domestication events occurring at least 200,000 years ago (Loftus et al., 1994a). Sequence polymorphisms among taurus and indicus breeds have also been detected in less variable regions of tRNA genes and in the origin of light strand replication (Pegoraro et al., 1996).
Zebu cattle were first brought from India to the American continent at the beginning of the nineteenth century. It is believed that a total of approximately 7,000 animals were brought to South America by Brazilian breeders during several imports lasting until the 1960's (Santiago, 1972; Magnabosco, 1997). While some females were included in shipments, most imported animals were males, which were mated to "native" Bos taurus cows originating in the Iberian Peninsula. The current registered population of Brazilian Zebu cattle subdivides into descendents of animals of imported origin (POI, purebred of imported origin) and those possibly derived locally by backcross matings to Zebu males. Mendelian segregation has purified the nuclear genotype of purebred origin (PO) animals to a point where they cannot be differentiated from the POI. However, cytoplasmic genes are generally inherited uniparentally and may be a significant source of polymorphism among this subspecies. As described early this century (Correns, 1909), cytoplasmic inheritance is controlled mainly through the expression of genes contained within the mitochondrion inherited almost exclusively through maternal lineage in mammals (Hutchinson et al., 1974; Giles et al., 1980). Bovine dairy and beef production traits have been shown to vary in different maternal lineages and likely originate from polymorphic mitochondrial genotypes (Tess et al., 1987; Brown et al., 1988; Schutz et al., 1993, 1994; Mannen et al., 1998). We hypothesize that the Zebu cattle population in the Americas is now composed of two groups according to mtDNA content; we demonstrate here that, among three breeds analyzed, only one third contain indicus mtDNA. These results suggest that a large proportion of American Zebu matrilineages were derived by backcrossing "native" females of taurine B. taurus origin to bulls imported from the Indian continent carrying B. indicus mitochondrial genotypes.
MATERIAL AND METHODS
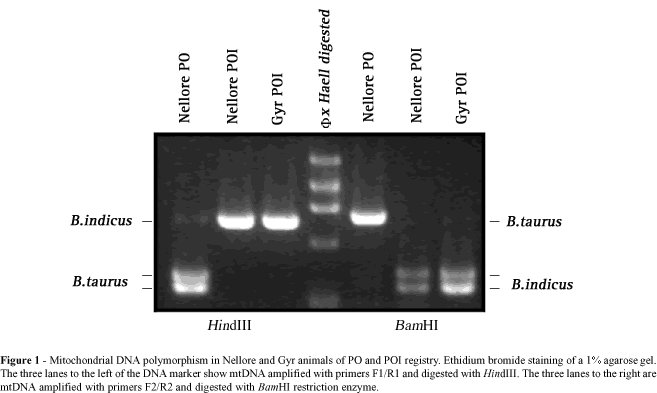
Animals with registered pedigrees from the Bos indicus breeds Nellore, Gyr and Brahman were chosen for this analysis. Animals from the Nellore and Gyr breeds are the most common breeds of Zebu cattle in South America and one of the first introduced whereas the Brahman breed is the most popular in North America. Both males and females were chosen for mtDNA analysis and all animals were grouped according to breed registry as PO or POI. Total DNA was extracted either from semen (males) or leukocytes (females) by standard procedures (Sambrook et al., 1989). Amplification of mtDNA was performed using these primers: 5´-CCCAACGAGGAA AATATACC-3´ (BosmtF1) and 5´-AACCGCAAACAACCTCTTCC-3´ (BosmtR1), which were synthesized to amplify a region of the ND5 gene of the mitochondrial genome (nucleotide 11770 to 12525 according to Anderson et al., 1982a). The PCR reaction was performed using 1.5 mM MgCl2 for 28 cycles of 30 s at 94°C, 45 s at 58°C and 90 s at 72°C. Amplified mtDNA was digested with HindIII, electrophoresed on an agarose gel and stained with ethidium bromide. The presence of the HindIII restriction site within the amplified region is indicative of Bos taurus mitochondrial contribution (Figure 1) (Loftus et al., 1994b). The amplified region was then sequenced as described by Pegoraro et al. (1996) to characterize the mutation generating this polymorphism.
Polymorphisms for B. indicus and B. taurus mtDNA were identified using two other RFLP markers (see restriction map, Figure 2). First, total DNA samples from several animals were re-amplified by PCR using primers 5´-CAAGGGCACCATACCCCAGC-3´ (BosmtF2) and 5´-CCCATAGGATCAGCCCGC-3´ (BosmtR2) which amplify part of the COIII gene, the tRNA glycine and a small part of the ND3 gene of mtDNA (nucleotide 9171 to 9948; Anderson et al., 1982a). Amplified mtDNA was then digested with BamHI enzyme that exclusively restricts B. indicus mtDNA (Figure 1). The last polymorphism marker used was obtained by amplification of part of the 16S rRNA from nucleotide 1692 to 2441 (Anderson et al., 1982a) with 5´-GCCCGAAACCAGACGAGCTAC-3´ (BosmtF3) and 5´-TTGTATGAATGGCCGCACGAGG-3´ (BosmtR3) primers. This fragment harbors either one (indicus) or two sites (taurus) for the HpaI. Restriction of the amplified fragment in both sites is indicative of B. taurus origin.
RESULTS AND DISCUSSION
A total of 124 animals of the Nellore, Gyr and Brahman breeds examined showed patterns of enzymatic restriction characteristic of either B. indicus or taurine mtDNA origin (Figure 1). None of these animals showed patterns of both B. indicus and B. taurus mtDNA, indicating absence of heteroplasmy for these mitochondrial genotypes. On average 58% of the analyzed Nellore and Gyr breeds carried mtDNA of taurine origin, indicating significant influence of taurine breeds in the formation of South American Zebu breeds. The proportion of animals with B. taurus mtDNA among PO (79 vs. 73%, P > 0.05) and POI (26 vs. 25%, P > 0.05) did not differ among the Nellore and Gyr breeds, suggesting similar formation patterns in both breeds. Females of imported origin and their descendents should contain exclusively B. indicus mtDNA, suggesting pedigree recording errors in approximately one quarter of the Nellore and Gyr POI matrilineages. Brahman animals carried exclusively mtDNA of taurine origin, indicating that participation of B. indicus breeds in the formation of the analyzed animals was entirely through paternal genes (Table I).
Among the markers used in this study only the loss of the HindIII restriction site leads to changes in amino acid sequence of a mitochondrial protein. The mutation was characterized as a T to G transversion in nucleotide 12178 that replaces a phenylalanine with a valine in the 24th amino acid of the ND5 subunit. This polymorphism, located in the most variable region of the ND5 amino acid sequence, suggests that it does not influence mitochondrial function (Anderson et al., 1981, 1982a,b; Bibb et al., 1981). Other polymorphisms present between indicus and taurus mtDNA could, however, influence energy supply to tissues due to changes in oxidative phosphorylation efficiency. Although this remains to be confirmed, the American Zebu breeds provide a unique opportunity for investigating participation of the cytoplasmic genome in production traits, since these breeds include animals with diverse mtDNA genotypes but similar nuclear genes.
In this work we also identified phenotypically similar B. indicus animals carrying mtDNA of either B. indicus or B. taurus origin; we postulate that this polymorphism arose during the introduction and establishment of Zebu cattle breeds in America. Since only animals from elite breeders were used in this study, the 78% of the PO cattle carrying B. taurus mtDNA probably underestimates the contribution of taurine matrilineages to the American Zebu, and although as yet unconfirmed, contribution of taurine mtDNA to nonregistered commercial Zebu herds is likely to be significantly higher than in the purebred registered herds here assessed. These findings will enable further analyses of mitochondrial genotypes participation in production traits in American Zebu breeds.
ACKNOWLEDGMENTS
The authors thank Mr. Antônio de Carvalho, and the Programa de Melhoramento Genético do Rebanho Nelore (PMGRN), for providing blood and semen samples of the Nellore breed, and Gyr breeders association (Assogir) for providing Gyr blood samples for DNA extraction. We also thank Mr. Mendelson Marzucato for excellent technical work. Financial support was provided by CAPES, FAPESP and FUNPEC-RP, Brazil. Publication supported by FAPESP.
RESUMO
O continente americano foi colonizado no século XVI por europeus que fizeram as primeiras introduções de bovinos, de origem taurina. Os registros das primeiras importações de Zebus para a América do Sul datam do século XIX e continuam até o século XX, constituídos na maioria por machos do sub-continente indiano. Neste artigo, demonstramos, através dos estudos de polimorfismos no DNA mitocondrial (mtDNA), uma participação majoritária de matriarcas de origem taurina na formação do Zebu PO americano (79% dos animais analisados da raça Nelore, 73% na Gir e 100% na Brahman). Ainda, criamos um mapa de restrição com os polimorfismos descritos de três enzimas de restrição. Os resultados estão discutidos em termos da origem do Zebu americano e da aplicação deste conhecimento no estudo dos efeitos do genoma citoplasmático, nas características produtivas dos bovinos.
(Received May 21, 1999)
- Anderson, S., Bankier, A.T., Barrell, B.G., de Bruijn, M.H., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., Sanger, F., Schreier, P.H., Smith, A.J., Staden, R. and Young, I.G. (1981). Sequence and organization of the human mitochondrial genome. Nature 290: 457-465.
- Anderson, S., de Bruijn, M.H.L., Coulson, A.R., Eperon, I.C., Sanger, F. and Young, I.G. (1982a). The complete sequence of bovine mitochondrial DNA: Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 156: 683-717.
- Anderson, S., Bankier, A.T., Barrell, B.G., de Bruijn, M.H., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., Sanger, F., Schreier, P.H., Smith, A.J., Staden, R. and Young, I.G. (1982b). Comparison of the human and bovine mitochondrial genomes. In: Mitochondrial Genes (Slonimski, P., Borst, P. and Attardi, G., eds.). Cold Spring Harbor, New York, pp. 5-43.
- Bibb, M.J., Van Etten, R.A., Wright, C.T., Walberg, M.W. and Clayton, D.A. (1981). Sequence and gene organization of mouse mitochondrial DNA. Cell 26: 167-180.
- Brown, D.R., George, M. and Wilson, A.C. (1988). Mitochondrial respiratory metabolism and performance in cattle. J. Anim. Sci. 66: 1347-1354.
- Correns, C. (1909). Vererbungsversuche mit blass (gelb) grünen und buntblattrigen Sippen bei Mirrabilis, Urtica, und Lunaria. Z. Vererbungsl. 1: 291-329.
- Epstein, H. (1971). Origin of the Domestic Animals of Africa Africana Publishing Cooperation, New York.
- Epstein, H. and Mason, I.L. (1984). Cattle. In: Evolution of Domesticated Animals (Mason, I.L., ed.). Longman, New York, pp. 6-27.
- Giles, R.E., Blanc, H., Cann, H.M. and Wallace, D.C. (1980). Maternal inheritance of human mitochondrial DNA. Proc. Nat. Acad. Sci. USA 77: 6715-6719.
- Hutchinson III, C.A., Newbold, J.E., Potter, S.S. and Edgell, M.H. (1974). Maternal inheritance of mammalian mitochondrial DNA. Nature 251: 536-538.
- Loftus, R.T., MacHugh, D.E., Bradley, D.G., Sharp, P.M. and Cunningham, P. (1994a). Evidence for two independent domestications of cattle. Proc. Nat. Acad. Sci. USA 91: 2757-2761.
- Loftus, R.T., MacHugh, D.E., Ngere, L.O., Balain, D.S., Badi, A.M., Bradley, D.G. and Cunningham, E.P. (1994b). Mitochondrial genetic variation in European, African, and Indian cattle populations. Anim. Genet. 25: 265-271.
- Magnabosco, A.U., Cordeiro, C.M.T., Trovo, J.B.F., Mariante, A.S., Lôbo, R.B. and Josahkian, L.A. (1997). Catálogo de Linhagens do Germoplasma Zebuíno: Raça Nelore Cenargen, Brasília, pp. 11-52.
- Mannen, H., Kojima, T., Oyama, K., Mukai, F., Ishida, T. and Tsuji, S. (1998). Effect of mitochondrial DNA variation on carcass traits of Japanese Black cattle. J. Anim. Sci. 76: 36-41.
- Manwell, C. and Baker, C.M.A. (1980). Chemical classification of cattle: 2. Phylogenetic tree and specific status of the Zebu. Animal and blood groups. Biochem. Genet.11: 151-162.
- Pegoraro, L., Yang, Z., Samaké, S., Meirelles, F.V., Bordignon, V., Moquin, L. and Smith, L.C. (1996). Sequence comparison of mitochondrial tRNA genes and origen of light strand replication in Bos taurus and Nellore (Bos indicus) breeds. Anim. Genet. 27: 91-94.
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Santiago, A.A. (1972). História do Zebu. In: O Gado Nelore Instituto de Zootecnia, Săo Paulo, pp. 69-196.
- Schutz, M.M, Freeman, A.E., Lindenberg, G.L. and Beitz D.C. (1993). Effect of maternal lineages grouped by mitochondrial genotypes on milk yield and composition. J. Dairy Sci. 76: 621-629.
- Schutz, M.M, Freeman, A.E., Lindenberg, G.L., Koheler, C.M. and Beitz, D.C. (1994). The effect of mitochondrial DNA on milk production and health of dairy cattle. Livest. Prod. Sci. 37: 283-295.
- Tess, M.V., Reodecha, C. and Robinson, O. (1987). Cytoplasmic genetic effects on preweaning growth and milk yield in Hereford cattle. Science 65: 675-684.
Publication Dates
-
Publication in this collection
17 Feb 2000 -
Date of issue
Dec 1999
History
-
Received
21 May 1999