Abstract
Karyotypic characteristics of three species of the genus serrasalmus ( S. altispinnis , S. gouldingi and S. Serrulatus ) from the middle and lower Negro River, Amazon Basin, were investigated using different staining techniques and Fluorescent in situ hybridization with 5S and 18S rDNA probes. The diploid number was invariably 2n = 60 and the fundamental number was FN = 110. Nevertheless, the karyotypes differed from each other in composition: 24m, 20sm, 6st, 10a in S. altispinnis ; 22m, 22sm, 6st, 10a in S. gouldingi and 20m, 22sm, 8st, 10a in S. serrulatus. The karyotype of S. altispinnis differed from the one previously described in a population from the Pitinga River. C-positive constitutive heterochromatin was mainly pericentromeric in the karyotypes of all species. Nucleolar organizer regions were multiple and preferentially located terminally on the short arms of the subtelocentric/acrocentric chromosomes, as evidenced by both silver nitrate staining and fluorescent in situ hybridization with the 18S rDNA probe. The maximum number of NORs varied among species, as did the NOR-bearing chromosomes. FISH with the 5S rDNA probe produced an interstitial signal on the long arms of the pair 7 in all species, coincident with a C-positive heterochromatic band. While some chromosome features were shared by the three species, some were species-specific and thus useful for cytotaxonomy.
chromosome banding; cytotaxonomy of piranhas; FISH; karyotype; ribosomal DNA
GENETICS
Mapping of ribosomal genes and chromosomal markers in three species of the genus Serrasalmus (Characidae, Serrasalminae) from the Amazon basin
Celeste M. Nakayama1
Eliana Feldberg1
Luiz Antonio C. Bertollo2
1Laboratório de Genética de Peixes, Coordenação de Pesquisas em Biologia Aquática, Instituto Nacional de Pesquisas da Amazônia, Manaus, AM, Brazil
2Departamento de Genética e Evolução, Universidade Federal de São Carlos, São Carlos, SP, Brazil
Karyotypic characteristics of three species of the genus serrasalmus (S. altispinnis, S. gouldingi and S. Serrulatus) from the middle and lower Negro River, Amazon Basin, were investigated using different staining techniques and Fluorescent in situ hybridization with 5S and 18S rDNA probes. The diploid number was invariably 2n = 60 and the fundamental number was FN = 110. Nevertheless, the karyotypes differed from each other in composition: 24m, 20sm, 6st, 10a in S. altispinnis; 22m, 22sm, 6st, 10a in S. gouldingi and 20m, 22sm, 8st, 10a in S. serrulatus. The karyotype of S. altispinnis differed from the one previously described in a population from the Pitinga River. C-positive constitutive heterochromatin was mainly pericentromeric in the karyotypes of all species. Nucleolar organizer regions were multiple and preferentially located terminally on the short arms of the subtelocentric/acrocentric chromosomes, as evidenced by both silver nitrate staining and fluorescent in situ hybridization with the 18S rDNA probe. The maximum number of NORs varied among species, as did the NOR-bearing chromosomes. FISH with the 5S rDNA probe produced an interstitial signal on the long arms of the pair 7 in all species, coincident with a C-positive heterochromatic band. While some chromosome features were shared by the three species, some were species-specific and thus useful for cytotaxonomy.
The subfamily Serrasalminae includes 15 genera and 80 species restricted to the Neotropical region and known as pacus and piranhas. All these genera occur in the Amazon Basin, six of them are also present in the Paraná-Paraguay Basin and three in the São Francisco Basin (Jégu, 2003).
The genus Serrasalmus is one of the most diversified Serrasalminae and has a wide distribution throughout South America. It includes highly specialized voracious fishes displaying a typical laterally compressed and deep body, with a series of middle-ventral abdominal spines (Géry, 1972; Machado-Allison and Fink, 1996). Besides the 24 recognized species of Serrasalmus, four species still require a detailed characterization and formal recognition (Jégu, 2003).
Ten Serrasalmus species have already been karyotyped and 2n = 60 was hypothesized as the basal diploid number (Cestari and Galetti, 1992a; Nakayama et al., 2002). The high intraspecific chromosome diversity of the genus is documented by the 17 distinct karyotypes that have already been reported (Nakayama et al., 2002). For instance, four distinct karyotypes were found in S. maculata (junior synonyme - S. spilopleura Jégu and Santos, 2001) in the confluence of the Negro and Solimões Rivers, Amazon Basin (Nakayama et al., 2000; Centofante et al., 2002), while three karyomorphs were reported in the Paraná Basin population (Cestari and Galetti, 1992b). While S. rhombeus presented a karyotype with 2n = 60; a criptic species presented 2n = 58 and a karyotype probably derived from the 2n = 60 by chromosome fusions (Nakayama et al., 2001), and a third new karyomorph was also reported (Teixeira et al., 2006).
In this work, the karyotypes of three species of serrasalmus (S. alispinnis, S. gouldingi and S. Serrulatus) were investigated after C-banding, silver nitrate staining of the nucleolus organizer regions (Ag-NORs) and fluorescent in situ hybridization (FISH) with the 5S and 18S rDNA probes.
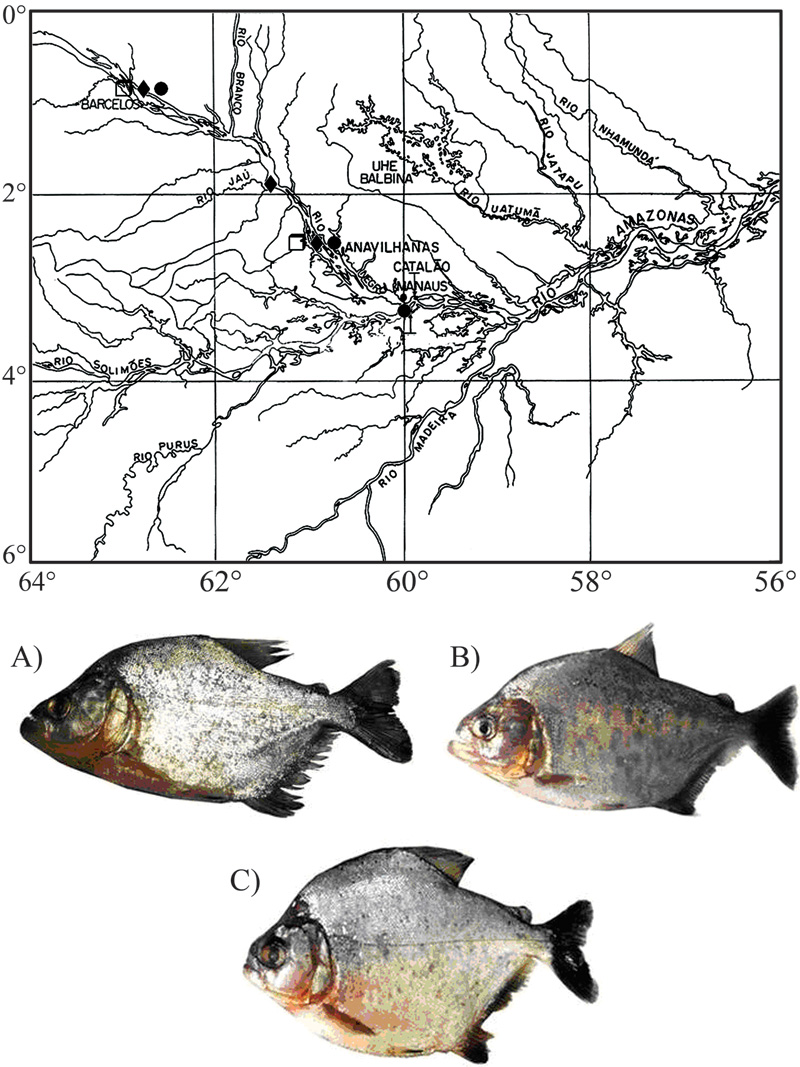
Figure 1 Map of the collection sites and Serrasalmus species analyzed: a) ◊ S. altispinnis (total length =16.0 cm); b) ♦ S. gouldingi (total length = 14.0 cm), and c) • S. serrulatus (total length = 15.5 cm).
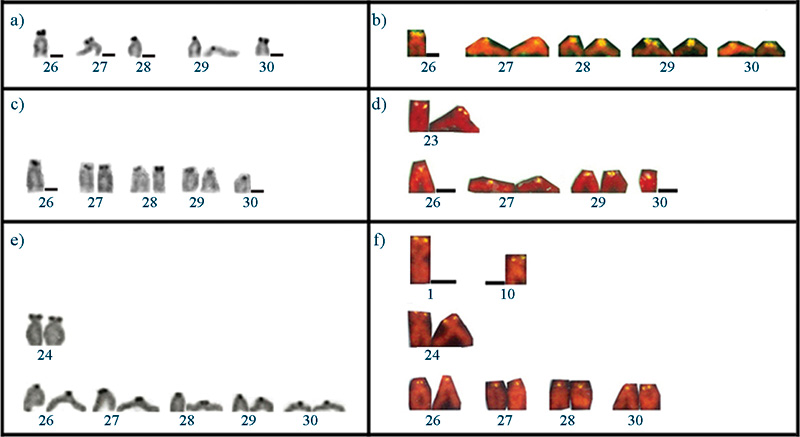
Figure 2 Karyotype of Serrasalmus altispinnis, S. gouldingi and S. serrulatus after: Giemsa-staining (a, c, e) and C-banding (b, d, f).
Figure 3 Metaphases of Serrasalmus altispinnis (a, b), S. gouldingi (c, d) and S. serrulatus (e, f), showing 18S rDNA (a, c, e) and 5S rDNA (b, d, f) positive sites after FISH.
Figure 4 Partial karyotypes of Serrasalmus altispinnis (a, b), S. gouldingi (c, d) and S.serrulatus (e, f) with Ag-NORs (a, c, e) and 18S rDNA sites (b, d, f).
Twenty-three specimens of Serrasalmus altispinnis, 18 specimens of S. gouldingi and 26 specimens of S. serrulatus from the middle and lower Negro River, Amazon Basin, state of Amazonas, Brazil, were analyzed (Figure 1; Table 1). The specimens were classified by Dr. Jansen Zuanon, from the Instituto Nacional de Pesquisas da Amazônia (INPA), where voucher specimens were deposited (Serrasalmus altispinnis: INPA 28887, 28890; S. gouldingi: INPA 28888, 28889; S. serrulatus: INPA 28884, 28885, 28886).
The chromosome preparations were obtained from kidney cells, according to the in vivo procedure described by Bertollo et al. (1978) and after mitotic stimulation with biological yeast, as described by Oliveira et al. (1988). About 30 metaphases per individual were analyzed. The chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) and acrocentric (a) and arranged by decreasing size. The fundamental number (FN), or number of chromosome arms was determined considering that metacentric (m), submetacentric (sm) and subtelocentric (st) chromosomes are biarmed and that acrocentric (a) chromosomes are uniarmed. C-positive constitutive heterochromatin was evidenced by C-banding (Sumner, 1972) and the active nucleolar organizer regions were identified after silver nitrate staining (Ag-NORs), according to Howell and Black (1980).
Fluorescent in situ hybridization (FISH) was performed to locate 18S and 5S DNA sites on the chromosomes, according to Pinkel et al. (1986). The 18S rDNA probe was obtained by PCR from the DNA of the fish Prochilodus argenteus (Hatanaka and Galetti, 2004), using the primers NS1 (5'-GTAGTCATATGCTTGTCT C-3') and NS8 (5'- TCCGCAGGTTCACCTACGGA-3'), according to White et al. (1990). The 5S rDNA probe was obtained from the fish Leporinus obtusidens (Martins and Galetti, 1999), using the primers A (5'-TACGCCCGATC TCGTCCGATC-3') and B (5'- CAGGCTGGTATGGCC GTAAGC-3'), according to Pendás et al. (1994). The probes were labeled by nick translation (BioNick Labeling System - Invitrogen) following the manufacturer's specifications. Denaturation was for 5 min in formamide 70%/2xSSC at 70 °C. Post-hybridization washes were performed in high stringency conditions (formamide 50%/2xSSC at 42 °C). The chromosomes were counterstained with propidium iodide (50 μg/mL and 200 μL of antifading solution) and analyzed in an epifluorescence microscope Olympus BX50. The images were documented using the software CoolSNAP-pro (Media Cybernetics).
The three species presented karyotypes with 2n = 60 and FN = 110. However, some differences regarding to karyotypic formula were observed (24m, 20sm, 6st, 10a for S. altispinnis; 22m, 22sm, 6st, 10a for S. gouldingi and 20m, 22sm, 8st, 10a for S. serrulatus). No heteromorphic sex chromosomes were identified when comparing male and female karyotypes of the three species (Table 2; Figure 2).
After C-banding, constitutive heterochromatin was located mainly at the pericentromeric chromosome regions of the three species. Some chromosome pairs had conspicuous pericentromeric heterochromatic blocks, on the short or on the long arms. This was the case of pairs 3, 4, 9, 14, 16 and 27 in S. altispinnis, of pairs 26, 27and 29 in S. gouldingi, and of pair 22 in S. serrulatus. Some remarkably dark regions seen on the short arms of one homologue of pairs 14 and 18 of S. serrulatus were actually technical artifacts (Figure 2 b, d, f). The metacentric pair 7 presented a remarkable C-band on its long arm close to the centromere in the three species (Figures 2 b, d, f, 5 a, c and eFigure 5 Pair 7 of Serrasalmus altispinnis (a, b), S. gouldingi (c, d) and S. serrulatus (e, f) showing the co-localization of the C-positive heterochromatic region (a, c, e) and the 5S rDNA positive site (b, d, f). ).
The multiple Ag-NORs were located at the terminal regions of the short arms of acrocentric/subtelocentric chromosomes. They varied inter- and intraindividually, as well as inter- and intraspecifically and their maximum numbers were nine in S. altispinnis, eight in S. goldingi and 12 in S. serrulatus (Table 2; Figure 4 a, c, e). Variations were also observed in the 18S rDNA sites. In S. altispinnis, the 18S rDNA signals were detected on the same acrocentric chromosomes that showed Ag-NORs. In S. gouldingi and S. serrulatus the 18S rDNA sites were found on subtelocentric and metacentric chromosomes, respectively, which were not evidenced as Ag-NORs-bearing chromosomes (Table 2; Figure 3 a, c, e; Figure 4 b, d, f). The 5S rDNA sites were present on the long arms of the metacentric pair 7, close to the centromeric region in the three species (Table 2; Figure 3 b, d, f; Figure 5 b, d, fFigure 5 Pair 7 of Serrasalmus altispinnis (a, b), S. gouldingi (c, d) and S. serrulatus (e, f) showing the co-localization of the C-positive heterochromatic region (a, c, e) and the 5S rDNA positive site (b, d, f). ).
Most of the ten Serrasalmus species previously analyzed showed 2n = 60 (Nakayama et al., 2002); except for S. hollandi with 2n = 64 (Muramoto et al., 1968) and a karyotypic variation of S. cf. rhombeus with 2n = 58 (Nakayama et al., 2001).
Our work presents new chromosomal data for the Serrasalmus genus. Indeed, there are no reported data for S. goulding and S. serrulatus and a new karyotypic form was described herein for S. altispinnis. The three species showed 2n = 60 and FN = 110, but they can be differentiated by their karyotypic formulas, which indicated that distinctive non-Robertsonian rearrangements have occurred during the evolution of each species. Nakayama et al. (2002) detected a karyotype composed of 20m, 28sm, 2st, 10a and FN = 110 in S. altispinnis from the Pitinga River (Uatumã Basin), which differs from the pattern presently described in the specimens from the middle Negro River (22m, 22sm, 6st, 10a and FN = 110). Therefore, this species shows chromosome variation in its geographical range. The fundamental number is the same (FN = 110) in all specimens, thus pericentric inversions, involving at least six chromosomes in one of the karyotypic forms must be the cause of this variation. The fixation of such chromosome rearrangements might have been facilitated by the lack of gene flow between both populations. Actually, there is no current connection between the Uatumã and the Negro Rivers, which were probably separated from each other by Late Tertiary or Early Quaternary (Nogueira and Sarges, 2001). Further associations of these chromosome data with morphologic and genetic features may help to elucidate whether these two karyotypic forms of S. altispinnis represent cryptic species or populations under different evolutionary paths.
C-banding was mainly located at the centromeric chromosome regions in the three species, although some bands were more conspicuous than others. A decreasing amount of heterochromatin seems to characterize the karyotypes of S. altispinnis, S. gouldingi and S. serrulatus, respectively. Furthermore, some conspicuous and easily identified species-specific C-bands were also observed, such as those located on long arms of the metacentric pairs 3 and 9 of S. altispinnis, on the long arms of the acrocentric pair 26 of S. gouldingi and on the long arms of the subtelocentric pair 22 of S. serrulatus. It is noteworthy that the three species had a positive C-band on the long arms of the metacentric pair 7, which agrees with the pattern observed in other Serrasalmus species (Nakayama et al., 2002; Centofante et al., 2002).
All species of Serrasalminae analyzed so far showed several rDNA sites, with inter- and intraindividual variations, ranging from 4 to 12 Ag-NORs. In species of Serrasalmus, the Ag-NORs were always located on the short arms of the acro-subtelocentric chromosomes (Galetti et al., 1985; Cestari and Galetti, 1992a, b; Martins-Santos et al., 1994; Nakayama et al., 2001, 2002; Centofante et al., 2002), coinciding with the pattern observed in the three species analyzed herein. Nonetheless, the maximum number of Ag-NORs differed among species (nine in S. altispinnis, eight in S. gouldingi and 12 in S. serrulatus). The maximum number of 18S rDNA sites and of Ag-NORs was the same in each species, but their localizations were not always coincident by FISH and by Ag-NOR, with the exception of S. altispinnis. S. gouldingi and S. serrulatus presented some subtelocentric and metacentric chromosomes with 18S rDNA sites, respectively, but not with Ag-NORs. Inversely, some Ag-NORs-bearing chromosomes presented no 18S rDNA signals.
The variation in the Ag-NORs could be explained by the differential activity among the distinct ribosomal sites within the species genome, since the silver nitrate staining can only detect NORs that were active in the preceding interphase (Miller et al., 1976). Actually, the inactivity of some ribosomal sites is a common feature observed in fish with multiple NORs, like Hoplias malabaricus (Born and Bertollo; 2000); Astyanax scabripinnis, A. parahybae, A. intermedius and A. giton (Kavalco and Moreira-Filho, 2003), or with single NORs, such as some Cichlidae and Curimatidae (Feldberg and Bertollo, 1985; Feldberg et al., 1992;). On the other hand, the lack of FISH signals in some Ag-NOR sites may be due to the small size of these regions. It is probable that the hybridization of the 18S rDNA was hindered or barely perceptible because there were only a few copies of the gene in these sites. Therefore, the real number of NORs in the karyotype of S. gouldingi and S. serrulatus should be higher than that detected by both silver nitrate staining and by FISH. However, NORs transposition among different chromosomes with the consequent variation in their location cannot be excluded.
The 5S rDNA is located in a single chromosome pair in many organisms. However, in amphibians (Lucchini et al., 1993) and fishes (Martins and Galetti, 1999; 2000 Martins C., Galetti Jr P.M. (2000) Conservative distribution of 5S rDNA loci in Schizodon (Pisces, Anostomidae) chromosomes. Chromosome Res 8:353-355. ), the 5S rDNA cistrons can be present in several chromosomes. Besides, the 45S and 5S rDNA loci may be syntenic (Morán et al., 1996) or located in distinct chromosome pairs (Martins and Galetti, 1999; Born and Bertollo, 2000), the last condition being commonly observed among fishes.The three Serrasalmus species showed a single 5S rDNA site, located on the same region of the medium-sized metacentric pair 7, coinciding with a C-positive heterochromatic band. Thus, the chromosome location of the 5S rDNA site and the heterochromatin band are features shared by the Serrasalmus species, representing additional important chromosome markers for this fish.
The evolutionary relationships among piranha species are still unclear and several studies have focused on this matter. Molecular and morphological data have been used in order to test the monophyletism of this group (Freeman et al., 2007). Similarly, chromosome data have revealed useful cytotaxonomic features and provided data on the relationships of these species. Therefore, some relatively conserved characteristics such as diploid and fundamental numbers, and some chromosome markers, support the relationship between S. altispinnis., S. gouldingi and S. serrulatus and other Serrasalmus species. On the other hand, distinctive features, such as the karyotype formulas, species-specific heterochromatic bands, and the number and location of the 18S rDNA represent helpful tools for the cytotaxonomy of these species, allowing the detection of possible cryptic species, as suggested for S. altispinnis.
Acknowledgments
The authors thank Dr. J.A.S. Zuanon for the specimens classification, A. C. dos Santos for help with the figures, and IBAMA for the license for fish capture (n. 049/2004 and 006/2004). This work was supported by the Instituto Nacional de Pesquisas da Amazônia (INPA) through the Research Institutional Projects (PPI 2-3450), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)/Programa de Apoio a Núcleos de Excelência em Ciência e Tecnologia (PRONEX), Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM)/Programa Integrado de Pesquisa e Inovação Tecnológica (PIPT). C.M. Nakayama was the recipient of a CNPq fellowship.
References
Bertollo L.A.C., Takahashi C.S., Moreira-Filho O. (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazil J Genet 1:103-120.
Born G.G., Bertollo L.A.C. (2000) An XX/XY sex chromosome system in a fish species Hoplias malabaricus with polymorphic NOR bearing X chromosome. Chromosome Res 8:11-118.
Centofante L., Porto J.I.R., Feldberg E. (2002) Chromosomal polymorphism in Serrasalmus spilopleura Kner, 1858 (Characidae, Serrasalminae) from Central Amazon Basin. Caryologia 55:37-45.
Cestari M.M., Galetti Jr P.M. (1992a) Chromosome evolution in the genus Serrasalmus and citotaxonomic considerations about Serrasalminae (Characidae, Pisces). Brazil J Genet 15:555-567.
Cestari M.M., Galetti Jr P.M. (1992b) Chromosome studies of Serrasalmus spilopleura (Characidae, Serrasalminae) from the Paraná-Paraguay rivers: Evolutionary and cytototaxonomic considerations. Copeia 1992:108-112.
Feldberg E., Bertollo L.A.C. (1985) Nucleolar organizer regions in some species of Neotropical cichlid fish (Pisces, Perciformes). Caryologia 38:319-324.
Feldberg E., Porto J.I.R., Bertollo L.A.C. (1992) Karyotype evolution in Curimatidae (Teleostei, Characiformes) of the Amazon region. I. Studies on the genera Curimata, Psectrogaster, Steindachnerina and Curimatella. Brazil J Genet 15:369-383.
Freeman B., Nico L., Osentoski M., Jelks H.L., Collins T.M. (2007) Molecular systematics of Serrasalmidae. Deciphering the identities of piranha species and unraveling their evolutionary histories. Zootaxa 1484:1-38.
Galetti Jr P.M., Silva E., Cerminaro R.T. (1985) Multiple NOR system in fish Serrasalmus spilopleura (Serrasalminae, Characidae). Brazil J Genet 8:479-484.
Géry J. (1972) Poisson Characoides des Guyanes. I. Généralités. II. Famille Serrasalmidae. Zool Verhand 122:1-250.
Hatanaka T., Galetti P.M. (2004) Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122:239-244.
Howell W.M., Black D.A. (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36:1014-1015.
Jégu M. (2003) Subfamily Serrasalminae (pacus and piranhas). Check List of Freshwater Fishes of South and Central AmericaReis R.E., Kullander S.O., Ferraris CJ JrPorto AlegreEdipucrs182-196.
Jégu M., Santos G.M. (2001) Mise au point à propos de Serrasalmus spilopleura Kner, 1858 et réhabilitation de S. maculatus Kner, 1858 (Characidae, Serrasalminae). Cybium 25:119-143.
Kavalco K.F., Moreira-Filho O. (2003) Cytogenetical analyses in four species of the genus Astyanax (Pisces, Characidae) from Paraíba do Sul river basin. Caryologia 56:453-461.
Lucchini S., Nardi I., BarsacchI G., Batistoni R., Andronico F. (1993) Molecular cytogenetics of the ribosomal (18S + 28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome 36:762-773.
Machado-Allison M., Fink W. (1996) Los Peces Caribes de Venezuela: Diagnosis, Claves, Aspectos Ecologicos y EvolutivosCaracasColección Monografías. Universidad Central de Venezuela149
Martins C., Galetti Jr P.M. (1999) Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res 7:363-367.
Martins-Santos I.C., Julio Jr H.F., Santos S.J. (1994) Chromosome study of two species of the genus Serrasalmus (Characidae, Serrasalminae) from the Paraná River. Caryologia 59:175-181.
Miller D.A., Devi V.G., Tantravahi R., Miller O.J. (1976) Supression of human nucleolus organizer in mouse-human somatic hybrid cells. Exp Cell Res 101:235-243.
Morán P., Martínez J.L., Garcia-Vazquez E., Pendás A.M. (1996) Sex chromosome linkage of 5S rDNA in rainbow trout. Cytogenet Cell Genet 75:145-150.
Muramoto J.I., Ohno S., Atkins N.B. (1968) On the diploid state of the fish order Ostariophysi. Chromosoma 24:59-66.
Nakayama C.M., Jégu M., Porto J.I.R., Feldberg E. (2001) Karyological evidence for a cryptic species of piranha within Serrasalmus rhombeus (Characidae, Serrasalminae) in the Amazon. Copeia 2001:866-869.
Nakayama C.M., Porto J.I.R., Feldberg E. (2000) Ocorrência de dois citótipos em Serrasalmus spilopreura Kner, 1958 (Characiformes, Serrasalmidae) na região de confluência dos rios Negro e Solimões, Amazonas, Brasil. Acta Amazonica 30:149-154.
Nakayama C.M., Porto J.I.R., Feldberg E. (2002) A comparative cytogenetic study of five piranha species (Serrasalmus, Serrasalminae) from the Amazon basin. Genetica 114:231-236.
Nogueira A.C.R., Sarges R.R. (2001) Characterization and genesis of waterfalls of the Presidente Figueiredo region, northeast State of Amazonas, Brazil. An Acad Bras Cienc 73:287-301.
Oliveira C., Almeida-Toledo L.F., Foresti F., Toledo-Filho S.A. (1988) Supernumerary chromosomes, Robertsonian rearrangement and multiple NORs in Corydoras aeneus (Pisces, Siluriformes, Callichthyidae). Caryologia 41:227-236.
Pendás A.M., Morán P., Freire J.P., Garcia-Vazquez E. (1994) Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet Cell Genet 67:31-36.
Pinkel D., Straume T., Gray J.W. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934-2938.
Sumner A.T. (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304-306.
Teixeira A.S., Nakayama C.M., Porto J.I.R., Feldberg E. (2006) Esterase-D and chromosome patterns in Central Amazon piranha (Serrasalmus rhombeus Linnaeus, 1766) from lake Catalão. Genet Mol Biol 29:498-502.
White T.J., Bruns T., Lee S., Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and ApplicationsNew YorkAcademic Press Inc315-322.
m = metacentric; sm = submetacentric; st = subtelocentric; a = acrocentric; p = short arm; q = long arm, t = terminal region; i = interstitial region; Ag-NOR = silver stained nucleolar organizer regions; 18S = maximum number of 18S rDNA sites; 5S = maximum number of 5S rDNA sites.
Received: September 6, 2007; Accepted: June 3, 2008
Celeste M. Nakayama. Instituto Nacional de Pesquisas da Amazônia (INPA), Coordenação de Pesquisas em Biologia Aquática, Caixa Postal 478, 69011-970 Manaus, AM, Brazil. E-mail: celnaka@inpa.gov.br.
Publication Dates
-
Publication in this collection
19 Nov 2008 -
Date of issue
2008
History
-
Accepted
3062 -
Received
6092