Abstract
Phenotypic plasticity is a hallmark of the caste systems of social insects, expressed in their life history and morphological traits. These are best studied in bees. In their co-evolution with angiosperm plants, the females of corbiculate bees have acquired a specialized structure on their hind legs for collecting pollen. In the highly eusocial bees (Apini and Meliponini), this structure is however only present in workers and absent in queens. By means of histological sections and cell proliferation analysis we followed the developmental dynamics of the hind legs of queens and workers in the fourth and fifth larval instars. In parallel, we generated subtractive cDNA libraries for hind leg discs of queen and worker larvae by means of a Representational Difference Analysis (RDA). From the total of 135 unique sequences we selected 19 for RT-qPCR analysis, where six of these were confirmed as differing significantly in their expression between the two castes in the larval spinning stage. The development of complex structures such as the bees’ hind legs, requires diverse patterning mechanisms and signaling modules, as indicated by the set of differentially expressed genes related with cell adhesion and signaling pathways.
social insect; caste development; imaginal disc; insect metamorphosis; transcript analysis
Introduction
Within the framework of a relatively fixed body plan, winged insects have become the most species-rich group in terrestrial ecosystems, especially so the holometalous orders, which have made their appearance about 350 mya (Misof et al., 2014Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG et al. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–67.). The first order to branch off as a clade in the Holometabola is the Hymenoptera (wasps, ants and bees). While most of the hymenopteran species and families have a carnivorous or parasitic life style, bees have switched to a plant-based diet, in particularly pollen and nectar. This switch, which is thought to have occurred in the early-to-mid Cretaceous (Michener, 2007Michener CD (2007) The Bees of the World. 2nd edition. Johns Hopkins University Press, Baltimore, 992 pp.), represents not only a major lifestyle novelty within this group, but also prompted a very strong co-evolutionary relationship with angiosperm plants. Not only are bees the major pollinators of angiosperm plants and, by doing so, guarantee the bio-diversity of most terrestrial ecosystems, they nowadays also provide multibillion dollar ecosystem services to agriculture worldwide. Notably, most of the high value crops (fruits, nuts, legumes) are strongly dependent on pollination by bees, especially so the corbiculate bees (Klein et al., 2006Klein AM, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C and Tscharntke T (2006) Importance of pollinators in changing landscapes for world crops. Proc Biol Sci 274:303–313.; Gallai et al., 2009Gallai N, Salles JM, Settele J and Vaissière BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68:810–821.).
In the corbiculate bees (corbiculate Apidae, sensuMichener, 2007Michener CD (2007) The Bees of the World. 2nd edition. Johns Hopkins University Press, Baltimore, 992 pp.), the female’s hind legs present a special set of structures adapted for pollen collection and transport, this being the corbicula and the pollen comb on the outside of the tibia, and rows of bristles on the enlarged basitarsus forming the pollen brush. The corbicula is a smooth area on each hind tibia that is surrounded by a fringe of stiff bristles, some of which are thought to be mechanoreceptors that allow the bee to estimate the amount of pollen carried during foraging (Ford et al., 1981Ford DM, Hepburn HR, Moseley FB and Rigby RJ (1981) Displacement sensors in the honeybee pollen basket. J Insect Physiol 27:339–346.; Proctor et al., 1996Proctor MCF, Yeo P and Lack A (1996) The Natural History of Pollination. Illustrated edition. Timber Press, Portland, 479 pp.).
While the presence of a corbicula is a synapomorphic character of this clade, its expression has undergone a considerable change in the highly eusocial honey bees and the stingless bees, where it makes its appearance in the worker caste only, but not in queens. Thus, although the presence of a corbicula is an ancestral character in the female sex of corbiculate bees (Cardinal and Danforth, 2011Cardinal S and Danforth BN (2011) The antiquity and evolutionary history of social behavior in bees. PloS One 6:e21086.), the secondary loss of these structures in queens apparently reverted their hind leg architecture to a more ancestral state of the Hymenoptera.
This interesting phenomenon is a result of caste determination in the premetamorphic larval stages of the highly eusocial bees, and this process is best understood in the honey bee, Apis mellifera, where caste differentiation is trigged by a switch in larval diets during the early developmental stages (for reviews see Haydak, 1970Haydak MH (1970) Honey bee nutrition. Annu Rev Entomol 15:143–156.; Leimar et al., 2012Leimar O, Hartfelder K, Laubichler MD and Page RE (2012) Development and evolution of caste dimorphism in honeybees - A modeling approach. Ecol Evol 2:3098–3109.; Hartfelder et al., 2015Hartfelder K, Guidugli-Lazzarini KR, Cervoni MS, Santos DE and Humann FC (2015) Old threads make new tapestry - rewiring of signaling pathways underlies caste phenotypic plasticity in the honey bee, Apis mellifera L.. Adv Insect Physiol 48:1–36.). Such triggers are the elevated sugar levels in royal jelly fed to queen larvae (Asencot and Lensky, 1988Asencot M and Lensky Y (1988) The effect of soluble sugars in stored royal jelly on the differentiation of female honeybee (Apis mellifera L.) larvae to queens. Insect Biochem 18:127–133.), and a specific protein moiety present in royal jelly, the MRJP1 monomer named Royalactin (Kamakura, 2011Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473:478–483.). While the former may affect caste development via the insulin/insulin-like signaling (IIS) pathway (Wheeler et al., 2006Wheeler DE, Buck N and Evans JD (2006) Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol 15:597–602.), the latter has been shown to drive queen differentiation via the Egfr pathway (Kamakura, 2011Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473:478–483.). These upstream nutritional factors and associated signaling pathways eventually converge onto a coordinated endocrine response resulting in caste-specifically modulated juvenile hormone (JH) and ecdysteroids hemolymph titers (Rachinsky et al., 1990Rachinsky A, Strambi C, Strambi A and Hartfelder K (1990) Caste and metamorphosis: hemolymph titres of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen Comp Endocrinol 79:31–38.). JH controls caste-specific cellular developmental programs in honey bee larvae, including programmed cell death in the ovaries (Schmidt-Capella and Hartfelder, 1998Schmidt-Capella IC and Hartfelder K (1998) Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J Insect Physiol 44:385–391.; Leimar et al., 2012Leimar O, Hartfelder K, Laubichler MD and Page RE (2012) Development and evolution of caste dimorphism in honeybees - A modeling approach. Ecol Evol 2:3098–3109.), and it also regulates the development of other tissues and organs via a modulatory action on the hemolymph ecdysteroid titer (Rachinsky and Engels, 1995Rachinsky A and Engels W (1995) Caste development in honeybees (Apis mellifera): Juvenile hormone turns on ecdysteroids. Naturwissenschaften 82:378–379.).
Throughout larval development, the locally acting signaling pathways [IIS (Wheeler et al., 2006Wheeler DE, Buck N and Evans JD (2006) Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol 15:597–602., 2014Wheeler DE, Buck NA and Evans JD (2014) Expression of insulin/insulin-like signalling and TOR pathway genes in honey bee caste determination. Insect Mol Biol 23:113–121.; Azevedo and Hartfelder, 2008Azevedo SV and Hartfelder K (2008) The insulin signaling pathway in honey bee (Apis mellifera) caste development - differential expression of insulin-like peptides and insulin receptors in queen and worker Larvae. J Insect Physiol 54:1064–1071.), Egfr (Kamakura, 2011Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473:478–483.), TOR (Patel et al., 2007Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K and Amdam GV (2007) The making of a queen: TOR pathway is a key player in diphenic caste development. PloS One 2:e509.) and hypoxia (Azevedo et al., 2011Azevedo SV, Caranton OA, de Oliveira TL and Hartfelder K (2011) Differential expression of hypoxia pathway genes in honey bee (Apis mellifera L.) caste development. J Insect Physiol 57:38–45.)], and globally acting morphogenetic hormones (JH and ecdysteroids) are thought to directly or indirectly affect gene expression patterns underlying the formation of the distinct queen and worker phenotypes (Evans and Wheeler, 2000Evans JD and Wheeler DE (2000) Expression profiles during honeybee caste determination. Genome Biol 2:RESEARCH0001.; Cristino et al., 2006Cristino AS, Nunes FMF, Lobo CH, Bitondi MMG, Simões ZLP, Costa LD, Lattorff HMG, Moritz RFA, Evans JD and Hartfelder K (2006) Caste development and reproduction: a genome-wide analysis of hallmarks of insect eusociality. Insect Mol Biol 15:703–714.; Barchuk et al., 2007Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP and Maleszka R (2007). Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol 7:e70.). While the above-cited transcriptome analyses were all done on RNA extracted from whole body preparations, little is known on expression profiles associated with tissue-specific development. First approaches towards elucidating the molecular underpinnings of developmental plasticity in specific tissues were done on ovaries (Hepperle and Hartfelder, 2001Hepperle C and Hartfelder K (2001) Differentially expressed regulatory genes in honey bee caste development. Naturwissenschaften 88:113–116.; Humann and Hartfelder, 2011Humann FC and Hartfelder K (2011) Representational difference analysis (RDA) reveals differential expression of conserved as well as novel genes during caste-specific development of the honey bee (Apis mellifera L.) ovary. Insect Biochem Mol Biol 41:602–612.), due to their fundamental differences associated with reproductive division of labor.
With respect to hind leg and specifically so corbicula development, a microarray screen on differentially expressed genes in hind legs of third instar worker larvae (Bomtorin et al., 2012Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.) drew attention to four genes (grunge, dachshund, cryptocephal, ataxin-2), all previously known as regulators of leg development and bristle morphology in Drosophila melanogaster. This study also investigated the localization pattern for two homeobox proteins, the honey bee Ultrabithorax (Ubx) and Abdominal A (AbdA) homologs, in the developing hind legs of honey bee pupae, showing Ubx-free spots in the tibia epidermis of worker pupae where the characteristic corbicula bristles are expected to be formed (Bomtorin et al., 2012Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.).
So as to further our understanding of the processes underlying the complex structural difference in the hind legs of queens and workers, we addressed this question through a histological analysis on the development of their hind legs throughout the critical stages of postembryonic development. Furthermore, we generated differential gene expression libraries for hind leg imaginal discs of last instar queen and worker larvae. This was done in a developmental stage when the imaginal discs just start to evaginate and form the adult compartments with their respective caste-specific structures, as seen in the histological analysis. The libraries were generated by means of a Representational Difference Analysis (RDA) protocol (Hubank and Schatz, 2000Hubank M and Schatz DG (2000) Representational difference analysis of cDNA. In: Hunt SP and Livesey FJ (eds) Functional Genomics. Oxford University Press, Oxford, pp 45–80.; Pastorian et al., 2000Pastorian K, Hawel L and Byus CV (2000) Optimization of cDNA representational difference analysis for the identification of differentially expressed mRNAs. Anal Biochem 283:89–98.) that had already been successfully adapted for studies in bees (Judice et al., 2006Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K and Pereira GAG (2006) Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol 15:33–44.; Colonello-Frattini and Hartfelder, 2009Colonello-Frattini NA and Hartfelder K (2009) Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie 40:481–495.; Humann and Hartfelder, 2011Humann FC and Hartfelder K (2011) Representational difference analysis (RDA) reveals differential expression of conserved as well as novel genes during caste-specific development of the honey bee (Apis mellifera L.) ovary. Insect Biochem Mol Biol 41:602–612.) and also in other social insects (Weil et al., 2007Weil T, Rehli M and Korb J (2007) Molecular basis for the reproductive division of labour in a lower termite. BMC Genomics 8:e198.; Oppelt et al., 2010Oppelt A, Humann FC, Fuessl M, Azevedo SV, Marco Antonio DS, Heinze J and Hartfelder K (2010) Suppression subtractive hybridization analysis reveals expression of conserved and novel genes in male accessory glands of the ant Leptothorax gredleri. BMC Evol Biol 10:e273.). Though considerably more labor intensive than microarray protocols, RDA has the advantage that it is not limited to a pre-existing platform. Furthermore, it specifically selects and enriches for differentially expressed transcripts, unlike RNAseq, which gives a general transcriptome result. Due to the sequential amplification steps, RDA does, however, not give strictly quantitative readings, thus requiring secondary confirmation of differential expression, generally done by quantitative PCR. Among the total of 135 contigs and singlets assembled for the two RDA libraries generated herein we selected 19 for RT-qPCR analysis, whereby six of these were confirmed as differing significantly in their expression levels among the two castes in the larval spinning stage. Two of these genes had Gene Ontology attributes related to cell adhesion and epithelial development, while one represented a putative microRNA.
Materials and Methods
Honey bee larva
Worker larvae were directly retrieved from brood combs of colonies kept in the Experimental Apiary of the University of São Paulo, Ribeirão Preto, Brazil. Queens were reared from first instar worker larvae transferred to queen cups to be raised in queenless hives, following standard apicultural procedures. The developmental stages were identified following the criteria of Rachinsky et al. (1990)Rachinsky A, Strambi C, Strambi A and Hartfelder K (1990) Caste and metamorphosis: hemolymph titres of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen Comp Endocrinol 79:31–38. and Michelette and Soares (1993)Michelette ERF and Soares AEE (1993) Characterization of preimaginal developmental stages in Africanized honey bee workers (Apis mellifera L). Apidologie 24:431–440..
Histology
For the histological analyses, larvae were collected from the fourth instar until the prepupal stage of the last, fifth instar. Imaginal discs from queens and workers larvae were dissected in Ringer solution and immersed in Bouin’s fixative (35% formaldehyde/saturated picric acid/glacial acetic acid 5:15:1). The samples were processed according to standard procedures, following inclusion in paraffin or methacrylate resin (Historesin, Leica). Briefly, tissues were dehydrated in a graded alcohol series (50–100%). For paraffin embedding they were further dehydrated in benzene, infiltrated in two baths of liquid paraffin (62 °C) for 1 h each, and a final bath under vacuum conditions for 30 min, before embedding in paraffin blocks. Embedding in Historesin was done directly after alcohol dehydration, following the manufacturer’s instructions. Sections of 5–6 μm thickness were stained with hematoxylin and eosin, and analyzed and photographed with a Zeiss microscope equipped with an AXIOCam-HR3 system (Zeiss). Images were processed with Adobe Photoshop CS software for brightness correction.
Cell proliferation assay
After dissection, imaginal discs were fixed in 4% paraformaldehyde solution (pH 7.2) for 2–3 h at room temperature. Samples were next incubated in PBS/glycine (0.1 M/0.1 M; pH 7.2) for 1 h, following three washing steps in PBS (15 min each). The tissues were then washed in PBS containing 2% Triton X-100 (PBST) for 15 min before an overnight (4 °C) permeabilization in this same solution. Subsequently, samples were treated with PBST supplemented with 1% bovine serum albumin (BSA) for 1 h at room temperature before incubation overnight at 4 °C in the mitosis marker Anti-phospho-Histone H3 (Ser10) antibody generated in rabbit (Upstate/Millipore-Merck), diluted at 10 μg/mL in PBST. After washing twice in PBS and once in PBST, they were then incubated overnight at 4 °C in a sheep anti-rabbit IgG Cy3-conjugated secondary antibody (Sigma-Aldrich) diluted 1:100 in PBST.
For immunofluorescence analysis, at least, three pairs of leg discs of each developmental stage were whole-mounted on slides in a drop of glycerol and viewed in a laser confocal microscope system (Leica TCS-SP2 and TCS-SP5). Images were captured with Leica LAS AF Lite software. Adobe Photoshop CS5 software was used for brightness adjustments. Negative controls were done with leg imaginal discs that were not incubated in primary antibody.
Representational Difference Analysis (RDA)
RNA extraction and preparation of double-stranded cDNA
A total of 25 pairs of hind leg discs were used to construct each cDNA library. The pairs of hind leg imaginal discs were dissected from early spinning-stage larvae (L5S1) and transferred to TRIzol reagent (Invitrogen) for RNA extraction following the manufacturer’s standard protocol. After treatment with RNase-free DNaseI (Fermentas), RNA quality was checked by electrophoresis in an agarose gel (1.2%) run under denaturing conditions, and RNA quantity was determined spectrophotometrically (Nanovue, GE Healthcare) at 260/280 nm. cDNA libraries were generated from 2 μg of total RNA by reverse transcription and long distance PCR using the SMARTPCR cDNA Synthesis (Clontech) protocol. The reverse transcription step was carried out using the SMART IIA and SMART CDS IIA primers (Clontech) and Superscript II (Life Technologies) enzyme. These primers contain a recognition sequence for MboI restriction enzyme. Double-stranded cDNA was then produced using the long distance primer PCR SMART IIA and Platinum Taq DNA Polymerase High Fidelity (Life Technologies) in a PTC 200 thermocycler (MJ Research) with a protocol of 94 °C for 5 s, 65 °C for 5 s and 68 °C for 8 min. Aliquots of the reaction mixture were collected at two cycle intervals between cycles 14 and 28 to find the optimal cycle number (clear bands and no signs of product overcycling). This optimal cycle number was then used in a mass-production PCR step of double-stranded cDNA.
Representational difference analysis
A Representational Difference Analysis (RDA) protocol (Pastorian et al., 2000Pastorian K, Hawel L and Byus CV (2000) Optimization of cDNA representational difference analysis for the identification of differentially expressed mRNAs. Anal Biochem 283:89–98.) optimized for honey bee studies (Judice et al., 2006Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K and Pereira GAG (2006) Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol 15:33–44.; Colonello-Frattini and Hartfelder, 2009Colonello-Frattini NA and Hartfelder K (2009) Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie 40:481–495.; Humann and Hartfelder, 2011Humann FC and Hartfelder K (2011) Representational difference analysis (RDA) reveals differential expression of conserved as well as novel genes during caste-specific development of the honey bee (Apis mellifera L.) ovary. Insect Biochem Mol Biol 41:602–612.) was employed. This suppression subtractive hybridization technique is based on two subsequent hybridization steps that enrich for differentially expressed transcripts. Briefly, double-stranded cDNA (1.5 μg) was digested with MboI restriction enzyme (Fermentas), and 1 μg of the product purified by means of a GFX Illustra PCR DNA and Gel band purification (GE Healthcare) protocol was ligated to R-adaptors (Judice et al., 2006Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K and Pereira GAG (2006) Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol 15:33–44.), followed by PCR amplification. This created the driver populations for L5S1 queen and worker imaginal discs. For production of the respective tester populations, the R-adaptors were removed from the PCR products and these were then ligated with two sets of new adaptors (J and N) in two successive rounds of PCR amplifications. In the first round, the tester cDNA was hybridized to a 1:100 excess of driver cDNA, in the second round the tester/driver ratio was set at a higher stringency of 1:800 (Hubank and Schatz, 2000Hubank M and Schatz DG (2000) Representational difference analysis of cDNA. In: Hunt SP and Livesey FJ (eds) Functional Genomics. Oxford University Press, Oxford, pp 45–80.). Before each round of hybridization, the adaptor-ligated cDNAs were purified using a GFX Illustra PCR DNA and Gel band purification kit (GE Healthcare). In the permutational hybridizations, tester cDNA from L5S1 worker hind legs was hybridized with driver cDNA from L5S1 queens and vice versa.
The differential products represented in each library were then ligated into pGEM-T Easy Vector (Promega) and used to transform E. coliDH5α chemocompetent cells. A total of 192 clones grown on LB agar plates in the presence of ampicillin were picked from the two libraries and the inserts purified in a miniprep protocol. Sequencing of the miniprep products amplified by a Big Dye Terminator Cycle Sequencing Ready Reaction (Applied Biosystems) protocol using M13 forward primer was done on an ABI-PRISM 3100 (Applied Biosystems) automated gene analyzer.
Bioinformatics analyses
A sequence analysis pipeline was customized using the E-Gene platform (Durham et al., 2005Durham AM, Kashiwabara AY, Matsunaga FTG, Ahagon PH, Rainonel F, Varuzza L and Gruber A (2005) EGene: A configurable pipeline generation system for automated sequence analysis. Bioinformatics 21:2812–2813.). Quality of the sequencing reads was evaluated by Phred before primer and vector sequences were removed. The trimmed reads were then filtered for quality and read size and removal of mitochondrial DNA and ribosomal RNA, and the resulting expressed sequence tags (ESTs) were assembled using Phrap and CAP3 (Huang and Madan, 1999Huang X and Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877.) software to produce the final list of contigs and singlets for each library. These unique sequences (US) were then fed into BLAST searches (Blastn and Blastx) against GenBank sequences. Sequences that did not match to any of these were entered into a new search for transcripts from other honey bee EST projects deposited in the NCBI sequence read archive (SRA) representing transcriptome shotgun assembly (TSA) reads. The US without matches to other organisms were also queried against the microRNA stem loop sequence database miRBase (Griffiths-Jones et al., 2008Griffiths-Jones S, Saini HK, van Dongen S and Enright AJ (2008) miRBase: Tools for microRNA genomics. Nucleic Acids Res 36(Database issue):D154–D158.). And finally, all US were mapped to the latest version of the A. mellifera whole genome assembly (Amel_4.5, available in BeeBase).
Quantitative RT-PCR
A subset of 19 genes represented in the two libraries was chosen for quantitative RT-PCR (RT-qPCR) analysis. Primers for these sequences were designed with Primer3 software and Primer Blast (NCBI), with predicted products ranging from 120 to 193 bp (Table 1). For calculation of relative expression ratios, two previously validated endogenous control genes were used, one encoding a cytoplasmic actin (act) and the other representing ribosomal protein 49 (rp49, also known as rpl32) (Lourenço et al., 2008Lourenço AP, Mackert A, Cristino AS and Simões ZLP (2008) Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative Real-Time RT-PCR. Apidologie 39:372–385.).
Primers used in quantitative RT-PCR analyses. Gene names and gene IDs are those attributed in BeeBase Official Gene Set v3.2. Unpredicted genes are named by the genomic scaffold numbers where the respective transcripts were mapped.
Total RNA was extracted from imaginal discs of queen and worker larvae with TRIzol reagent (Invitrogen), treated with RNase-free DNase I (Fermentas) to remove any contaminating genomic DNA, and then purified with the RNeasy kit (Qiagen). Aliquots of 2 μg of RNA were reverse transcribed using SuperScript II enzyme (Life Technologies). The RT-qPCR assays were done using a Maxima SYBR Green/RoxqPCR Master Mix (Fermentas) protocol in an ABI-Prism 7500 Real-Time PCR System (Applied Biosystems). Quantification assays to determine primer efficiency were prepared from standard curves of serial cDNA dilutions (1:5 to 1:500). Dissociation curves were analyzed to verify product specificity. The amplification protocol comprised one cycle at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Three biological replicates were prepared, each composed of 10 pairs of imaginal discs, and each biological replicate was assayed as technical triplicates. The cycle threshold values for each biological replicate were used as inputs to the Relative Expression Software Tool (REST) for statistical analyses (Pfaffl et al., 2002Pfaffl MW, Horgan GW and Dempfle L (2002) Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in Real-Time PCR. Nucleic Acids Res 30:e36.). A p ≤ 0.05 was considered as statistically significant.
Results
Imaginal disc histology and cell proliferation analysis
The legs of honey bees develop from imaginal discs. Sitting in the ventral midline of the larval body they are contiguous with the body wall epidermis, representing pockets of thickened epidermis beneath the ventral thoracic cuticle (Figure 1B,C,F). We analyzed leg disc development in queens and workers from the fourth instar larvae (L4) until the prepupal stage (L5PP) and all subsequent descriptions equally suit both castes, as there were no obvious histological differences between the two.
Hind leg imaginal discs development in honey bee larvae. (A–B) In the fourth larval instar (L4,) the disc occupies the peripodial sac (PS and arrow in A). Mesenchymal cells (MC) are observed inside the disc, which is covered by the larval cuticle (C). (C–E) During the feeding stage of the fifth instar (L5F) the discs grow considerably and the leg compartments start to become apparent, but the epithelium (E) remains unchanged. Muscles cells (M) are adjacent to the epithelium where they will establish future connections. (F) In the legs of early spinning stage (L5S) larva, segmentation is more apparent, but the epithelium still remains the same. A, B - L4 queen; C - L5F1 worker; D - L5F2 worker; E - L5F3 worker; F, L5S1 queen.; C, cuticle; E, epidermis; FB, fat body; MC, mesenchymal cells; M, muscle; PS, peripodial sac; Tr, trachea; TG, thoracic ganglion. Scale bars: A, C, D, E, 100 μm; B, 50 μm; F, 200 μm.
As in other holometabolous insects, the honey bee leg discs develop within a peripodial space, limited by a peripodial membrane, which is an epithelial monolayer continuous with body wall epidermis on the one hand and with the thickened disc epithelium on the other (Figure 1A–E). The disc epithelium is extensively folded within the peripodial sac (PS), giving it a multilayered appearance (Figure 1). Inside the disc pouch, small mesenchymal cells are visible (Figure 1A–D).
During the feeding stage of the last larval instar (L5F) the discs grow and the leg compartments start to appear (Figure 1C–E), but the epithelial structure remains the same as that seen in the more cup-shaped discs of fourth instar larvae. This growth occurs through cell proliferation, detected in the entire structure, including the peripodial membrane, mesenchymal cells and disc epithelium (Figure 2). During this larval stage the most distal portion of the leg disc, the future tarsal region, folds and starts to project towards the posterior portion of the respective thoracic segment. Furthermore, myoblasts that originated from mesenchymal cell precursors start to aggregate close to the epithelium, where they form the future muscle fibers (Figure 1E,F). At this point, tracheae were observed to invade the internal disc space (Figure 1D).
Cell proliferation detection in the hind leg imaginal discs from queens and workers by anti-phospho-histone H3 immunofluorescence (yellow). Cell proliferation was evidenced during all stages analyzed, but it appears to be more prominent during the feeding stage. (A) L5F2 queen; (B) L5F3 queen; (C) L5F2 worker; (D) negative control. Scale bars: A, B, D, 100 μm; C, 50 μm.
During the larval-pupal transition, here represented as the L5S1 phase (Figure 1F), the leg discs evaginate out of the peripodial space and expand beneath the larval cuticle where they gradually acquire the pupal leg shape. The leg segments can now clearly be distinguished, but the cuticle that covers the leg is still the larval one and, thus, does not yet present any bristles. Being characteristics of the adult legs, these will only develop later, during the pupal-adult transition.
As far as caste differences during larval development are concerned, the leg imaginal discs of queens and workers did not show any overt differences in overall structure, nor were there any noteworthy differences with respect to cell proliferation. Nonetheless, the fourth and early fifth instars are the stages when caste fate of the legs and other caste-specific structures is decided (Dedej et al., 1998Dedej S, Hartfelder K, Aumeier P, Rosenkranz P and Engels W (1998) Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J Apic Res 37:183–190.).
Differential gene expression - RDA library characteristics
So as to gain insight into molecular underpinnings of these caste-specific differentiation processes we performed a Representational Difference Analysis. From the 192 sequencing reads representing the queen imaginal disc RDA library, 138 were considered of good quality (Phred quality ≥ 20). After E-Gene pipeline and CAP3 analysis, these were assembled into 11 contiguous sequences (contigs) and 60 single sequences (singlets). Blastn and Blastx analyses against the GenBank non-redundant database revealed significant similarities for 62 sequences, considering a cutoff E-value of 1.0 × e−5. Among these, 25 sequences had significant similarity to Drosophila genes (Table 2), and for most of these, Gene Ontology attributes could be retrieved from Flybase. Another five sequences were similar to stem loop microRNA sequences deposited in miRBase (Table 2).
Unique sequences assembled from expressed sequence tags of the queen imaginal discs RDA library. Listed are genes with similarity to Drosophila melanogaster genes or stem-loop micro RNAs. The genomic scaffold (Group in Amel_4.5) and predicted CDS (GB number of Official Gene Set v3.2). Gene Ontology terms for Biological Process and Molecular Function are from Flybase and given as inferred from electronic annotation (IEA), inferred from sequence or structural similarity (ISS), inferred from physical interaction, (IPI), or inferred from direct assay (IDA).
The same analysis performed on the sequencing reads for the worker RDA library resulted in 131 quality ESTs, which assembled into 19 contigs and 45 singlets, with 56 sequences presenting significant matches to GenBank sequences. Among these, 15 had best matches with Drosophila genes (Table 3) and six sequences to stem loop microRNAs (Table 3). As in the queen library, some of the worker RDA library sequences also did not present similarities with nr database sequences.
Unique sequences assembled from expressed sequence tags of the worker imaginal discs RDA library with similarity to Drosophila sequences or stem-loops micro RNAs. Listed are the genomic scaffold (Group in Amel_4.5) and predicted CDS (GB number of Official Gene Set 3). Gene Ontology terms for Biological Process and Molecular Function are from Flybase and given as inferred from electronic annotation (IEA), inferred from sequence or structural similarity (ISS), inferred from physical interaction, (IPI), or inferred from direct assay (IDA).
For validation of origin, Blastn and Blastx analyses were done on all library sequences against the nucleotide and the consensual protein databases of predicted A. mellifera genes (Amel Official Gene Set v3.2). Sequences without BLAST matches to predicted honey bee genes were mapped to the assembled honey bee genome (Amel 4.5). Thus, all sequences could be grouped into six categories: (i) sequences with similarity in the nr database, represented herein as similar to Drosophila, since this is the best annotated insect genome; (ii) sequences corresponding to predicted honey bee genes, including those from the bee Transcriptome Shotgun Assembly (TSA) database; (iii) sequences with similarity to stem loop microRNAs; (iv) sequences mapping in intronic regions of predicted coding sequences in the honey bee genome; (v) sequences mapping close to predicted coding sequences in the honey bee genome, thus indicating possible location in an untranslated region (5′ or 3′ UTR); and (vi) sequences without information in public databases, including predicted honey bee genes, but mapping to the honey bee genome. Figure 3 shows the relative representation of sequences for the two libraries within these six categories.
Characteristics of the queen and worker RDA libraries prepared from queen and worker larval hind leg discs. The assembled sequences were grouped into (i) genes similar (E ≤ e−5)to Drosophila melanogaster, (ii) genes similar to predicted or exclusive transcripts for A. mellifera and other bees represented in the Transcriptome Shotgun Assembly (TSA) database, (iii) genes similar to stem-loop microRNAs (miRBase), (iv) genes with intronic location, (v) genes with a putative UTR location, and (vi) sequences representing unpredicted genes but mapping to the honey bee genome sequence.
Quantitative RT-PCR analyses
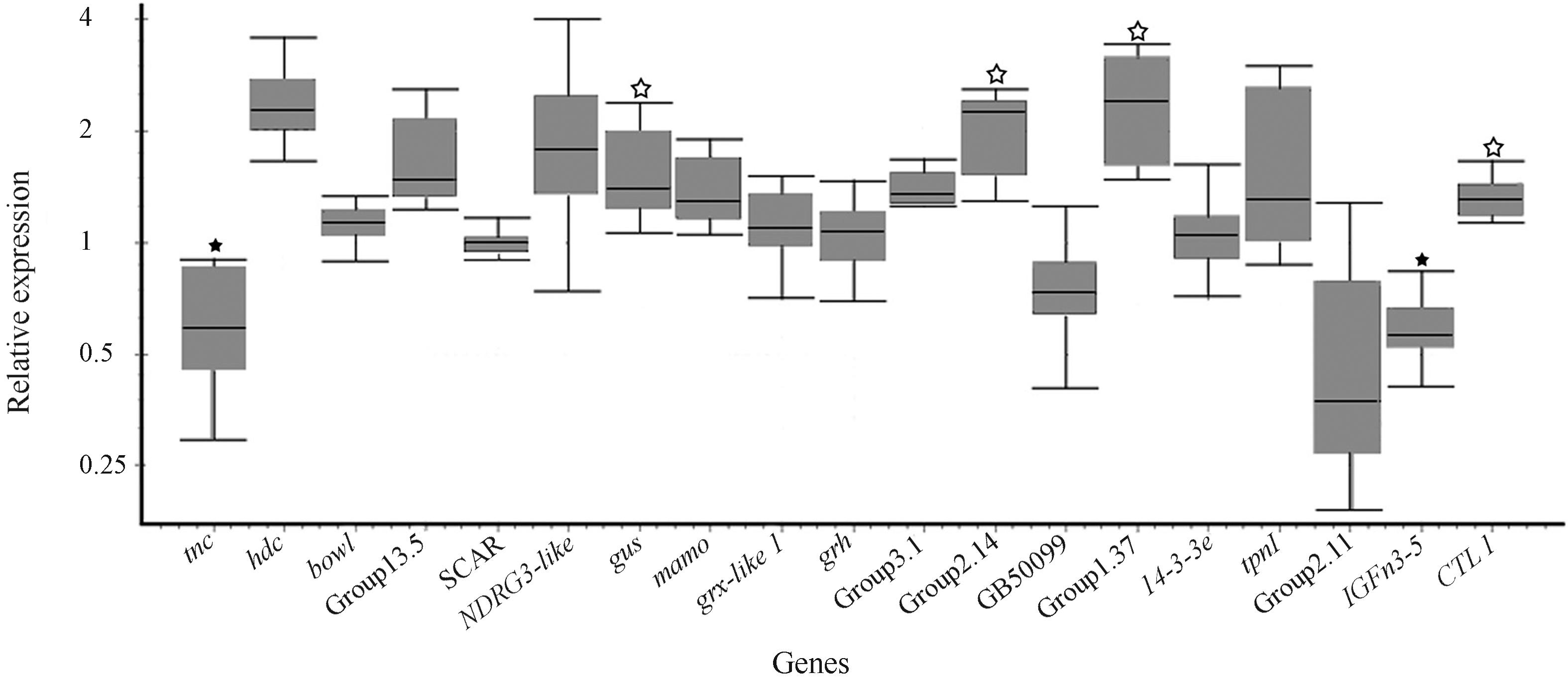
We designed primers for 19 sequences that were selected based on the following criteria: representation in the libraries (high number of ESTs per unique sequence) and similarity with Drosophila genes related with appendage development and/or body size. The genes selected for this analysis are listed in Table 1, and the REST analysis results for their differential expression in hind legs of worker vs. queen spinning phase larvae (L5S1) are shown in Figure 4. In this analysis, a statistically significant difference in expression was confirmed for six genes, four of which overexpressed in workers and two in queens. These were: Group2.14, Group1.37, gustavus, and C-type lectin 1, and tenectin and IGFn3-5, respectively.
Relative expression levels measured by RT-qPCR of genes selected from the RDA libraries. Each whisker plot represents worker gene expression relative to queens, the latter set at equal to 1. The plot was generated by REST analysis (Pfaffl et al., 2002Pfaffl MW, Horgan GW and Dempfle L (2002) Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in Real-Time PCR. Nucleic Acids Res 30:e36.). Black asterisks represent genes validated as overexpressed in queens and white ones those overexpressed in workers (random test, REST, p ≤ 0.05).
Discussion
In the grub-like larvae of holometabolous insects, the adult legs, like other appendages, develop from ectodermal cells set aside during embryonic development to form larval imaginal discs. Under the endocrine milieu prevailing during larval development, these epidermal imaginal discs proliferate but do not secrete cuticle (Nijhout, 1994Nijhout HF (1994) Insect Hormones. 1st edition. Princeton University Press, Princeton, 267 pp; Truman and Riddiford, 2007Truman JW and Riddiford LM (2007) The morphostatic actions of juvenile hormone. Insect Biochem Mol Biol 37:761–770.). Their high regenerative potential was fundamental for the transplantation studies that led to the concept of homeosis and discovery of homeotic genes in Drosophila melanogaster. Leg development is, thus, a complex process that involves several patterning genes coordinating proper development along the three axes. Most of these genes were functionally identified in Drosophila melanogaster (Campbell and Tomlinson, 1995Campbell G and Tomlinson A (1995) Initiation of the proximodistal axis in insect legs. Development 121:619–628.; Diaz-Benjumea et al., 1994Diaz-Benjumea FJ, Cohen B and Cohen SM (1994) Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature 372:175–179.; Couso and Bishop, 1998Couso JP and Bishop SA (1998) Proximo-distal development in the legs of Drosophila. Int J Dev Biol 42:345–352.; Kubota et al., 2003Kubota K, Goto S and Hayashi S (2003) The role of Wg signaling in the patterning of embryonic leg primordium in Drosophila. Dev Biol 257:117–126.; Brook, 2010Brook WJ (2010) T-box genes organize the dorsal ventral leg axis in Drosophila melanogaster. Fly 4:159–162.; Giorgianni and Mann, 2011Giorgianni MW and Mann RS (2011) Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless. Dev Cell 20:455–468.).
In contrast, most studies on imaginal disc development in bees are rather old (Myser, 1954Myser WC (1954) The larval and pupal development of the honeybee, Apis mellifera Linnaeus. Ann Entomol Soc Am 47:683–711.) or not very detailed. The most complete study is, to our knowledge, a description of imaginal disc development in the stingless bee Scaptotrigona postica (Cruz-Landim, 2009Cruz-Landim C (2009) Abelhas: Morfologia e Função de Sistemas. 1st edition. UNESP Press, São Paulo, 408 pp.), but this one is more concerned with differences in developmental timing between the leg and other imaginal discs, than with caste-related processes. Information on molecular underpinnings of caste-specific leg development is also lacking, as differential gene expression in caste development has mostly been assessed through whole body analyses, which may mask mechanisms underlying the development of individual caste-specific structures. The current study thus aimed at analyzing hind leg development of honey bee larvae, both from a morphological, as well as a gene expression perspective.
Morphology of the developing hind leg of the honey bee
During leg disc development in A. mellifera, the imaginal disc epithelium grows within a peripodial sac, as in Drosophila(Ursprung, 1972Ursprung H (1972) The fine structure of imaginal discs. In: Ursprung H and Nöthiger R (eds) The Biology of Imaginal Disks. Springer Verlag, Heidelberg, pp 93–107.), and although consisting of a monolayer it has a multilayered appearance due to extensive folding inside the peripodial space (Figure 1). The leg discs rapidly increase in cell number during the larval feeding stage (here shown by labeling of the mitosis marker phospho-histone H3, Figure 2). Subsequently, during the spinning stage, the leg compartments start to differentiate, while cell proliferation continues. Mesenchymal cells in the luminal side of leg discs were described in Drosophila (Ursprung, 1972Ursprung H (1972) The fine structure of imaginal discs. In: Ursprung H and Nöthiger R (eds) The Biology of Imaginal Disks. Springer Verlag, Heidelberg, pp 93–107.) and S. postica (Cruz-Landim, 2009Cruz-Landim C (2009) Abelhas: Morfologia e Função de Sistemas. 1st edition. UNESP Press, São Paulo, 408 pp.) as precursors of neural cells and muscles. Progressive compartimentalization of the leg imaginal discs became evident during the fifth larval instar, culminating with the evagination of the legs and other imaginal disc-derived appendages during the larval-pupal molt (Figure 1F).
As expected, in the larval stages studied here, the imaginal discs did not present obvious morphological differences related to caste-specific structures, such as a corbicula or bristle patterning. These structures become discernible in brown-eyed pupae only (Bomtorin et al., 2012Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.), i.e. at the beginning of the pharate-adult stage, and are concomitant with general bristle and trichome development in the bee’s epidermis. These events are synchronized by the hemolymph ecdysteroid titer which shapes the cuticle structure and pigmentation of pharate-adult honey bees (Elias-Neto et al., 2009Elias-Neto M, Soares MPM and Bitondi MMG (2009) Changes in integument structure during the imaginal molt of the honey bee. Apidologie 40:29–39.; Soares et al., 2011Soares MPM, Silva-Torres FA, Elias-Neto M, Nunes FMF, Simões ZLP and Bitondi MMG (2011) Ecdysteroid-dependent expression of the tweedle and peroxidase genes during adult cuticle formation in the honey bee, Apis mellifera. PloS One 6:e20513.). While these processes of pupal/adult cuticle structuring are now reasonably well understood, both in terms of morphology and gene expression, comprehension of the developmental processes underlying appendage formation and differentiation in the larval stages of the honey bee is still lacking. The current study is the first one to investigate the morphological dynamics of leg development during the larval/pupal transition, a stage that is critical for the caste-specific characteristics of these structures (Dedej et al., 1998Dedej S, Hartfelder K, Aumeier P, Rosenkranz P and Engels W (1998) Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J Apic Res 37:183–190.).
Differential gene expression in honey bee leg development
Although the leg imaginal discs of queens and workers do not show any notable differences in their morphologies in the larval stages, the caste-related endocrine millieu (Rembold, 1987Rembold H (1987) Caste specific modulation of juvenile hormone titers in Apis mellifera. Insect Biochem 17:1003–1006.; Hartfelder and Engels, 1998Hartfelder K and Engels W (1998) Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol 40:45–77.) and the gene expression environment differ considerably (Barchuk et al., 2007Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP and Maleszka R (2007). Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol 7:e70.; Bomtorin et al., 2012Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.). For our investigation on molecular events underlying the differentiation of caste-specific hind leg structure we, thus, chose a developmental stage right at the onset of metamorphosis. At this stage, growth of the discs ended and disc eversion begins to take place.
Overall, the queen and worker larval hind leg libraries were similar in terms of the total number of ESTs and the numbers of contigs and singlets. Furthermore, the hind leg libraries comprised only a relatively small number of unpredicted Apis mellifera genes, i.e. without similarity to non-redundant sequences in public databases, which is in contrast with RDA honey bee libraries generated earlier for other tissues (male accessory glands, Colonello-Frattini and Hartfelder, 2009Colonello-Frattini NA and Hartfelder K (2009) Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie 40:481–495.; and larval ovaries, Humann and Hartfelder, 2011Humann FC and Hartfelder K (2011) Representational difference analysis (RDA) reveals differential expression of conserved as well as novel genes during caste-specific development of the honey bee (Apis mellifera L.) ovary. Insect Biochem Mol Biol 41:602–612.). These previous libraries had, however, been analyzed against an earlier gene prediction set (Amel Official Gene Set 2.0), which comprised an underestimated number of genes (10,157). This was subsequently corrected through Transcriptome Shotgun Assembly data and is now updated in a revised honey bee genome version which comprises over 15,000 protein coding genes (Elsik et al., 2014Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. (2014) Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics 15:e86.). Some of the still unpredicted sequences probably represent regulatory long non-coding RNAs, which represent a considerable part of the mammalian genomes. For the honey bee, only few such regulatory RNAs are known, including two that have recently been revealed in an RDA study on ovary development in honey bee larvae (Humann et al., 2013Humann FC, Tiberio GJ and Hartfelder K (2013). Sequence and expression characteristics of long noncoding RNAs in honey bee caste development-potential novel regulators for transgressive ovary size. PloS One 8:e78915.).
The RT-qPCR analyses revealed six sequences among the 19 selected for validation as significantly differentially expressed (Figure 4). The two sequences that were more expressed in queens were tenectin and immunoglobulin-like and fibronectin type III domain containing 5 (IGFn3-5). IGF n3-5, known as echinoid in D. melanogaster, is predicted as GB49260 in the honey bee genome. The protein is related with cell adhesion, playing a role in lateral inhibition of proneural clusters and the Notch signaling pathway. It has been described as an Egfr antagonist in Drosophila, acting during bristle patterning (Escudero et al., 2003Escudero LM, Wei SY, Chiu WH, Modolell J and Hsu JC (2003) Echinoid synergizes with the Notch signaling pathway in Drosophilamesothorax bristle patterning. Development 130:6305–6316.) and compound eye development (Fetting et al., 2009Fetting JL, Spencer SA and Wolff T (2009) The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye. Development 136:3323–3333.). It also acts upon actin/myosin structures during epithelial development and modulates cell adhesion (Bogdan and Klämbt, 2001Bogdan S and Klämbt C (2001) Epidermal growth factor receptor signaling. Curr Biol 11:R292–R295.; Tepass and Harris, 2006; Laplante and Nilson, 2011Laplante C and Nilson LA (2011) Asymmetric distribution of Echinoid defines the epidermal leading edge during Drosophila dorsal closure. J Cell Biol 192:335–348.).
tenectin is predicted in the honey bee Official gene Set 3.2 as GB53632, located in the genomic scaffold 3.9. It encodes an integrin binding protein related to developmental processes and is regulated by ecdysone during wing development. Transcripts for this gene were recently identified in Drosophila leg discs, eye and brain (Fraichard et al., 2010Fraichard S, Bougé AL, Kendall T, Chauvel I, Bouhin H and Bunch TA (2010) Tenectin is a novel αPS2βPS integrin ligand required for wing morphogenesis and male genital looping in Drosophila. Dev Biol 340:504–517.).
Thus, interestingly, the genes that had their differential expression confirmed in queens were both cell adhesion molecules and related to certain signaling pathways. The possible responsiveness to ecdysone and involvement in the Egfr pathway furthermore indicate that these proteins not only maintain the adhesive properties of the disc epithelium, but may also coordinate or integrate upstream or downstream signals in leg differentiation. The role of ecdysone in caste development is well established (Rachinsky et al., 1990Rachinsky A, Strambi C, Strambi A and Hartfelder K (1990) Caste and metamorphosis: hemolymph titres of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen Comp Endocrinol 79:31–38.; Hartfelder et al., 1995Hartfelder K, Köstlin K and Hepperle C (1995) Ecdysteroid-dependent protein synthesis in caste-specific development of the larval honey bee ovary. Rouxs Arch Dev Biol 205:73–80.; Mello et al., 2014Mello TRP, Aleixo AC, Pinheiro DG, Nunes FMF, Bitondi MMG, Hartfelder K, Barchuk AR and Simões ZLP (2014) Developmental regulation of ecdysone receptor (EcR) gene expression and its targets in the honeybee, Apis mellifera. Frontiers Genet 5:e445.), and the Egfr pathway has recently been revealed as a major player in caste fate determination (Kamakura, 2011Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473:478–483.).
Among the four sequences that were confirmed by RT-qPCR as differentially expressed in workers, only two had a prediction in the public database. The first one encodes a C-type lectin I, predicted as GB49260 in the honey bee genome. It encodes a calcium-dependent lectin that belongs to a large family of endocytic receptors, proteoglycans, colectins and selectins. This lectin type presents high evolutionary divergence from the ones found in vertebrates, which suggests that it may actually have functions other than those typically assigned to lectins (Dodd and Drickamer, 2001Dodd RB and Drickamer K (2001) Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11:71R–79R.).
GB55797 encodes the honey bee SOCS box 1 protein containing an SPRY domain, similar to the D. melanogaster gene gustavus(CG2944). According to its Gene Ontology attributes, the gustavus protein is related to dorsal appendage formation, oocyte anterior/posterior axis specification and intracellular signal transduction. SOCS-box proteins act as E3 ubiquitin ligases.
Also overexpressed in workers were the transcripts Group2.14 and Group1.37. These two sequences did not have a prediction in the A. melliferaOfficial Gene Set (v3.2), but all could precisely be mapped to the genome sequence. While no further information exists for the Group2.14 sequence, Group1.37 has similarity to stem-loop microRNAs. This microRNA, however, does not belong to any of the known miR families listed in miRBase for the honey bee. Nonetheless, not all honey bee microRNAs have been identified so far. Alternatively, stem loops are also frequently found as structural elements of long non-coding RNAs, as seen in the two honey bee long non-coding RNAs identified in ovary development (Humann et al., 2013Humann FC, Tiberio GJ and Hartfelder K (2013). Sequence and expression characteristics of long noncoding RNAs in honey bee caste development-potential novel regulators for transgressive ovary size. PloS One 8:e78915.).
Genes previously associated with honey bee leg development (Barchuk, et al., 2007Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP and Maleszka R (2007). Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol 7:e70.) and the patterning genes analyzed by Bomtorin et al. (2012)Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111. did not appear in the RDA libraries. The developmental stages analyzed by Barchuk et al. (2007)Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP and Maleszka R (2007). Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol 7:e70., as well as the analysis method used, were however different from those of the current study. The tissue-specific quantitative analyses on the leg development candidate genes abdominal-A, cryptocephal and ultrabithorax on the other hand did not reveal expression differences in queens and workers during the last instar spinning stage (Bomtorin et al., 2012Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.), which was the developmental stage in focus in the present study.
Among the other honey bee genes with reads in the RDA libraries, several are similar to Drosophila melanogaster genes and with Gene Ontology (GO) attributes, related to muscle development, cell adhesion, wing imaginal disc development and epithelial morphogenesis. The gene encoding the protein SCAR (suppressor of cyclic AMP receptor) was represented in the queen library. SCAR is an actin nucleation factor that acts on myoblasts migration and fusion during D. melanogaster embryogenesis (Gildor et al., 2009Gildor B, Massarwa R, Shilo BZ and Schejter ED (2009) The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep 10:1043–1050.). The Drosophila gene bowl (brother of odd with entrails limited, also known as bowel) regulates joint formation, with mutants presenting fusion of legs segments. It belongs to the odd-skipped family and acts downstream of the Notch pathway during leg joint morphogenesis (Hao et al., 2003Hao I, Green RB, Dunaevsky O, Lengyel JA and Rauskolb C (2003) The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev Biol 263:282–295.; Kojima, 2004Kojima T (2004) The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ 46:115–129.).
Another interesting finding was the identification of 14-3-3ε and ypsilon-schachtel transcripts in the queen library, as both had previously been identified using the RDA methodology on honey bee male accessory glands (Colonello-Frattini and Hartfelder, 2009Colonello-Frattini NA and Hartfelder K (2009) Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie 40:481–495.). Furthermore, a proteomic approach had also identified a 14-3-3ε protein as overexpressed in 72 and 120 hours workers larvae (Li et al., 2010Li J, Wu J, Rundassa DB, Song F, Zheng A and Fang Y (2010) Differential protein expression in honeybee (Apis mellifera L.) larvae underlying caste differentiation. PloS One 5:e13455.). 14-3-3 proteins are a family of conserved regulatory molecules that participate in a wide range of cellular processes, such as cell proliferation, cancer progression, apoptosis, cell cycle control, actin cytoskeleton regulation. Since several isoforms are present in all eukaryotes, some of them could have overlapping functions (Freeman and Morrison, 2011Freeman AK and Morrison DK (2011) 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol 22:681–687.; Gardino and Yaffe, 2011Gardino AK and Yaffe MB (2011) 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol 22:688–695.; Aitken, 2011Aitken A. (2011) Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin Cell Dev Biol 22:673–680.). Furthermore, another 14-3-3ε function appears to be that of a modulator of signal integration by FoxO (Nielsen et al., 2008Nielsen MD, Luo X, Biteau B, Syverson K and Jasper H (2008) 14-3-3 epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell 7:688–699.), downstream in the insulin/TOR signaling pathway. Especially the latter is of importance in honey bee caste development (Patel et al., 2007Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K and Amdam GV (2007) The making of a queen: TOR pathway is a key player in diphenic caste development. PloS One 2:e509.). The fact that 14-3-3 proteins represent a large family of relatively conserved proteins may have been a reason why we could not find statistical differences in the RT-qPCR assays.
ypsilon-schachtel (yps) is a Y-box protein component of an ovarian ribonucleoprotein complex that acts as translational regulator of oskar mRNA in Drosophila (Mansfield et al., 2002Mansfield JH, Wilhelm JE and Hazelrigg T. 2002. Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development 129:197–209.). Again, the quantitative real time PCR did not validate it as differentially expressed in queens and workers. Nevertheless, the recurrent appearance of 14-3-3ε and yps in honey bee RDA libraries generated for different tissues and developmental processes indicates that theses genes may be redundantly involved in developmental pathways common to both castes and sexes.
Another gene related to epithelial development revealed in the worker RDA library was the transcription factor grainy head.In Drosophila development it plays a role as an ecdysone-response gene in cuticle differentiation (Gangishetti et al., 2012Gangishetti U, Veerkamp J, Bezdan D, Schwarz H, Lohmann I and Moussian B (2012) The transcription factor Grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster. Insect Mol Biol 21:283–295.). In bees it has previously been denoted as differentially expressed in caste development of the stingless bee Melipona quadrifasciata (Judice et al., 2006Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K and Pereira GAG (2006) Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol 15:33–44.) and it was also identified as a major factor promoting thelytokous reproduction in the Cape honey bee Apis mellifera capensis (Lattorff et al., 2007Lattorff HMG, Moritz RFA, Crewe RM and Solignac M (2007) Control of reproductive dominance by the thelytoky gene in honeybees. Biol Lett 3:292–295.).
In conclusion, our histological analysis of honey bee hind leg development, represents the first detailed description of general growth and differentiation processes that occur during the larval developmental stages critical for determining the caste fate of these structures (Dedej et al., 1998Dedej S, Hartfelder K, Aumeier P, Rosenkranz P and Engels W (1998) Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J Apic Res 37:183–190.). As comprehensively shown in the fruit fly, development of the body appendages depends on several patterning mechanisms and signaling modules. In honey bee development, an additional layer of complexity is added to these processes due to the caste-specificity in anatomical specializations, such as the corbicula and pollen brush on the hind legs of workers. In the differential gene expression analysis by means of RDA library sequencing we could see this reflected in the detection of a series of genes predicted as related to cell adhesion and signaling pathways, as well as a set of reads mapping to genome sequences for which no gene function has been predicted so far. Obviously, especially this set of novel genes should deserve attention in future studies.
Acknowledgments
We thank Luiz R. Aguiar for expert technical assistance in the apiary and Fernanda Carvalho Humann for help with the RDA protocol. Laser confocal microscopy analyses were done on systems installed in the Faculdade de Medicina de Ribeirão Preto (FAPESP grant 2004/08868-0). The project received financial support from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, grants 2008/09505-0, 2011/0371-5 and 2012/01808-9).
-
Associate Editor: Igor Schneider
References
- Aitken A. (2011) Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin Cell Dev Biol 22:673–680.
- Asencot M and Lensky Y (1988) The effect of soluble sugars in stored royal jelly on the differentiation of female honeybee (Apis mellifera L.) larvae to queens. Insect Biochem 18:127–133.
- Azevedo SV and Hartfelder K (2008) The insulin signaling pathway in honey bee (Apis mellifera) caste development - differential expression of insulin-like peptides and insulin receptors in queen and worker Larvae. J Insect Physiol 54:1064–1071.
- Azevedo SV, Caranton OA, de Oliveira TL and Hartfelder K (2011) Differential expression of hypoxia pathway genes in honey bee (Apis mellifera L.) caste development. J Insect Physiol 57:38–45.
- Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP and Maleszka R (2007). Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera BMC Dev Biol 7:e70.
- Bogdan S and Klämbt C (2001) Epidermal growth factor receptor signaling. Curr Biol 11:R292–R295.
- Bomtorin AD, Barchuk AR, Moda LM and Simoes ZLP (2012) Hox gene expression leads to differential hind leg development between honeybee castes. PLoS One 7:e40111.
- Brook WJ (2010) T-box genes organize the dorsal ventral leg axis in Drosophila melanogaster Fly 4:159–162.
- Campbell G and Tomlinson A (1995) Initiation of the proximodistal axis in insect legs. Development 121:619–628.
- Cardinal S and Danforth BN (2011) The antiquity and evolutionary history of social behavior in bees. PloS One 6:e21086.
- Colonello-Frattini NA and Hartfelder K (2009) Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie 40:481–495.
- Couso JP and Bishop SA (1998) Proximo-distal development in the legs of Drosophila Int J Dev Biol 42:345–352.
- Cristino AS, Nunes FMF, Lobo CH, Bitondi MMG, Simões ZLP, Costa LD, Lattorff HMG, Moritz RFA, Evans JD and Hartfelder K (2006) Caste development and reproduction: a genome-wide analysis of hallmarks of insect eusociality. Insect Mol Biol 15:703–714.
- Cruz-Landim C (2009) Abelhas: Morfologia e Função de Sistemas. 1st edition. UNESP Press, São Paulo, 408 pp.
- Dedej S, Hartfelder K, Aumeier P, Rosenkranz P and Engels W (1998) Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J Apic Res 37:183–190.
- Diaz-Benjumea FJ, Cohen B and Cohen SM (1994) Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature 372:175–179.
- Dodd RB and Drickamer K (2001) Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11:71R–79R.
- Durham AM, Kashiwabara AY, Matsunaga FTG, Ahagon PH, Rainonel F, Varuzza L and Gruber A (2005) EGene: A configurable pipeline generation system for automated sequence analysis. Bioinformatics 21:2812–2813.
- Elias-Neto M, Soares MPM and Bitondi MMG (2009) Changes in integument structure during the imaginal molt of the honey bee. Apidologie 40:29–39.
- Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. (2014) Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics 15:e86.
- Escudero LM, Wei SY, Chiu WH, Modolell J and Hsu JC (2003) Echinoid synergizes with the Notch signaling pathway in Drosophilamesothorax bristle patterning. Development 130:6305–6316.
- Evans JD and Wheeler DE (2000) Expression profiles during honeybee caste determination. Genome Biol 2:RESEARCH0001.
- Fetting JL, Spencer SA and Wolff T (2009) The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye. Development 136:3323–3333.
- Ford DM, Hepburn HR, Moseley FB and Rigby RJ (1981) Displacement sensors in the honeybee pollen basket. J Insect Physiol 27:339–346.
- Fraichard S, Bougé AL, Kendall T, Chauvel I, Bouhin H and Bunch TA (2010) Tenectin is a novel αPS2βPS integrin ligand required for wing morphogenesis and male genital looping in Drosophila Dev Biol 340:504–517.
- Freeman AK and Morrison DK (2011) 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol 22:681–687.
- Gallai N, Salles JM, Settele J and Vaissière BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68:810–821.
- Gangishetti U, Veerkamp J, Bezdan D, Schwarz H, Lohmann I and Moussian B (2012) The transcription factor Grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster Insect Mol Biol 21:283–295.
- Gardino AK and Yaffe MB (2011) 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol 22:688–695.
- Gildor B, Massarwa R, Shilo BZ and Schejter ED (2009) The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep 10:1043–1050.
- Giorgianni MW and Mann RS (2011) Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless. Dev Cell 20:455–468.
- Griffiths-Jones S, Saini HK, van Dongen S and Enright AJ (2008) miRBase: Tools for microRNA genomics. Nucleic Acids Res 36(Database issue):D154–D158.
- Hao I, Green RB, Dunaevsky O, Lengyel JA and Rauskolb C (2003) The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev Biol 263:282–295.
- Hartfelder K and Engels W (1998) Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honeybee. Curr Top Dev Biol 40:45–77.
- Hartfelder K, Köstlin K and Hepperle C (1995) Ecdysteroid-dependent protein synthesis in caste-specific development of the larval honey bee ovary. Rouxs Arch Dev Biol 205:73–80.
- Hartfelder K, Guidugli-Lazzarini KR, Cervoni MS, Santos DE and Humann FC (2015) Old threads make new tapestry - rewiring of signaling pathways underlies caste phenotypic plasticity in the honey bee, Apis mellifera L.. Adv Insect Physiol 48:1–36.
- Haydak MH (1970) Honey bee nutrition. Annu Rev Entomol 15:143–156.
- Hepperle C and Hartfelder K (2001) Differentially expressed regulatory genes in honey bee caste development. Naturwissenschaften 88:113–116.
- Huang X and Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877.
- Hubank M and Schatz DG (2000) Representational difference analysis of cDNA. In: Hunt SP and Livesey FJ (eds) Functional Genomics. Oxford University Press, Oxford, pp 45–80.
- Humann FC and Hartfelder K (2011) Representational difference analysis (RDA) reveals differential expression of conserved as well as novel genes during caste-specific development of the honey bee (Apis mellifera L.) ovary. Insect Biochem Mol Biol 41:602–612.
- Humann FC, Tiberio GJ and Hartfelder K (2013). Sequence and expression characteristics of long noncoding RNAs in honey bee caste development-potential novel regulators for transgressive ovary size. PloS One 8:e78915.
- Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K and Pereira GAG (2006) Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata Insect Mol Biol 15:33–44.
- Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473:478–483.
- Klein AM, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C and Tscharntke T (2006) Importance of pollinators in changing landscapes for world crops. Proc Biol Sci 274:303–313.
- Kojima T (2004) The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ 46:115–129.
- Kubota K, Goto S and Hayashi S (2003) The role of Wg signaling in the patterning of embryonic leg primordium in Drosophila Dev Biol 257:117–126.
- Laplante C and Nilson LA (2011) Asymmetric distribution of Echinoid defines the epidermal leading edge during Drosophila dorsal closure. J Cell Biol 192:335–348.
- Lattorff HMG, Moritz RFA, Crewe RM and Solignac M (2007) Control of reproductive dominance by the thelytoky gene in honeybees. Biol Lett 3:292–295.
- Leimar O, Hartfelder K, Laubichler MD and Page RE (2012) Development and evolution of caste dimorphism in honeybees - A modeling approach. Ecol Evol 2:3098–3109.
- Li J, Wu J, Rundassa DB, Song F, Zheng A and Fang Y (2010) Differential protein expression in honeybee (Apis mellifera L.) larvae underlying caste differentiation. PloS One 5:e13455.
- Lourenço AP, Mackert A, Cristino AS and Simões ZLP (2008) Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative Real-Time RT-PCR. Apidologie 39:372–385.
- Mansfield JH, Wilhelm JE and Hazelrigg T. 2002. Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development 129:197–209.
- Mello TRP, Aleixo AC, Pinheiro DG, Nunes FMF, Bitondi MMG, Hartfelder K, Barchuk AR and Simões ZLP (2014) Developmental regulation of ecdysone receptor (EcR) gene expression and its targets in the honeybee, Apis mellifera Frontiers Genet 5:e445.
- Michelette ERF and Soares AEE (1993) Characterization of preimaginal developmental stages in Africanized honey bee workers (Apis mellifera L). Apidologie 24:431–440.
- Michener CD (2007) The Bees of the World. 2nd edition. Johns Hopkins University Press, Baltimore, 992 pp.
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG et al. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–67.
- Myser WC (1954) The larval and pupal development of the honeybee, Apis mellifera Linnaeus. Ann Entomol Soc Am 47:683–711.
- Nielsen MD, Luo X, Biteau B, Syverson K and Jasper H (2008) 14-3-3 epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila Aging Cell 7:688–699.
- Nijhout HF (1994) Insect Hormones. 1st edition. Princeton University Press, Princeton, 267 pp
- Oppelt A, Humann FC, Fuessl M, Azevedo SV, Marco Antonio DS, Heinze J and Hartfelder K (2010) Suppression subtractive hybridization analysis reveals expression of conserved and novel genes in male accessory glands of the ant Leptothorax gredleri BMC Evol Biol 10:e273.
- Pastorian K, Hawel L and Byus CV (2000) Optimization of cDNA representational difference analysis for the identification of differentially expressed mRNAs. Anal Biochem 283:89–98.
- Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K and Amdam GV (2007) The making of a queen: TOR pathway is a key player in diphenic caste development. PloS One 2:e509.
- Pfaffl MW, Horgan GW and Dempfle L (2002) Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in Real-Time PCR. Nucleic Acids Res 30:e36.
- Proctor MCF, Yeo P and Lack A (1996) The Natural History of Pollination. Illustrated edition. Timber Press, Portland, 479 pp.
- Rachinsky A and Engels W (1995) Caste development in honeybees (Apis mellifera): Juvenile hormone turns on ecdysteroids. Naturwissenschaften 82:378–379.
- Rachinsky A, Strambi C, Strambi A and Hartfelder K (1990) Caste and metamorphosis: hemolymph titres of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen Comp Endocrinol 79:31–38.
- Rembold H (1987) Caste specific modulation of juvenile hormone titers in Apis mellifera Insect Biochem 17:1003–1006.
- Schmidt-Capella IC and Hartfelder K (1998) Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J Insect Physiol 44:385–391.
- Soares MPM, Silva-Torres FA, Elias-Neto M, Nunes FMF, Simões ZLP and Bitondi MMG (2011) Ecdysteroid-dependent expression of the tweedle and peroxidase genes during adult cuticle formation in the honey bee, Apis mellifera PloS One 6:e20513.
- Tepass U and Harris KP (2007) Adherens junctions in Drosophila retinal morphogenesis Trends Cell Biol 17:26–35.
- Truman JW and Riddiford LM (2007) The morphostatic actions of juvenile hormone. Insect Biochem Mol Biol 37:761–770.
- Ursprung H (1972) The fine structure of imaginal discs. In: Ursprung H and Nöthiger R (eds) The Biology of Imaginal Disks. Springer Verlag, Heidelberg, pp 93–107.
- Weil T, Rehli M and Korb J (2007) Molecular basis for the reproductive division of labour in a lower termite. BMC Genomics 8:e198.
- Wheeler DE, Buck N and Evans JD (2006) Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera Insect Mol Biol 15:597–602.
- Wheeler DE, Buck NA and Evans JD (2014) Expression of insulin/insulin-like signalling and TOR pathway genes in honey bee caste determination. Insect Mol Biol 23:113–121.
Internet Resources
- Beebase, http://www.hymenopteragenome.org/beebase/ (January 12, 2014).
» http://www.hymenopteragenome.org/beebase/ - GenBank, http://blast.ncbi.nlm.nih.gov/Blast.cgi (Dezember 15, 2014).
» http://blast.ncbi.nlm.nih.gov/Blast.cgi - Flybase, http://www.flybase.org (May 5, 2014).
» http://www.flybase.org - miRBase, http://www.mirbase.org (May 5, 2014).
» http://www.mirbase.org - Long Noncoding RNA Database v2.0, http://www.lncrnadb.org (May 5, 2014).
» http://www.lncrnadb.org - Primer3 software, http://frodo.wi.mit.edu/primer3 (May 10, 2012).
» http://frodo.wi.mit.edu/primer3
Publication Dates
-
Publication in this collection
21 Aug 2015 -
Date of issue
July-Sept 2015
History
-
Received
30 Dec 2014 -
Accepted
25 Mar 2015