Abstract
The role of myogenic enhancer transcription factor 2a (MEF2A) in avian muscle during fetal development is unknown. In this work, we cloned the duck MEF2A cDNA sequence (GenBank accession no. HM460752) and examined its developmental expression profiles in cardiac muscle, non-vascular smooth muscle and skeletal muscle. Duck MEF2A cDNA comprised 1479 bp encoding 492 amino acid residues. In silico analysis showed that MEF2A contained MADS (MCM1, AGAMOUS, DEFICIENS and SRF -serum response factor), MEF2 and mitogen-activated protein kinase (MAPK) transcription domains with high homology to related proteins in other species. Modified sites in these domains were conserved among species and several variants were found. Quantitative PCR showed that MEF2A was expressed in all three muscles at each developmental stage examined, with the expression in smooth muscle being higher than in the other muscles. These results indicate that the conserved domains of duck MEF2A, including the MADS and MEF2 domains, are important for MEF2A transcription factor function. The expression of MEF2A in duck smooth muscle and cardiac muscle suggests that MEF2A plays a role in these two tissues.
duck MEF2A; expression profile; in silico analysis; molecular cloning; muscle tissues
Molecular cloning and in silico analysis of the duck (Anas platyrhynchos) MEF2A gene cDNA and its expression profile in muscle tissues during fetal development
Hehe Liu, Jiwen Wang; Jianmin Si; Jing Jia; Liang Li; Chunchun Han; Kailiang Huang; Hua He; Feng Xu
Key Laboratory of Animal Genetic Resources, College of Animal Science and Technology, Sichuan Agricultural University, Ya'an, Sichuan, P.R. China
Send correspondence to Send correspondence to: Jiwen Wang. College of Animal Science and Technology, Sichuan Agricultural University 625014 Ya'an, Sichuan, P.R. China. Email: wjw2886166@163.com
ABSTRACT
The role of myogenic enhancer transcription factor 2a (MEF2A) in avian muscle during fetal development is unknown. In this work, we cloned the duck MEF2A cDNA sequence (GenBank accession no. HM460752) and examined its developmental expression profiles in cardiac muscle, non-vascular smooth muscle and skeletal muscle. Duck MEF2A cDNA comprised 1479 bp encoding 492 amino acid residues. In silico analysis showed that MEF2A contained MADS (MCM1, AGAMOUS, DEFICIENS and SRF -serum response factor), MEF2 and mitogen-activated protein kinase (MAPK) transcription domains with high homology to related proteins in other species. Modified sites in these domains were conserved among species and several variants were found. Quantitative PCR showed that MEF2A was expressed in all three muscles at each developmental stage examined, with the expression in smooth muscle being higher than in the other muscles. These results indicate that the conserved domains of duck MEF2A, including the MADS and MEF2 domains, are important for MEF2A transcription factor function. The expression of MEF2A in duck smooth muscle and cardiac muscle suggests that MEF2A plays a role in these two tissues.
Key words: duck MEF2A, expression profile, in silico analysis, molecular cloning, muscle tissues. Received: May 2, 2011; Accepted: November 23, 2011.
Introduction
Animals possess three major types of muscle, namely, cardiac, skeletal and smooth muscle that are essential for movement (Byers et al. (, 1991; Handschin et al. (, 2007; Leong et al. (, 2008; MacDonald, 2008). The developmental stages from embryo formation to birth are important for muscle development (Rehfeldt, 2005). The skeletal muscle mass of adult animals is determined early in development and, in most animals, there is no post-natal increase in the number of myofibers. Post-natal muscle hypertrophy involves only myofiber lengthening and thickening (Chauvigne, 2005). Although the heart is one of the earliest organs formed during embryogenesis (Forouhar et al., 2006), subsequent development is nevertheless important because heart muscle development is completed during this stage (Noorlander et al. (, 2008). The fetal stage is also important for smooth muscle maturation and the initiation of physiological functions (Olson, 2006). Muscle development in this stage is regulated by many genes such as muscle regular factors (MRFs) and myostatin (MSTN) (Owens et al. 2004; Olson, 2006; Potthoff and Olson, 2007). However, only the myogenic enhancer transcription factor 2a (MEF2A) gene has been reported to be expressed in all three muscle tissues (Edmondson et al., 1994; Wang et al., 2000).
MEF2A is a muscle-specific factor that belongs to the MEF2 gene family. In mammals, four members of the gene family have been identified, i.e., MEF2A, -B, -C and -D. MEF2A is involved in a variety of muscle responses, including skeletal muscle myofiber differentiation, smooth muscle maturation, and normal development of cardiac muscle (Edmondson et al. (,, 1994; Wang et al. (,, 2000; Potthoff and Olson, 2007). In addition to muscle, MEF2A is expressed in neuronal cells and at low levels in a wide range of cell types during embryogenesis and adulthood. In skeletal muscle, MEF2A plays a cooperative role with the MRF gene family in inducing muscle hypertrophy (Molkentin et al., 1995; Black and Olson, 1998), and its polymorphisms are associated with growth traits in Chinese indigenous cattle breeds (Chen et al. (, 2010) and with muscle growth in chickens (Zhou et al. (, 2010). MEF2A is also highly expressed in the heart during zebrafish embryo-genesis (Wang YX et al. (, 2005). Mice lacking MEF2A disdie suddenly (Naya et al. (, 2002). MEF2A polymorphisms have been associated with several cardiac diseases in human and may account for 1.93% of such diseases (Wang Q et al. (, 2005). The MEF2A gene also plays a role in smooth muscle cell proliferation and contraction and is expressed in smooth muscle mainly during adulthood (Firulli et al. (, 1996). Almost all of the studies of MEF2A have focused on in its functions during embryogenesis and in adulthood. In contrast, there have been no studies of the roles of MEF2A in muscle tissues during fetal development.
Most studies of MEF2A have used mammals as a model, although MEF2A and MEF2D have also been studied in birds (Buchberger and Arnold, 1999; Xue et al., 2000). Buchberger and Arnold (1999) reported that chicken MEF2A has a role avian in skeletal muscle and cardiac muscle, although the precise functions of this protein are still unclear. In this work, we examined the functions of MEF2A in muscle tissue from the Pekin duck, a famous meat poultry breed. The coding sequence of duck MEF2A was cloned and its expression profile in muscle during fetal development was determined using real-time PCR. These results provide essential data for elucidating the roles of MEF2A in muscle tissue and may also provide new clues for improving muscle production in duck breeding.
Materials and Methods
Tissues collection and total RNA extraction
All procedures were approved by the Guidelines on Humane Treatment of Laboratory Animals (2006). Twenty five Pekin duck (Anas platyrhynchos) embryos (embryonic age: 10, 14, 18, 22 or 27 days old, designated E10d, E14d, E18d, E22d and E27d, respectively) and five neonates (one week post-natal, designated as p7d) were used. All of the embryos and birds were from the Sichuan Agricultural University Waterfowl Breeding Experimental Farm. The hatching embryos and birds were maintained under the same conditions. For each embryo and newborn bird, approximately 2 g of tissue was collected from the heart, small intestine and leg muscle. The samples were immediately frozen in liquid nitrogen and stored at -86 °C for RNA extraction. Total RNA was extracted using Trizol (Invitrogen, USA) according to the manufacturer's instructions and then measured spectrophotometrically. First-strand cDNA was obtained from 10 /g of total RNA using a cDNA synthesis kit (Takara, Japan) according to the manufacturer's instruction.
Cloning and sequencing of the duck MEF2A CDS
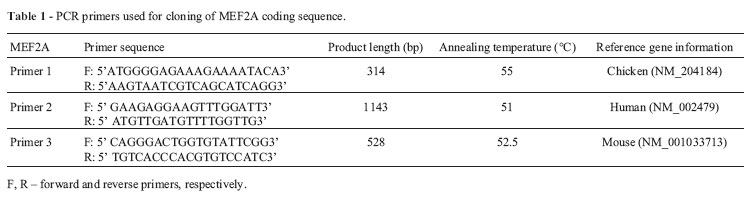
All of the primers were designed using Primer Premier 5 software and were synthesized by the Shanghai Yingjun Biology Company. The primers used to amplify the duck MEF2A gene coding sequence (CDS) are shown in Table 1. Two microliters of the cDNA mix was used as a template in a 20 /L PCR reaction volume. The reaction conditions included denaturation for 5 min at 94 °C followed by 30 cycles of amplification at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR products were separated by electrophoresis on a 1% agarose gel and purified using a gel extraction kit (Watson Biomedical Inc., Shanghai, China). The purified PCR products were ligated into the pGEM19T vector (Invitrogen). Positive clones were selected and sequenced by the Shanghai Yingjun Biology Company. The cDNA segments obtained from sequencing were edited and assembled with Editseq and Seqman in DNAstar software (DNASTAR Inc., Madison, WI, USA)
Real-time PCR
The levels of MEF2A mRNA expression in duck muscles at different developmental stages were measured by real-time PCR using a 96-well iCycler IQ5 (BioRad, USA) and Takara ExTaq RT-PCR kits with SYBR green (Takara, Dalian, China). One sample was collected from each muscle type, each bird and each stage, and the real-time PCR were repeated three times together for each sample. Duck f-actin (EF667345) and duck glyceraldehyde3-phosphate dehydrogenase (GAPDH, AY436595), the sequences of which were obtained from GenBank, were used as internal controls. The real-time PCR primers were designed using Primer 5 software (Primer Biosoft International, USA) and are shown in Table 2. The primers were synthesized by the Shanghai Yingjun Biology Company. Amplicons corresponding to each target gene were examined by agarose gel electrophoresis to confirm the presence of a single band of the expected size. The procedure for real-time PCR was as follows: 10 s of pre-denaturation at 95 °C, followed by 45 cycles of 95 °C for 5 s and 62 °C for 30 s. PCR products were diluted 16-fold and used to generate the calibration curve and determine the PCR amplification efficiency (Eff) for each gene (MEF2A, f-actin and GAPDH, provided in Table 2).
The relative mRNA expression for target genes was determined with the following formula after obtaining the Ct value of the gene of interest (MEF2A) and the Ct value of the internal control genes:
The relative quantity (Rel. quantity) is the multiple of the differential expression of the gene of interest (GOI) between the test samples and control samples. The normalizer (Norm) is the reference gene and the PCR amplification efficiency is represented by Eff.
Statistical analysis
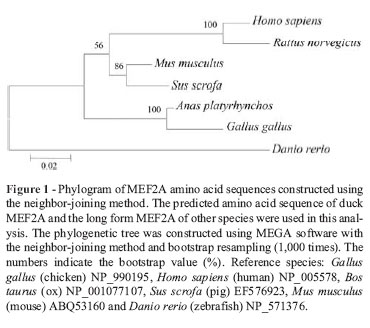
BLASTn (NCBI) was used to identify the gene. Analysis of the nucleotide and amino acid sequences, prediction of secondary structure and multiple sequence alignment were done with Editseq, Protean, and MegAlign in DNAstar, respectively. Conserved domains in the protein were analyzed using the CD-search tool available from NCBI. Phylogenetic trees were constructed with MEGA 4.0 software by the neighbor-joining method and bootstrap sampling 1,000 times.
The real-time PCR data were subjected to analysis of variance (ANOVA) and the means were compared for significance by Tukey's test. The ANOVA and t-tests were done with SAS (SAS Institute, Cary, NC, USA) software. A p-value of p < 0.05 was considered statistically significant.
Results
Comparison of duck MEF2A with other species
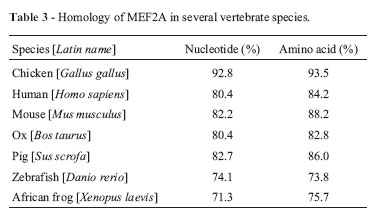
After sequencing and assembling of the fragments, the total coding sequence of duck MEF2A was obtained and submitted to GenBank (GenBank accession no. HM460752). This consisted of 1479 nucleotides encoding 492 amino acids and showed high similarity with the chicken sequence (92.8% similarity at the nucleotide level and 93.5% at the amino acid level; Table 3).
Phylogenetic tree construction
The phylogenetic tree constructed based on vertebrate MEF2A amino acid sequences showed that the sequences formed three main groups: mammalian MEF2A, fish MEF2A and avian MEF2A (Figure 1). A CD-search on NCBI revealed that duck MEF2A had three conserved domains in the peptide strands, i.e., the MADS (aa 3-57), MEF2 (aa 58-86) and MAPK (mitogen-activated protein kinase) binding domains (aa 264-285), but lacked a beta domain that is present in other organisms, except for zebrafish. There were no amino acid variants in the three conserved domains of the duck and chicken proteins. These three regions were also highly conserved between birds and mammals. Only two variant residues were observed in the porcine MEF2A, MADS and MEF2 domains compared with other mammals, in addition to a single residue change from 'N' in avian sequences to 'S' in mammalian and fish (zebrafish) sequences; the latter also showed two additional amino acid changes compared to the avian sequences.
Several modified residues were identified in MEF2A in addition to the conserved domains of MEF2A. Most of these were conserved among species, including acetyllysine sites at positions 117, 248, 253 and 394, phosphoserine sites at positions 98, 221, 233, 346, 399 and 439, and phosphothreonine sites at positions 302 and 310. Several modified sites differed between birds and mammals, such as the phosphoserine sites at positions 148, 153, 175, 176, 208 and 267. The modified sites could be grouped into four regions, identified as regions 1, 2 and 3 (D1, D2, and D3, respectively) and a region of variants 1 (V1) (Figure 3). The D1-D3 regions had the same characteristics as avian (duck and chick) MEF2A, which differed from those of mammals. The V1 region varied in all of the species examined. The secondary structures of the D1-D3 and V1 regions were predicted using the protean program in DANstar, and the results showed that the secondary structure distributions in the D1-D3 regions differed between birds and mammals (Figure 3).
Expression profiles of duck MEF2A in muscles during fetal development
Figures 4-6 show the expression profiles of duck MEF2A. MEF2A was expressed in skeletal, smooth and cardiac muscle at each developmental stage examined. The expression level was higher in smooth muscle than in the other two muscles. In cardiac muscle, MEF2A showed a significant change (increase) only at embryo stage 27d, when the eggs were close to hatching. In skeletal muscle, MEF2A expression increased from embryo stage 10d to 18d, after which it decreased from embryo stage 22d onwards; at postnatal 7d the expression was lower than in any of the preceding periods. In smooth muscle, there was a progressive increase in MEF2A expression up to 22d, followed by a slight decline in later periods.
Discussion
Identification and functional prediction of duck MEF2A
MEF2A is one of the MEF2 gene family members and in birds has so far been cloned only from chicken (Buchberger and Arnold, 1999). Duck MEF2A is the second member of avian origin to be cloned. It shared > 90% identity with the chicken protein at the nucleotide and amino acid levels. An in silico analysis showed that duck MEF2A contained a MADS domain in its N-terminal, followed by a MEF2 domain. This structural organization was similar to the corresponding chicken and mammalian proteins (Buchberger and Arnold, 1999; Huang et al. (, 2000; Wu et al. (, 2010). The MADS domain is reportedly involved in specific DNA binding and dimerization while the MEF2 domain increases DNA binding and mediates the interaction of MEF2 proteins with other cofactors such as GATA, ERK5 and HDAC. The duck MEF2A gene also had an evolutionarily conserved regulatory region for interaction with MAPKs. The three conserved domains (MADS, MEF2 and MAPK) show few variants among different organisms, which suggests that duck MEF2A plays a similar role to the corresponding protein in other species.
Developmental expression of MEF2A in duck muscle during the muscle development stage
Previous studies of MEF2A have focused on its functions during embryogenesis and in adulthood. Our results show that duck MEF2A was expressed in cardiac, smooth and skeletal muscle during fetal development. In skeletal muscle, MEF2A expression increased to reach a peak at embryonic day 18, followed by a subsequent decrease. Buchberger and Arnold (1999) reported that in chickens MEF2A expression was not detected in newly formed so-mites until the muscle-specific transcription factors MyoD and myogenin were present, indicating that activation of the MEF2A gene in skeletal muscle was dependent on these basic helix-loop-helix transcription factors. We have examined the developmental expression of duck MyoG and MRF4 in skeletal muscle during the fetal stage and found that MyoG and MRF4 are highly expressed in the leg muscles at embryonic days 14d and 18d, respectively (Liu et al. (, 2010), which is before the peak expression of MEF2A. Our results are consistent with those of Buchberger and Arnold (1999) and suggest that activation of the MEF2A gene in skeletal muscle is dependent on these basic helix-loophelix transcription factors.
Myocardial muscle develops mainly during early embryogenesis and MEF2A is highly expressed in zebrafish heart during embryogenesis (Mathavan et al. (, 2005; Wang Q et al. (, 2005). In chickens, MEF2A expression is first observed in the precardiac mesoderm of HH stage 8 embryos, and expression in heart and skeletal muscle continues into adulthood, when it is also observed in the intestinal mesenchyme and brain. During later embryonic development, MEF2A expression continues in the heart tube and later in the atrium and ventricle (Buchberger and Arnold, 1999; Czubryt and Olson, 2004). Our results support the above findings since MEF2A was found to be expressed at a steady level up to 22d, with a peak at 27d. This change in expression as the egg approaches hatching may enable the duckling to adapt to a new environment when the heart will mature further.
MEF2A is thought to be important for the activation and expression of contractile proteins and other muscle-specific factors in skeletal and cardiac muscle (Czubryt and Olson, 2004). Firulli et al. ( (1996) showed that MEF2A is highly expressed in smooth cells producing contractile proteins. Duck MEF2A was highly expressed in smooth muscle at almost all of the fetal stages, with greater expression than in skeletal and cardiac muscle. The elevated expression of duck MEF2A in smooth muscle during these stages suggests that this protein has an important role in smooth muscle development.
Relationship between MEF2A expression and protein structure
MEF2 binds directly to the promoters or enhancers of the majority of muscle-specific genes (Naya and Olson, 1999), and virtually all muscle genes, including MRFs, have MEF2 binding sites in their regulatory regions (Messina et al. (, 2010). The MADS box and adjacent MEF2 domain of MEF2 proteins play a cooperative role in binding to the regulatory regions of muscle genes (Potthoff and Olson, 2007; Ramachandran et al. (, 2008) to activate skeletal muscle differentiation. However, unlike MRFs, MEF2 factors cannot activate myogenesis alone but, instead, combine with MRFs through protein-protein interactions in the MADS box and bHLH domain to vastly improve the efficiency of the myogenic program (Kaushal et al. (, 1994). The MEF2A domains in poultry are homologous to those of mammals. Duck MEF2A lacks a beta domain region that is present in chickens and other species; this domain may be alternatively spliced in humans (Yu et al. (, 1992). The RNA transcripts of all MEF2 members undergo alternative splicing, with some of these being limited to specific cell types (Yu, 1996). The significance of these alternative splicing events is not yet understood.
The analysis of human tissues has shown that MEF2A is expressed only in skeletal and cardiac muscle and in the brain, while two isoforms of MEF2A that lack a beta domain region are expressed in all tissues examined (Pollock and Treisman, 1991). The duck MEF2A sequence cloned here was expressed in all three types of muscle, a finding similar to that for the two MEF2A isoforms in humans. We suggest that the duck MEF2A sequence cloned here may be only one of several alternatively spliced isoforms, and that the tissue expression of duck MEF2A may differ from that of other organisms. These possibilities warrant further study.
The modified sites in MEF2A are important for regulating gene expression and function. The phosphorylation status of MEF2A is a critical determinant of its tissue-specific functions and can be directly regulated by tissue-specific pathways acting on selected amino acid residues (Wang et al. (, 2004). Pathways such as p38 MAPK signaling have an important role in regulating cardiac and smooth muscle function (Dentel et al. (, 2005). As shown here, there are several important differences in the amino acid sequences of avian and mammalian MEF2A. Although the influence of amino acid alterations on MEF2A function, especially in the D1-D3 and V1 regions, was not examined here, these novel modified sites may be involved in the differential regulation of gene expression and function in avian and mammalian muscle.
In conclusion, duck MEF2A contains MADS, MEF2 and MAPK domains that share high similarity with related domains in other species. These conserved domains are important for maintaining protein functions. The expression of MEF2A in duck cardiac, smooth and skeletal muscle suggests an important role for this protein in muscle development.
Acknowledgments
The work was supported by the National High Technology Research Development Program of China (No. 2010AA10A109) and China Agriculture Research Service (No. CARS-43-6).
Associate Editor: Ricardo Guelerman P. Ramos
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Angelelli C, Magli A, Ferrari D, Ganassi M, Matafora V, Parise F, Razzini G, Bachi A, Ferrari S and Molinari S (2008) Differ-entiation-dependent lysine 4 acetylation enhances MEF2C binding to DNA in skeletal muscle cells. Nucleic Acids Res 36:915-928.
- Barsyte-Lovejoy D, Galanis A, Clancy A and Sharrocks AD (2004) ERK5 is targeted to myocyte enhancer factor 2A (MEF2A) through a MAPK docking motif. Biochem J 381:693-699.
- Black BL and Olson EN (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14:167-196.
- Buchberger A and Arnold HH (1999) The MADS domain containing transcription factor cMef2a is expressed in heart and skeletal muscle during embryonic chick development. Dev Genes Evol 209:376-381.
- Byers TJ, Kunkel LM and Watkins SC (1991) The subcellular distribution of dystrophin in mouse skeletal, cardiac, and smooth muscle. J Cell Biol 115:411-421.
- Chauvigne FCCR (2005) Muscle fiber differentiation in fish embryos as shown by in situ hybridization of a large repertoire of muscle-specific transcripts. Dev Dyn 233:659-666.
- Chen F, Chen H, Wang JQ, Niu H, Lan XY, Hua LH, Li ZJ, Lei CZ and Fang XT (2010) MEF2A gene polymorphisms are associated with growth traits in Chinese indigenous cattle breeds. J Anim Vet Adv 9:814-819.
- Cox DM, Du M, Marback M, Yang EC, Chan J, Siu KW and McDermott JC (2003) Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J Biol Chem 278:15297-15303.
- Czubryt MP and Olson EN (2004) Balancing contractility and energy production: The role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog Horm Res 59:105-124.
- Dentel JN, Blanchard SG, Ankrapp DP, McCabe LR and Wise-man RW (2005) Inhibition of cross-bridge formation has no effect on contraction-associated phosphorylation of p38 MAPK in mouse skeletal muscle. Am J Physiol Cell Physiol 288:C824-C830.
- Edmondson DG, Lyons GE, Martin JF and Olson EN (1994) Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120:1251-1263.
- Firulli AB, Miano JM, Bi W, Johnson AD, Casscells W, Olson EN and Schwarz JJ (1996) Myocyte enhancer binding factor-2 expression and activity in vascular smooth muscle cells. Association with the activated phenotype. Circ Res 78:196-204.
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C and Greenberg ME (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311:10081012.
- Forouhar AS, Liebling M, Hickerson A, Nasiraei-Moghaddam A, Tsai HJ, Hove JR, Fraser SE, Dickinson ME and Gharib M (2006) The embryonic vertebrate heart tube is a dynamic suction pump. Science 312:751-753.
- Friedman A, Bar-shira E and Sklan D (2003) Ontogeny of gut associated immune competence in the chick. World Poultry Sci J 59:209-219.
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J and Mao Z (2003) Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38:33-46.
- Han J and Molkentin JD (2000) Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc Med 10:19-22.
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z and Spiegelman BM (2007) Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1a muscle-specific knock-out animals. J Biol Chem 282:30014-30021.
- Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD and Clore GM (2000) Solution structure of the MEF2A-DNA complex: Structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J 19:2615-2628.
- Kato Y, Zhao M, Morikawa A, Sugiyama T, Chakravortty D, Koide N, Yoshida T, Tapping RI, Yang Y and Yokochi T (2000) Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J Biol Chem 275:18534-18540.
- Kaushal S, Schneider JW, Nadal-Ginard B and Mahdavi V (1994) Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science 266:12361240.
- Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ and Adams JL (2000) Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacology 47:185-201.
- Leong XF, Aishah A, Nor AU, Das S and Jaarin K (2008) Heated palm oil causes rise in blood pressure and cardiac changes in heart muscle in experimental rats. Arch Med Res 39:567-572.
- Liu HH, Wang JW, Han CC, Jia J, Si JM, Huang KL, Li L and Xu F (2010) Molecular cloning of the duck myog and mrf4 genes cds and their differential expression patterns in the breast and leg muscle during fetal development. Can J Anim Sci 90:179-188.
- MacDonald JA (2008) Smooth muscle phenotypic plasticity in mechanical obstruction of the small intestine. Neurogastroenterol Motil 20:737-740.
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH and Yang H (2005) Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1:260-276.
- Messina G, Biressi S, Monteverde S, Magli A, Cassano M, Perani L, Roncaglia E, Tagliafico E, Starnes L and Campbell CE (2010) Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell 140:554-566.
- Molkentin JD, Black BL, Martin JF and Olson EN (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136.
- Naya FJ and Olson E (1999) MEF2: A transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol 11:683-688.
- Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA and Olson EN (2002) Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8:1303-1309.
- Noorlander CW, Ververs FF, Nikkels PG, van Echteld CJ, Visser GH and Smidt MP (2008) Modulation of serotonin transporter function during fetal development causes dilated heart cardiomyopathy and lifelong behavioral abnormalities. PLoS One 3:e2782.
- Olson EN (2006) Gene regulatory networks in the evolution and development of the heart. Science 313:1922-1927.
- Owens GK, Kumar MS and Wamhoff BR (2004) Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84:767-801.
- Pollock R and Treisman R (1991) Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev 5:2327-2341.
- Potthoff MJ and Olson EN (2007) MEF2: A central regulator of diverse developmental programs. Development 134:41314140.
- Ramachandran B, Yu G, Li S, Zhu B and Gulick T (2008) Myocyte enhancer factor 2A is transcriptionally autoregulated. J Biol Chem 283:10318-10329.
- Rehfeldt C (2005) Prenatal events that determine the number of muscle fibres are important for lean growth and meat quality in pigs. Arch Tierz 48(Special Issue):11-22.
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW and Bonni A (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311:1012-1017.
- Wang Q, Sigmund CD and Lin JJ (2000) Identification of cis elements in the cardiac troponin T gene conferring specific expression in cardiac muscle of transgenic mice. Circ Res 86:478-484.
- Wang JQ, Tang Q, Parelkar NK, Liu Z, Samdani S, Choe ES, Yang L and Mao L (2004) Glutamate signaling to Ras-MAPK in striatal neurons: Mechanisms for inducible gene expression and plasticity. Mol Neurobiol 29:1-14.
- Wang Q, Rao S and Topol EJ (2005) Miscues on the "lack of MEF2A mutations" in coronary artery disease. J Clin Invest 115:1399-1401.
- Wang YX, Qian LX, Yu Z, Jiang Q, Dong YX, Liu XF, Yang XY, Zhong TP and Song HY (2005) Requirements of myocytespecific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett 579:4843-4850.
- Wu Y, Dey R, Han A, Jayathilaka N, Philips M, Ye J and Chen L (2010) Structure of the MADS-box/MEF2 domain of MEF2A bound to DNA and its implication for myocardin recruitment. J Mol Biol 397:520-533.
- Xue ZG, Xue JX, Roncier B, Chamagne AM and Portier MM (2000) Isolation of quail qMEF2D gene and its expression pattern in the developing central nervous system. Biochim Biophys Acta 1492:543-547.
- Yu YT (1996) Distinct domains of myocyte enhancer binding factor-2A determining nuclear localization and cell type-specific transcriptional activity. J Biol Chem 271:24675-24683.
- Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V and Nadal-Ginard B (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev 6:1783-1798.
- Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ and Han J (1999) Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol 19:21-30.
- Zhou Y, Liu Y, Jiang X, Du H, Li X and Zhu Q (2010) Polymorphism of chicken myocyte-specific enhancer-binding factor 2A gene and its association with chicken carcass traits. Mol Biol Rep 37:587-594.
- Internet Resources CD-search tool available from NCBI, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
Publication Dates
-
Publication in this collection
16 Feb 2012 -
Date of issue
2012
History
-
Received
02 May 2011 -
Accepted
23 Nov 2011