Abstract
We used simple sequence repeat (SSR) markers to investigate the genetic diversity of 78 sweet potato (Ipomoea batatas) accessions (58 landraces and 20 putative clones) from traditional agricultural households from 19 local communities in the Vale do Ribeira, São Paulo, Brazil. Eight SSR loci were assessed using 6% (w/v) polyacrylamide gels stained with silver nitrate and the accessions genotyped considering the presence or absence of bands. The results were subjected to analysis of molecular variance (AMOVA), and cluster and principal coordinate analyses. Spatial structure was assessed using Mantel's test to compare genetic and geographic distances. Each primer pair generated between three and ten clearly scorable polymorphic fragments. Cluster analyses showed a Jaccard's index from 0.3 to 1.0, indicating high genetic and intravarietal diversity. Accessions from all 19 communities were not spatially structured (r = 0.15, p < 0.054), with AMOVA indicating that most of the variability (58.2%) was distributed within households and only 18.1% of the variability was distributed between households within communities. The outcrossing mating system of sweet potato, and anthropic factors such as selection of different varieties and their maintenance within household small plots and home gardens, as well as an extensive exchange system between agriculturists, may all be contributing to these results.
genetic variability; landraces; SSR; traditional agriculture; Vale do Ribeira
PLANT GENETICS
RESEARCH ARTICLE
Genetic diversity in Brazilian sweet potato (Ipomoea batatas (L.) Lam., Solanales, Convolvulaceae) landraces assessed with microsatellite markers
Elizabeth Ann VeaseyI; Aline BorgesI; Mariana Silva RosaI; Jurema R. Queiroz-SilvaI; Eduardo de Andrade BressanI; Nivaldo PeroniII
IDepartamento de Genética, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Piracicaba, SP, Brazil
IIInstituto de Biologia, Museu de História Natural, Universidade Estadual de Campinas, Campinas, SP, Brazil
Send correspondence to Send correspondence to: EA Veasey Departamento de Genética, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo Caixa Postal 83 13400-970 Piracicaba, SP, Brazil E-mail: eaveasey@esalq.usp.br
ABSTRACT
We used simple sequence repeat (SSR) markers to investigate the genetic diversity of 78 sweet potato (Ipomoea batatas) accessions (58 landraces and 20 putative clones) from traditional agricultural households from 19 local communities in the Vale do Ribeira, São Paulo, Brazil. Eight SSR loci were assessed using 6% (w/v) polyacrylamide gels stained with silver nitrate and the accessions genotyped considering the presence or absence of bands. The results were subjected to analysis of molecular variance (AMOVA), and cluster and principal coordinate analyses. Spatial structure was assessed using Mantel's test to compare genetic and geographic distances. Each primer pair generated between three and ten clearly scorable polymorphic fragments. Cluster analyses showed a Jaccard's index from 0.3 to 1.0, indicating high genetic and intravarietal diversity. Accessions from all 19 communities were not spatially structured (r = 0.15, p < 0.054), with AMOVA indicating that most of the variability (58.2%) was distributed within households and only 18.1% of the variability was distributed between households within communities. The outcrossing mating system of sweet potato, and anthropic factors such as selection of different varieties and their maintenance within household small plots and home gardens, as well as an extensive exchange system between agriculturists, may all be contributing to these results.
Key words: genetic variability, landraces, SSR, traditional agriculture, Vale do Ribeira.
Introduction
The genus Ipomoea contains 600 to 700 species, most of which are concentrated in the Americas (Austin, 1988). Sweet potato (Ipomoea batatas (L.) Lam. Solanales, Convolvulaceae) was originally domesticated in the New World (Austin, 1988; Zhang et al., 2004) and is now grown between latitudes 42° N and 35° S, from sea level up to an altitude of 3000 m. Over 95 percent of the global sweet potato crop is produced in developing countries, where it is the fifth most important food crop on a fresh weight basis after rice, wheat, maize and cassava. Asia is the world's largest sweet potato producing region, with China alone producing 90% of the total world production (CIP, 2008).
In Brazil, sweet potato is one of the most important food crops and is cultivated in practically every state, the northeastern states of Pernambuco and Paraíba and the southern states of Paraná, Santa Catarina and Rio Grande do Sul being the largest producers (IBGE, 1999). Sweet potato is also one of the principal crops cultivated by traditional farmers in Brazilian home gardens and slash and burn clearings (swiddens). A common characteristic of such cultivation systems is the cultivation of a heterogeneous group of species (Martins, 2001), permitting the coexistence of high inter and intraspecific diversity. Traditional farmers have a fundamental role in the conservation and generation of diversity in cultivated species and have been studied in many parts of the world for more than a century (Martins, 1994; Brush, 2000). A series of factors considered important in choosing the species in traditional agriculture are cultural factors and the abundance, availability and ease of acquisition of the species (Cleveland et al., 1994). Quality in terms of flavor, nutritional value and durability is often the determining factor (Nazarea, 1998), but the adaptation capacity of a species or landrace to climatic, geographic and cultural variants is also crucial, since traditional farmers are interested in the diversity and population structure that enables them to maximize local adaptation (Soleri and Smith, 1995).
Although sweet potato has an outcrossing mating system, it is propagated vegetatively with each cultivar considered a clone (Prakash et al., 1996). Self-incompatibility in the flowers (Martin, 1965) results in allogamy, increasing genetic heterozygosity (Thompson et al., 1997). Sexual compatibility is related to a multiallelic sporophytic self-incompatibility system expressed in the stigma (Diaz et al., 1996).
Genetic markers have shown important and critical application in the assessment and conservation of genetic variation. Amplified fragment length polymorphism (AFLP) has been used for studying the historic dispersal of sweet potato (Zhang et al., 2004) as well as for assessing the genetic diversity of cultivars and landraces (Zhang et al., 2000; Fajardo et al., 2002). Huang and Sun (2000) and Hu et al. (2003) used inter-simple sequence repeat (ISSR) and restriction analysis of chloroplast DNA to investigate the genetic relationships between cultivated sweet potato and its wild relatives. Microsatellite or simple sequence repeat (SSR) markers exhibit high levels of polymorphism, and several such markers have been developed for sweet potato (Jarret and Bowen, 1994; Buteler et al., 1999; Hu et al., 2004) and used successfully for determining the genetic relationship between cultivars derived from hybrid or polycross breeding programs (Hwang et al., 2002) and for analyzing the genetic diversity of Latin American and East African sweet potato landraces (Zhang et al., 2001; Gichuru et al., 2006).
In the study described in this paper, microsatellite markers were used to assess the genetic diversity of sweet potato landraces from the Vale do Ribeira in the Brazilian state of Sao Paulo. This is the first report assessing sweet potato accessions with microsatellite markers in Brazil. The questions addressed in this study were: a) is there high genetic diversity among sweet potato landraces grown under the traditional agriculture system of the Vale do Ribeira?; b) What is the genetic structure of these landraces regarding distribution of the genetic variability within and between households and communities of Vale do Ribeira?; and c) Are these landraces genetically structured in space? This data is important for our knowledge of the genetic variability status of these landraces in the region and to assist future ex situ, in situ and on farm sweet potato conservation programs.
Materials and Methods
Plant materials
We evaluated 78 accessions of sweet potato (I. batatas), consisting of 58 landraces and 20 putative clones (Table 1), collected from 32 households distributed in 19 local communities practicing traditional agriculture in Atlantic Forest areas in the Vale do Ribeira (Figure 1) in the municipalities of Iguape (24°42' S, 47°33' W), Cananéia (25°00' S, 47°55' W), Ilha Comprida (24°53' S, 47°47' W) and Eldorado (24°31' S, 48°06' W) (Bressan et al., 2005). In Table 1, accessions with a letter (e.g., 3.1 and 3.2) represent replicate material from different vines or tubers of the same accession and can be considered as putative clones. The accessions are held as part of an ex situ collection in the Genetics Department of ESALQ, São Paulo University, in Piracicaba, São Paulo, Brazil.
The collection strategy consisted of visiting the households at random in the principal communities of the municipalities and applying semi-structured interviews based on questionnaires about the agricultural system and the crops currently grown. These interviews focused on the use, origin and location of the crop plantings and whether swidden, plots or home gardens were used (Bressan et al., 2005). Each household provided one or more tubers or vines from each sweet potato variety and, on average, 1.8 sweet potato landraces were collected per household, distributed as follows: 16 households provided one landrace each, 11 provided two landraces each, three provided three landraces each, one (from Pontal de Icapara) provided four landraces and one (from Agrossolar) seven landraces. More than one vine or tuber was collected from landraces in the Iguape municipality and were considered replicates or clones (called clones in the text) for intravariety variability assessment. The accessions were initially multiplied in pots in a greenhouse and then planted in plots in a field at the Genetics Department, ESALQ, São Paulo University, in Piracicaba, Sao Paulo (22°42' S, 47°38' W) with 1.5 m spacing between plants and 1.5 m between rows to maintain an ex situ collection.
Isolation of DNA and polymerase chain reaction (PCR) analysis
We isolated total DNA from young sweet potato leaves using liquid nitrogen and the cetyltrimethylammonium bromide (CTAB) method described by Doyle and Doyle (1987) with some modifications. The raw DNA was incubated at 65 °C for 45 min in 0.7 mL of 2% CTAB buffer (containing 20 mM EDTA, 0.1 M tris-HCL pH 8.0, 1.4 M NaCl and 1% polyvinylpyrrolidone (PVP) supplemented with 0.4% 2-mercaptoethanol added just before use) and 0.5 mL of 24:1 (v/v) chloroform-isoamyl alcohol was added and the samples centrifuged for 10 min at 12,000 revs min-1. After centrifugation about 0.6 mL of the supernatant was transferred to a fresh tube with 0.5 mL of 24:1 (v/v) chloroform-isoamyl alcohol, this procedure being repeated twice. After the final centrifugation, DNA was precipitated by adding isopropanol, and the crude DNA rinsed with 70% (w/v) ethanol and then dissolved in 0.1 mL of TE buffer (10 mM Tris-HCL pH 8.0, 1 mM EDTA pH 8.0) containing 5 µL of a 10 mg mL-1 RNAse solution (Sigma, Italy) and incubated at 37 °C for 1 h, after which the DNA samples were stored at -20 °C until use.
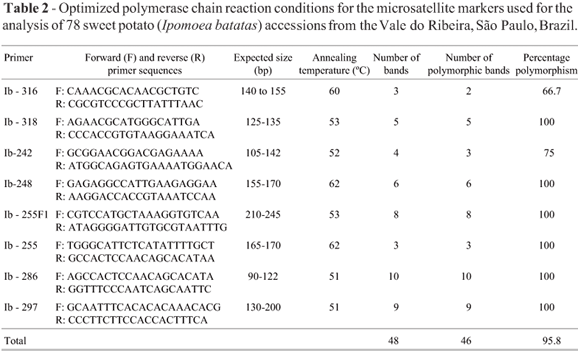
For PCR analysis we screened eleven SSR microsatellite primer pairs designed for sweet potato, three (Ib2-38, Ib2-45A and Ib2-66) described by Jarret and Bowen (1994) and eight by Buteler et al. (1999). However, only the eight from Buteler et al. (1999) resulted in high resolution banding patterns and were used in our study (Table 2). The reaction conditions were optimized to improve the banding patterns for each primer. The total reaction volume for amplification was 10.2 µL, consisting of 0.2 µL Taq polymerase (50 units/µL), 1.0 µL 10X Amplification Buffer (Mg+ Free), 1.0 µL MgCl2 (50 mM), 0.5 µL Forward Primer (5 pmoles/µL), 0.5 µL Reverse Primer (5 pmoles/µL), 1.0 µL dNTP's (2.5 mM each), 3 µL de Milli-Q H2O and 3 µL DNA (5 ng/µL). The PCR amplifications were carried out in a Gradient MyCycler thermal cycler (BioRad) under the following conditions: 3 min at 95 °C followed by 5 cycles of 45 s at 94 °C, 15 s for the annealing temperature (Table 2) and 45 s at 72 °C then 20 cycles of 1 min at 90 °C, 1 min for the annealing temperature and 1 min at 72 °C, with a final 7 min extension at 72 °C (Buteler et al., 1999). The products were separated by electrophoreses on a 6% (w/v) polyacrylamide gel using a running buffer composed of 1X TBE buffer [100 mL 10X TBE (0.89 M Tris base, 0.89 M Boric acid, 20 mM EDTA pH 8.0) and 900 mL distilled water] and an initial voltage of 60 V for 30 min and 120 V for 1.5 h. Amplified fragments were visualized using a silver staining procedure (Bassam et al., 1991).
Data analysis
Sweet potato is a hexaploid species (Ozias-Akins and Jarret, 1994) and, therefore, produces many different band patterns. In our case, we scored the banding patterns phenotypically for the presence or absence of bands as if each marker were dominant. The binary data was subjected to analysis of molecular variance (AMOVA) for dominant markers using the Arlequin software (Schneider et al., 2000). Cluster analysis using Jaccard similarity coefficients and the unweighted pair group method with arithmetic mean (UPGMA) clustering criteria and principal coordinate analysis (PCO) were conducted using MVSP (Kovach, 1999), NTSYSpc (Rohlf, 1992) and CANOCO (ter Braak and Smilauer, 2002) programs. To confirm the group structure found in the PCO scatter graph, a second AMOVA analysis was conducted. To verify if the landraces were spatially structured we used the NTSYS-pc program to calculate the Pearson correlation coefficient (r) between the average Jaccard dissimilarity coefficient matrix for the 19 communities and the geographic distances matrix between communities, the latter indicating the shortest distance between two given points on the map. The significance of these correlations was tested using the Mantel Z statistic (Mantel, 1967) and 1000 random permutations.
Results
Levels of SSR polymorphism and molecular variance analysis (AMOVA)
From a total of 11 SSR primer pairs designed for sweet potato and tested for microsatellite polymorphisms in this study, the three pairs Ib2-38, Ib2-45A and Ib2-66 (Jarret and Bowen, 1994) provided weak resolution and were not used in our study but the eight pairs described by Buteler et al. (1999) provided banding patterns with good resolution (Table 2). In the 78 sweet potato accessions tested with the eight loci evaluated we scored 46 polymorphic bands (95.8%) and a total of 48 bands, with an average of 6 bands per primer and a range from 3 to 10 bands per primer (Table 2). The high level of polymorphism indicated high genetic diversity. Primer Ib-286 produced the highest number of bands (10) while primers Ib-316 and Ib-255 showed the lowest number of band (3 each).
The AMOVA results indicated that the largest contribution to genetic variability was due to differences between varieties within households (58.2%, p < 0.0001) (Table 3). The difference between households within communities was also significant (p < 0.0001) but contributed to only 18.1% of the total variance, followed by the difference between communities (13.6%, p < 0.05).
Genetic relationship among sweet potato accessions
The 78 accessions, including 58 landraces and 20 possible clones, investigated using cluster analysis resulted in 70 genotypes that did not form clear groups according to the geographic proximity of the communities or municipalities. The landraces from Iguape were clustered with the landraces from Cananéia and Ilha Comprida. However, the landraces from the Iguape communities of Icapara, Pontal de Icapara and Praia do Leste did form a large cluster at the upper side of the dendrogram containing the accessions DGB 1.0 to DGB 9.0 plus the DGB 65 and DGB 70 landraces from Sapatu in Eldorado municipality, these latter two landraces being grouped as outsiders (Figure 2). This group was also clustered on the right side of the first principal coordinate in the scatter graph (Figure 3), while all the other accessions were scattered on the left side of this coordinate which explained 14.5% of total variation. In addition, there was a particular cluster that grouped landraces and putative clones DGB 15, 16, 17, 18, 19, 20, 21.1, 21.2, 22, 23.1, 23.2 and 24, from the Iguape communities Momuna, Cavalcanti and Peropava (Figure 2). In the principal coordinate scatter graph, landraces and clones DGB 21.1, 21.2, 22, 23.1, 23.2 and 24 were clustered in the upper quadrant of the second principal coordinate (Figure 3). Thus, three main groups were found in the principal coordinate scatter graph, one from the Iguape communities Icapara, Pontal de Icapara and Praia do Leste on the right side of the first principal coordinate, a second group from the communities Momuna (three accessions), Peropava and Cavalcante, and a third group with all the other accessions from Iguape, Cananéia, Ilha Comprida and Eldorado in the center. This group structure was confirmed in the AMOVA analysis presented in Table 4, where 21.85% (p < 0.0001) of total variation was due to the between group variability component.
As mentioned above, the high genetic variability was confirmed by examining the wide range of the Jaccard similarity coefficient (0.31 to 1.0), indicating a variation of up to 69% among the Vale do Ribeira accessions (Figure 2). Some of the extra vines were considered clones of a particular landrace because they presented the same banding pattern and appeared as duplicates in the dendrogram, examples being DGB 6.2 and 6.3, DGB 5.3, 5.4, 5.6, 5.7 and 5.8, DGB 9.2 and 9.5, and DGB 14.1 and 14.2. However, some different accessions collected from separate households were genetically identical, an example being clones DGB 6.1 and 7.1 and DGB 7.3 and 8.0, all collected at the Pontal de Icapara community (Table 1). However, several potential clones from the same sampled unit identified by the agriculturists by the same folk name formed different banding patterns, indicating the existence of high intraspecific genetic variability.
When comparing the average Jaccard dissimilarity coefficients for the 19 communities with the geographic distances between communities, there was a small but positive and non-significant correlation (r = 0.147; p = 0.054), indicating that the landraces were not spatially structured. This result is in accordance with the general patterns observed in the dendrogram and the scatter graph, where undefined groups were formed according to the origin of the landraces. However, on examining the AMOVA analysis in Table 4, a small space structuring was observed between specific Vale do Ribeira accessions (landraces and clones), independent of their origin.
Discussion
Genetic diversity
A high level of polymorphism was observed in this study, with an average of six polymorphic bands per SSR primer. Hwang et al. (2002) used eight SSR primers to analyze 22 sweet potato landraces as well as cultivars derived from hybrids or polycross breeding in Taiwan and reported 17 polymorphic bands, corresponding to 85% of total polymorphism.
The high variation (69%) observed among the accessions (landraces and putative clones) in the dendrogram (Figure 2) shows the capacity of the traditional farmers of the Vale do Ribeira to maintain high levels of sweet potato genetic variation in their households and communities. Veasey et al. (2007) also reported high variability for morphological traits in this same set of Vale do Ribeira landraces, with a high range for the Jaccard similarity index (0.12 to 1.0). The same farmers are known to maintain a high diversity of species, as well as several varieties within a particular species, as reported by Peroni and Hanazaki (2002) for crops such as cassava (Manihot esculenta, 62 varieties), yams (Dioscorea spp., 29 varieties) and sweet potato (29 varieties) as well as for rice (Oryza sativa, 26 varieties), bananas (Musa paradisiaca, 24 varieties), sugarcane (Saccharum officinarum, 22 varieties), beans (Phaseolus vulgaris, 16 varieties) and other crops with five or less varieties.
The existence of intravarietal polymorphism was also evidenced when analyzing the vines collected as clones of a particular landrace, some showing different banding patterns. The assumption that these vines collected from plants adjacent to a particular landrace would be clones is due to the vegetative propagation of this crop. Sweet potato is asexually propagated via stem cuttings and adventitious buds arising from storage roots, which may result in the accumulation of random mutations. Therefore, this intravariety variability may be related to the high somatic mutation reported in this crop (Hernandez et al., 1964). However, it may also be due to the identification of each landrace by the local farmers or by the degree of perception the farmer has regarding the morphological variation that constitute a landrace as an identification unit (Veasey et al., 2007). This variation can be identified by the farmer and depends on their local knowledge and experience with the plants. Identification by the local farmers of vegetatively propagated crop species could be considered a special human selection model, known as ''selection for perceptual distinctiveness'', where the maintenance of a variety depends on morphological variations that can distinguish it from others (Boster, 1985). Intravarietal polymorphism related to vegetative crop species has also been reported for potato (Solanum tuberosum) by Quiroz et al. (1990) and cassava (M. esculenta) by Colombo et al. (2000) and Sambatti et al. (2001).
Genetic structure and analysis of molecular variance
The AMOVA results indicated that the high diversity discussed above was mostly distributed within the evolutionary units (households) for sweet potato accessions (landraces and putative clones) from the Vale do Ribeira. A similar result (64.4% variation within households) was obtained by Veasey et al. (2007) when studying morphological traits of the same accessions. This same pattern of a greater portion of variation being due to differences between accessions within households has also been reported for cassava (Sambatti et al., 2000), as well as within groups or regions (Faraldo et al., 2000; Cabral et al., 2002). Other studies with sweet potatoes using AMOVA have also shown higher diversity within rather than between regions, which could, on a much smaller scale, be compared to the higher diversity found within rather than between households and communities. Zhang et al. (2000) used AFLP to study the genetic diversity of 69 sweet potato landraces from tropical America collected in 13 countries, reporting a high variation (90%) within regions and a statistically significant variation of 10% between regions (Central America, Colombia-Venezuela, Caribbean, Peru-Ecuador). Furthermore, Zhang et al. (2004) found a highly significant variation of 86.7% within regions and 12.4% among regions when studying 75 sweet potato landraces from Latin America and Pacific areas with AFLP markers.
One of the factors leading to the high degree of variation between the accessions within households found in this study may be related to the mating system of sweet potato, a cross pollinating and hexaploid species (Ozias-Akins and Jarret, 1994). New plants or new genotypes, arising from seeds resulting from cross pollinations, will originate as part of this genetic variation. A difficulty with this hypothesis is that the farmers affirmed they have noticed flowers of sweet potato, but not seeds or fruits (Veasey et al., 2007). However, we must consider that the small seeds of sweet potato are not the main concern of the farmers, who are more interested in the roots. Also, as many of the households visited had only one landrace, this could imply the absence of seed production due to the auto-incompatibility of each variety. Therefore, the mating system could not be the main factor for the maintenance of the genetic variability. A high incidence of random mutations in sweet potato, as discussed above, and anthropic factors such as the selection of different types and the maintenance of these within the households are also factors that may be contributing for this higher variability within the households. The exchange of plants among the farmers and the chances of new genotypes emerging from seed banks arising from other plantings in the past, were probably the most important factors in the maintenance and amplification of the genetic diversity for this crop in the Vale do Ribeira.
This intensive exchange of sweet potato landraces between farmers, especially between neighbors and relatives, may explain not only the absence of defined groups in the dendrogram and scatter graph (Figure 2 and Figure 3) from a particular community or municipality but also the lack of correlation between genetic distances as measured by the Jaccard dissimilarity index and geographic distances, indicating a non-space structuring of the variability.
This is a past colonization region that has been relatively isolated from other more urbanized regions until the 1970's, with a local population characterized by a macro region of low mobility and intraregional high mobility (Hogan et al., 1999). Due to strong indigenous influence, local populations maintain their varieties within their families and also try out new varieties. Many agriculturists do not remember the origin of the varieties, reporting them to be of local origin. However, many varieties have been brought to the place where they are currently cultivated together with the migratory process of the family units. Considering that in this region varieties of different species are carried around jointly in a migratory event, it can be expected that the age of the oldest varieties exceeds 75 years, or three generations, as occurs with cassava varieties in this region (Peroni et al., 2007).
The lack of correlation between genetic and geographic distances when studying landraces has also been reported for cassava (Colombo et al., 2000; Zacarias et al., 2004) and yams (Dioscorea trifida, D. alata, D. cayenensis and D. bulbifera) (Bressan EA, MSc. Dissertation, ESALQ, 2005). These authors have concluded that this lack of genetic structuring is due to the common practice of exchanging botanical materials between farmers, resulting in the same genotype bearing different names in different places, since farmers name their new varieties according to their own experience, and also due to recombination among genotypes. However, a weak (r = 0.157) but significant correlation (p < 0.032) between genetic and geographic distances has been found for cassava landraces from the Vale do Ribeira (Peroni, 2007) and was quite similar to the correlation reported in our study (r = 0.147), indicating that the influence of genetic distance is small but should be taken into consideration. The significant variation found between the three groups in our PCO analysis also suggests a little spatial structuring in the Vale do Ribeira sweet potato accessions studied.
In summary, our data supports the existence of high genetic variability between sweet potato landraces maintained by the traditional Vale do Ribeira farmers, with a higher proportion of this variation occurring within households, possibly due to factors such as the sweet potato mating system or somatic mutations but mainly to management and the exchange system between farmers. Furthermore, the fact that the genetic variability lacked significant spatial structure may be attributable to the practice of exchanging biological material between the farmers neighbors and relatives.
Acknowledgments
The authors wish to thank the Brazilian programs Biota/FAPESP and CNPq for the financial support and grants given for this research, as well as technicians Ronaldo Jose Rabello and Domingos de Salvio Amaral for their contribution in the field experiments and field collections. We particularly wish to thank the agriculturists of the Vale do Ribeira who contributed the plant material from their households and provided valuable information acquired from years of practical experience. We also appreciated the relevant contributions of two anonymous reviewers.
Internet Resources
Received: September 14, 2007; Accepted: March 4, 2008.
Associate Editor: Everaldo Gonçalves de Barros
- Austin DF (1988) The taxonomy, evolution and genetic diversity of sweetpotatoes and related wild species. In: Gregory P (ed) Exploration, Maintenance and Utilization of Sweet Potato Genetic Resources. International Potato Center, Lima, pp 27-60.
- Bassam BJ, Caetano-Anolles G and Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80-83.
- Boster JS (1985) Selection for perceptual distinctiveness: Evidence from Aguaruna cultivars of Manihot esculenta Econ Bot 39:310-325.
- Bressan EA, Veasey EA, Peroni N, Felipim AP and Santos KMP (2005) Collecting yam (Dioscorea spp.) and sweet potato (Ipomoea batatas) germplasm in traditional agriculture small-holdings in the Vale do Ribeira, São Paulo, Brazil. Plant Genet Resour Newsl 144:8-13.
- Brush SB (2000) The issues of in situ conservation of crop genetic resources. In: Brush SB (ed) Genes in the Field: On-Farm Conservation of Crop Diversity. 1st ed. IPGRI/IDRC/Lewis Publishers, Rome, pp 3-26.
- Buteler MI, Jarret RL and Labonte DR (1999) Sequence characterization of microsatellites in diploid and polyploid Ipomoea Theor Appl Genet 99:123-132.
- Cabral BLR, Souza JAB, Ando A, Veasey EA and Cardoso EMR (2002) Isoenzymatic variability of cassava accessions from different regions in Brazil. Sci Agric 59:521-527.
- Cleveland DA, Soleri D and Smith SE (1994) Do folk crop varieties have a role in sustainable agriculture? BioScience 44:740-751.
- Colombo C, Second G and Charrier A (2000) Diversity within American cassava germ plasm based on RAPD markers. Genet Mol Biol 23:189-199.
- Diaz J, Schmiediche P and Austin DF (1996) Polygon of crossability between eleven species of Ipomoea: Section Batatas (Convolvulaceae). Euphytica 88:189-200.
- Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 191:11-15.
- Fajardo DS, La Bonte DR and Jarret RL (2002) Identifying and selecting for genetic diversity in Papua New Guinea sweetpotato Ipomoea batatas (L.) Lam. germplasm collected as botanical seed. Genet Resour Crop Evol 49:463-470.
- Faraldo MIF, Silva RM and Martins PS (2000) Variabilidade genética de etnovariedades de mandioca em regiões geográficas do Brasil. Sci Agric 57:499-505 (Abstract in English).
- Gichuru V, Aritua V, Lubega GW, Edema R, Adipula E and Rubaihayo PR (2006) A preliminary analysis of diversity among East African sweet potato landraces using morphological and simple sequence repeats (SSR) markers. Acta Hortic 703:159-164.
- Hernandez TP, Hernandez T and Miller JC (1964) Frequency of somatic mutations in several sweet potato varieties. Proc Am Soc Hortic Sci 85:430-434.
- Hogan DJ, Carmo RL, Alves HPF and Rodrigues IA (1999) Sustentabilidade no Vale do Ribeira (SP): Conservação ambiental e melhoria das condições de vida da população. Ambient Soc 3-4:151-175 (Abstract in English).
- Hu J, Nakatani M, Lalusin AG, Kuranouchi T and Fujimura T (2003) Genetic analysis of sweet potato and wild relatives using inter-simple sequence repeats (ISSRs). Breed Sci 53:297-304.
- Hu J, Nakatani M, Mizuno K and Fujimura T (2004) Development and characterization of microsatellite markers in sweetpotato. Breed Sci 54:177-188.
- Huang JC and Sun M (2000) Genetic diversity and relationship of sweetpotato and its wild relatives in Ipomoea series batatas (Convolvulaceae) as revealed by inter-simple sequence (ISSR) and restriction analysis of chloropalst DNA. Theor Appl Genet 100:1050-1060.
- Hwang SY, Tseng YT and Lo HF (2002) Application of simple sequence repeats in determining the genetics relationships of cultivars used in sweet potato polycross breeding in Taiwan. Sci Hortic 93:215-224.
- Fundação Instituto Brasileiro de Geografia e Estatística (1999) Anuário Estatístico do Brasil. IBGE, Rio de Janeiro.
- Jarret RL and Bowen N (1994) Simple sequence repeats (SSRs) for sweetpotato germplasm characterization. Plant Genet Resour Newsl 100:9-11.
- Kovach WL (1999) MVSP: A Multivariate Statistical Package for Windows. User's Manual, v. 3.1. Kovach Computing Services, Pentraeth, 133 pp.
- Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209-220.
- Martin FW (1965) Incompatibility in the sweetpotato, a review. Econ Bot 19:406-415.
- Martins PS (1994) Biodiversity and agriculture: Patterns of domestication of Brazilian native plant species. An Acad Bras Cienc 66:219-226.
- Martins PS (2001) Dinâmica evolutiva em roças de caboclos amazônicos. In: Vieira ICG, Silva JMC, Oren DC and D'Incao MA (eds) Diversidade Biológica e Cultural na Amazônia. Museu Paraense Emílio Goeldi, Belém, pp 369-384 (Abstract in English).
- Nazarea VD (1998) Cultural Memory and Biodiversity. The University of Arizona Press, Tucson, 189 pp.
- Ozias-Akins P and Jarret RL (1994) Nuclear DNA content and ploidy levels in the genus Ipomoea J Am Soc Hortic Sci 119:110-115.
- Peroni N (2007) Manejo e domesticação de mandioca por caiçaras da Mata Atlântica e ribeirinhos da Amazônia. In: Boef WS, Thijssen MH, Ogliari JB and Sthapit BR (eds) Biodiversidade e Agricultores: Fortalecendo o manejo comunitário. v. 1. L&PM, Porto Alegre, pp 234-242.
- Peroni N and Hanazaki N (2002) Current and lost diversity of cultivated varieties, especially cassava, under swidden cultivation systems in the Brazilian Atlantic Forest. Agric Ecosyst Environ 92:171-183.
- Peroni N, Kageyama YP and Begossi A (2007) Molecular differentiation, diversity, and folk classification of "sweet"' and "bitter'' cassava (Manihot esculenta) in Caiçara and Caboclo management systems (Brazil). Genet Resour Crop Evol 54:1333-1349.
- Prakash CS, He G and Jarret RL (1996) DNA marker-based study of genetic relatedness in United States sweetpotato cultivars. J Am Soc Hortic Sci 121:1059-1062.
- Quiroz CF, Brush SB, Douches DS, Zimmerer KS and Huestis G (1990) Biochemical and folk assessment of variability of Andean cultivated potatoes. Econ Bot 44:254-266.
- Rohlf FJ (1992) NTSYS-pc: Numerical taxonomy and multivariate analysis system. v. 1.70. Stony Brook, New York.
- Sambatti JBM, Ando A and Martins PS (2000) Distribuição das diversidades isoenzimática e morfológica da mandioca na agricultura autóctone de Ubatuba. Sci Agric 57:75-80 (Abstract in English).
- Sambatti JBM, Martins PS and Ando A (2001) Folk taxonomy and evolutionary dynamics of cassava: A case study in Ubatuba-Brazil. Econ Bot 55:93-105.
- Soleri D and Smith SE (1995) Morphological and phenological comparisons of two hopi maize varieties conserved in situ and ex situ Econ Bot 49:56-77.
- ter Braak CJF and Smilauer P (2002) CANOCO Reference Manual and CanocoDraw for Windows User's Guide: Software for Canonical Community Ordination, v. 4.5. Microcomputer Power, New York, 500 pp.
- Thompson PG, Hong LL, Ukoskit K and Zhu S (1997) Genetic linkage of randomly amplified polymorphic DNA (RAPD) markers in sweetpotato. J Am Soc Hortic Sci 122:79-82.
- Veasey EA, Silva JRQ, Borges A, Bressan EA and Peroni N (2007) Phenology and morphological diversity of sweet potato (Ipomoea batatas) landraces of the Vale do Ribeira. Sci Agric 64:416-427.
- Zacarias AM, Botha AM, Labuschagne MT and Benesi IRM (2004) Characterization and genetic distance analysis of cassava (Manihot esculenta Crantz) germplasm from Mozambique using RAPD fingerprinting. Euphytica 138:49-53.
- Zhang DP, Carbajulca D, Ojeda L, Rossel G, Milla S, Herrera C and Ghislain M (2001) Microsatellite analysis of genetic diversity in sweetpotato varieties from Latin America. In: International Potato Centre Program Report 1999-2000, International Potato Centre, Lima, pp 295-301.
- Zhang DP, Cervantes J, Huamán Z, Carey E and Ghislain M (2000) Assessing genetic diversity of sweet potato (Ipomoea batatas (L.) Lam.) cultivars from tropical America using AFLP. Genet Resour Crop Evol 47:659-665.
- Zhang DP, Rossel G, Kriegner A and Hijmans R (2004) AFLP assessment of diversity in sweetpotato from Latin America and the Pacific region: Its implications on the dispersal of the crop. Genet Resour Crop Evol 51:115-120.
- CIP - Centro Internacional de la Papa (2008) Sweetpotato I. http://www.cipotato.org/sweetpotato (June 11, 2008).
- Schneider S, Roessli D and Excoffier L (2000) Arlequim: A software for population data analysis. University of Geneva, Genetic and Biometry Laboratory, Geneva. http:// anthropologie.unige.ch/arlequin (October 20, 2004).
Send correspondence to:
Publication Dates
-
Publication in this collection
18 Aug 2008 -
Date of issue
2008
History
-
Accepted
03 Apr 2008 -
Received
14 Sept 2007