Abstracts
This study was developed to evaluate selection effects on gene frequencies in a synthetic beef cattle cross (5/8 Charolais, 3/8 Zebu) named Canchim. A sample of 154 Canchim animals, representing three generation classes, was analyzed for seven molecular markers. Thirty-six Charolais cattle were also studied for comparison. A highly significant variation (P < 0.01), with a linear increase of the allele coding for valine at position 127 of the growth hormone peptide, was observed through generations. This allele was found in the Charolais sample but is not observed in Brazilian Bos indicus breeds. Four alleles were found for a microsatellite in the 5' flanking region of insulin-like growth factor I gene in Canchim. Allele sizes ranged from 231 to 225 bp. There was a significant (P < 0.05) nonlinear increase in the 225-bp allele frequency. This allele was not observed in Charolais. Six alleles were observed for microsatellite CSFM50 in the Canchim population. A significant (P < 0.05) linear reduction of the 168-bp allele was observed over the generations. The hypothesis of preferential mating for growth hormone polymorphism was supported by Wright's F-statistics. Estimated FIS value for growth hormone was 0.59 (P < 0.01). These results suggest that selection influenced three out of seven loci analyzed. The simultaneous increases of a Bos taurus-specific growth hormone allele and a Bos indicus-specific insulin-like growth factor I allele indicate that phenotypic selection favored different regions of both genomes.
Este estudo foi desenvolvido com o objetivo de avaliar as freqüências gênicas em diferentes gerações de bovinos da raça sintética Canchim (5/8 Charolês, 3/8 Zebu). Uma amostra de 154 animais, representando três classes de gerações de um rebanho das raça Canchim, foi analisada para sete marcadores moleculares. Uma amostra da raça Charolesa (N = 36) foi incluída nas análises para permitir comparações. Observou-se um aumento linear, altamente significativo (P < 0,01), na freqüência do alelo que codifica o aminoácido valina na posição 127 do hormônio de crescimento, ao longo das gerações de Canchim. Este alelo foi observado na amostra da raça Charolesa e não é encontrado em raças zebuínas nacionais. Quatro alelos foram observados para o microssatélite localizado na região 5' não transcrita do gene do fator de crescimento semelhante à insulina do tipo I (IGF-I), com tamanhos variando de 231 a 225 pb. A freqüência do alelo de 225 pb apresentou um aumento significativo (P < 0,05) não linear ao longo das gerações. Este alelo não foi observado na raça Charolesa e é predominante nas raças zebuínas. Para o microssatélite CSFM50, seis alelos foram observados na raça Canchim, tendo sido verificada uma redução linear (P < 0,05) na freqüência do alelo de 168 bp. A hipótese de acasalamento preferencial com relação ao polimorfismo do hormônio de crescimento foi reforçada pelos resultados da análise de estatística F de Wright. O valor estimado de FIS para este loco foi 0,59 (P < 0,01). O aumento simultâneo de um alelo do hormônio de crescimento característico de Bos taurus e de um alelo de IGF-I característico de Bos indicus sugere que a seleção fenotípica tenha favorecido regiões diferentes de ambos os genomas que entraram na formação dessa raça.
SELECTION FOR BREED-SPECIFIC GROWTH HORMONE AND IGF-I ALLELES IN A SYNTHETIC BEEF CATTLE CROSS, CANCHIM
L.C.A. Regitano1, J.L. Azevedo2, R. Vencovsky2, I.U. Packer3, P.F. Barbosa1, A.J.M. Rosa3, N.A. Silva3, M.A.L. Etchegaray3 and L.L. Coutinho3
1Empresa Brasileira de Pesquisa Agropecuária, Centro de Pesquisa de Pecuária do Sudeste, Caixa Postal 339, 13560-970 São Carlos, SP, Brasil. Send correspondence to L.C.A.R. Fax: +55-16-272-5754 E-mail: luciana@cppse.embrapa.br
Departamentos de 2Genética and 3Zootecnia, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Caixa Postal 9, 13400-970 Piracicaba, SP, Brasil.
ABSTRACT
This study was developed to evaluate selection effects on gene frequencies in a synthetic beef cattle cross (5/8 Charolais, 3/8 Zebu) named Canchim. A sample of 154 Canchim animals, representing three generation classes, was analyzed for seven molecular markers. Thirty-six Charolais cattle were also studied for comparison. A highly significant variation (P < 0.01), with a linear increase of the allele coding for valine at position 127 of the growth hormone peptide, was observed through generations. This allele was found in the Charolais sample but is not observed in Brazilian Bos indicus breeds. Four alleles were found for a microsatellite in the 5' flanking region of insulin-like growth factor I gene in Canchim. Allele sizes ranged from 231 to 225 bp. There was a significant (P < 0.05) nonlinear increase in the 225-bp allele frequency. This allele was not observed in Charolais. Six alleles were observed for microsatellite CSFM50 in the Canchim population. A significant (P < 0.05) linear reduction of the 168-bp allele was observed over the generations. The hypothesis of preferential mating for growth hormone polymorphism was supported by Wright's F-statistics. Estimated FIS value for growth hormone was 0.59 (P < 0.01). These results suggest that selection influenced three out of seven loci analyzed. The simultaneous increases of a Bos taurus-specific growth hormone allele and a Bos indicus-specific insulin-like growth factor I allele indicate that phenotypic selection favored different regions of both genomes.
INTRODUCTION
Molecular markers based on unique sequences are widely used for population structure analysis. The recent development of saturated microsatellite maps for the bovine genome (Bishop et al., 1994; Barendse et al., 1994, 1997) now allows population analysis with markers linked to specific genes or spread over the entire genome. Canchim is a synthetic beef cattle breed, produced by crosses between Charolais (Bos taurus) and several Bos indicus breeds (hereafter Zebu). With a genetic composition of 5/8 Charolais + 3/8 Zebu, Canchim combines growth ability associated with a Bos taurus breed and tolerance to tropical environments that is attributed to Zebu breeds. The herd used in this study was formed in 1953 and has been maintained on pastures and subjected to phenotypic selection for yearling weight and fertility. There was no introduction of outside animals, and the average inbreeding coefficient was kept low by avoiding consanguineous mating (Alencar et al., 1981).
This study was developed to evaluate change in gene frequencies over four generations of this herd. The main objective was to investigate if selection for growth in a tropical environment would favor specific regions of Charolais and Zebu genomes, based on the premise that performance is a combination of the expression of favorable genes for growth and adaptation.
MATERIAL AND METHODS
Animals
The herd used in this study was developed and maintained at the Centro de Pesquisa de Pecuária do Sudeste - Embrapa, São Carlos, SP, Brazil. Animals were maintained on pastures formed essentially of Digitaria decumbens, Panicum maximum, Melinis minutiflora and Hyparrhenia rufa, (Ness) Stapf. until 1977. Thereafter, pastures were improved by the introduction of Brachiaria decumbens, Panicum maximum c.v. "Makueni" and "Green panic" and Pennisetum purpureum.
Male selection was based on three criteria: morphological parameters according to the breed standards, andrological evaluation and yearling weight. Annual selection intensity was close to 10%. From 1953 to 1976 the only selection applied to females was to cull old or ill cows. After 1976, female selection was based on the following criteria: registry in the herd-book, reproductive health and weight of at least 310 kg at the first service, with an annual culling of 20%.
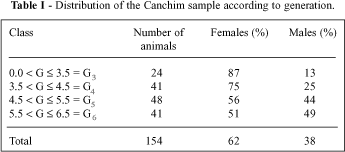
The generation number of each animal in this Canchim herd was determined using the following formula: (GS + GD + 2)/2, where GS is the generation of the sire and GD is the generation of the dam (Brinks et al., 1961). Foundation animals were considered generation 0. Blood was collected from a sample of 154 animals grouped into four classes (Table I). Semen or blood was also collected from a sample of Charolais cattle (N = 36) maintained at artificial insemination centers or at the Instituto Agronômico do Paraná (Ponta Grossa, PR, Brazil).
Marker analysis
Animals were scored for three RFLP markers and four microsatellites. k-casein (CSN3), b-lactoglobulin (LGB) and growth hormone (GH) genotypes were determined by digestion of PCR products according to procedures described by Medrano and Aguilar-Cordova (1990), Ron et al. (1994) and Schlee et al. (1994), respectively. Microsatellite loci IGF-I, CSFM50, BM1224 and INRA006 were amplified from 100 ng genomic DNA in 50 mM KCl, 1.5 mM MgCl2, 20 mM Tris-HCl, pH 8.4, 0.16 mM forward primer end labelled with 32P, 0.16 mM reverse primer, 0.2 mM dNTP and 0.5 units Taq DNA polymerase. Primers were synthesized according to published sequences (Bishop et al., 1994; Moore et al., 1994; Vaiman et al., 1995). Thermocycling conditions were: initial denaturing at 94°C for 2 min followed by 25 cycles of 1 min at 94°C, 30 s at annealing temperature and 1 min at 72°C. An additional 4 min extension was used at the end of the cycles. PCR products were mixed with an equal volume of loading buffer (95% formamide, 20 mM EDTA, pH 8.0, 0.05% bromophenol blue, 0.05% xylene cyanol) and denatured for 5 min at 90°C. After electrophoresis in 6% denaturing polyacrylamide gels, genotypes were determined in autoradiographies. Allele sizes were estimated by comparison to a 10-bp ladder molecular weight marker.
Data analysis
Gene and genotype frequencies were determined for each Canchim generation class and for the Charolais sample. Variation in allele frequencies over generations was tested by chi-square for contingency tables of allele counts per generation (with three degrees of freedom). In multiallelic loci, each allele was tested against the sum of alternative alleles at the same locus. Rare alleles (counts < 5 in all generations or 0 in one generation) were not tested. Whenever a significant test (P < 0.05) was obtained, a chi-square test for trends in proportions was used, according to Snedecor and Cochran (1967). Deviations from Hardy-Weinberg equilibrium were tested by conventional chi-square statistics for all loci with genotype counts greater than five or expected counts greater than one. Exact probabilities were calculated using the computer program BIOSYS-1 (Swofford and Selander, 1989). This program was also used to calculate Wright´s F-statistics. Significance of parameters FIS and FIT was tested by chi-square according to Li and Horvitz (1953). Significance of FST for each locus was tested by chi-square for contingency tables, according to Weir (1996). Considering that all loci were independent, the sum of chi-squares was used to test for significance of average FST over the seven loci.
RESULTS
Allele frequency variation
Allele frequencies and standard errors were calculated for each locus in all samples (Table II). Two alleles at each RFLP marker were present in both Canchim and Charolais. Although previous reports indicate the influence of k-casein and b-lactoglobulin on maternal effects in beef cattle (Moody et al., 1994), no significant variation of allele frequencies was observed over generations of Canchim. A highly significant variation (P < 0.01) was observed for GH AluI polymorphism, with a linear increase of the allele coding for valine (V) at position 127 of this peptide (Figure 1). Fixation of the allele coding for leucine at the same position has been observed in all Brazilian Zebu populations (Coutinho, L.L., unpublished results) indicating that the V allele found in Canchim came from Charolais.
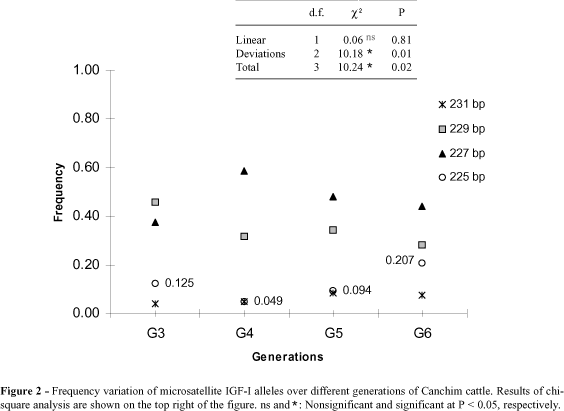
Four IGF-I microsatellite alleles ranging in size from 225 to 231 bp were observed in Canchim. Significant variation (P < 0.05) was only observed for the 225-bp allele. The trend observed through generations was not linear (Figure 2) and could be the result of dispersion in allele frequencies due to drift or selection associated to drift. This allele was not present in Charolais, and only three alleles were reported for this microsatellite in other populations of European cattle (Kirkpatrick, 1992). However, the 225-bp allele was predominant in Nelore (Rosa, 1997), which is a Zebu breed.
A significant variation in allele frequencies was observed for one out of six alleles at the microsatellite CSFM50. This microsatellite maps to a region of chromosome 2 that has been associated with weight gain from weaning to maturity in the Angleton reference families and is a candidate marker for selection, since it does not affect birth weight (Davis and Taylor, 1995). The linear reduction in frequency of the 168-bp allele over generations of Canchim (Figure 3) suggests that selection may be acting on this locus.
Hardy-Weinberg equilibrium
Deviations from Hardy-Weinberg expected genotype proportions were significant (P £ 0.01) for GH in all generations of Canchim except G3 (Table III). Disequilibrium for the IGF-I microsatellite was observed in generation 5 of Canchim and the total population (P < 0.01). Although a significant variation of frequencies for the 168-bp allele of CSFM50 was observed, this marker showed no significant departure from Hardy-Weinberg equilibrium. This finding is contrary to the hypothesis of selection against an allele at this locus and should be investigated in more detail. Despite the significant chi-square value obtained for BM1224 in the sample representing generations < 3, it was not supported by the exact probability test and could be an effect of sample size. All markers were in Hardy-Weinberg equilibrium in the Charolais sample.
F-statistics analysis
Fixation indexes most currently referred to as F-statistics were proposed by Wright (1951) to describe the properties of a subdivided population. Parameters FIT and FIS are the correlation between two uniting gametes with respect to the whole population and to gametes of subpopulations, respectively. Parameter FST is the correlation between random gametes from different individuals within subpopulations with respect to the total population and is a measure of the differentiation of subpopulations. If there is no significant selective advantage of different alleles, FIS can be interpreted as a measure of inbreeding. The high positive estimates of FIS and FIT for growth hormone (Table IV) indicate an excess of homozygotes (P < 0.01). This would not be expected by inbreeding alone since the mean inbreeding coefficient calculated for this sample of the population by pedigree analysis was 0.008.
Homozygote excess could be the result of preferential mating with regard to alleles at the growth hormone locus, which would be in agreement with the linear pattern of frequency variation observed. Significant FST values (P < 0.05) were obtained for this locus and for microsatellite IGF-I, as was expected from the results obtained for variation in allele frequencies. No significant FST value (0.018, P > 0.05) was observed for CSFM50 when considering all six alleles at this locus. The mean FST (0.016) value for all seven loci was also significant (P < 0.05), indicating gene differentiation over generations for these loci.
DISCUSSION
Synthetic breeds based on crosses between Bos taurus and Bos indicus are an alternative to overcome environmental stress in the tropics. The cross-bred population studied here has been the subject of recurrent phenotypic selection for weight at one year of age for nearly 50 years. Although much information is available on the development of this breed, results of selection on specific loci had never been investigated.
Analysis of allele frequencies over generations indicated significant variation at three out of seven loci. The linear increase of the V allele of growth hormone suggests selection. This hypothesis was supported by significant departures from Hardy-Weinberg proportions and high positive estimates of Wright's FIS. Variation of allele frequencies over generations was also significant for the 225-bp allele of microsatellite IGF-I, but did not follow a linear trend. This microsatellite is located at the 5' untranscribed region of IGF-I gene. It has been associated with differences in expected progeny differences (EPDs) for weaning weight, yearling weight and maternal EPD for milk in Hereford cattle (Moody et al., 1994).
Using molecular markers, we were able to identify breed-specific polymorphisms that may be useful to follow introgression of European genes into Zebu cattle. Simultaneous variation at GH and IGF-I alleles observed in Canchim is intriguing when considering the increase in frequency of a Bos taurus-specific allele at the first locus and a Bos indicus-specific allele at the second locus. As pointed out by Frisch (1981), growth under moderate to high environmental stress is directly related to stress tolerance instead of growth potential itself. Selection for growth rate under these conditions would favor genotypes that have a lower maintenance requirement. In view of the physiological relations between these two hormones and their relationship with maintenance requirements, new genotypic combinations with respect to these two loci resulting from crossing Bos taurus and Bos indicus breeds could be of some benefit to growth under suboptimal environmental conditions. Further investigations on the effects of these new combinations on economic traits such as yearling weight and growth rate should be conducted to provide the necessary background for marker assisted selection in this population.
ACKNOWLEDGMENTS
The authors thank the Centrais de Inseminação Artificial Genética Avançada, Lagoa da Serra, Pecplan-Bradesco for providing semen samples; Carlos Leskiu, from the Instituto Agronômico do Paraná for providing Charolais blood samples, and Prof. Dr. Henrique Nunes de Oliveira, from UNESP, Botucatu for calculating inbreeding coefficients. Research supported by Embrapa. Publication supported by FAPESP.
RESUMO
Este estudo foi desenvolvido com o objetivo de avaliar as freqüências gênicas em diferentes gerações de bovinos da raça sintética Canchim (5/8 Charolês, 3/8 Zebu). Uma amostra de 154 animais, representando três classes de gerações de um rebanho das raça Canchim, foi analisada para sete marcadores moleculares. Uma amostra da raça Charolesa (N = 36) foi incluída nas análises para permitir comparações. Observou-se um aumento linear, altamente significativo (P < 0,01), na freqüência do alelo que codifica o aminoácido valina na posição 127 do hormônio de crescimento, ao longo das gerações de Canchim. Este alelo foi observado na amostra da raça Charolesa e não é encontrado em raças zebuínas nacionais. Quatro alelos foram observados para o microssatélite localizado na região 5' não transcrita do gene do fator de crescimento semelhante à insulina do tipo I (IGF-I), com tamanhos variando de 231 a 225 pb. A freqüência do alelo de 225 pb apresentou um aumento significativo (P < 0,05) não linear ao longo das gerações. Este alelo não foi observado na raça Charolesa e é predominante nas raças zebuínas. Para o microssatélite CSFM50, seis alelos foram observados na raça Canchim, tendo sido verificada uma redução linear (P < 0,05) na freqüência do alelo de 168 bp. A hipótese de acasalamento preferencial com relação ao polimorfismo do hormônio de crescimento foi reforçada pelos resultados da análise de estatística F de Wright. O valor estimado de FIS para este loco foi 0,59 (P < 0,01). O aumento simultâneo de um alelo do hormônio de crescimento característico de Bos taurus e de um alelo de IGF-I característico de Bos indicus sugere que a seleção fenotípica tenha favorecido regiões diferentes de ambos os genomas que entraram na formação dessa raça.
(Received May 27, 1998)
- Alencar, M.M., Silva, A.H.G. and Barbosa, P.F. (1981). Efeitos da consanguinidade sobre o peso ŕ desmama de bezerros da raça Canchim. Rev. Soc. Bras. Zootec. 10: 156-172.
- Barendse, W., Armitage, S.M., Kossarek, L.M., Shalom, A., Kirkpatrick, B.W., Ryan, A.M., Clayton, D., Li, L., Neiberg, H.L., Zhang, N., Grosse, W.M., Weiss, J., Creighton, P., McCarthy, F., Ron, M., Teale, A.J., Fries, R., McGraw, R.A., Moore, S.S., Georges, M., Soller, J.E., Womack, J.E. and Hetzel, D.J.S. (1994). A genetic linkage map of the bovine genome. Nat. Genet. 6: 227-244.
- Barendse, W., Vaiman, D., Kemp, S.J., Sugimoto, Y., Armitage, J.L., Williams, J.L., Sun, H.S., Eggen, A., Agaba, M., Aleyasin, S.A., Band, M., Bishop, M.D., Buitkamp, J., Byrne, K., Collins, F., Cooper, L., Copettiers, W., Denys, B., Drinkwater, R.D., Easterday, K., Elduque, C., Ennis, S., Erhardt, G., Ferretti, L., Flavin, N., Gao, Q., Georges, M., Gurung, R., Harzilius, B., Hawkins, G., Hetzel, J., Hirano, T., Hulme, D., Jorgensen, C., Kessler, M., Kirkpatrick, B.W., Konfortov, B., Kostia, S., Kuhn, C., Lenstra, J.A., Leveziel, H., Lewin, H.A., Leyhe, B., Lil, L., Burriel, I.M., Mcgraw, R.A., Miller, J.R., Moody, D.E., Moore, S.S., Nakane, S., Nijman, I.J., Olsaker, I., Pomp, D., Rando, A., Ron, M., Shalom, A., Teale, A.J., Thieven, U., Urquhart, B.G.D., Vage, D.I., Van De Weghe, A., Varvio, S., Velmala, R., Vilkki, J., Weikard, R., Woodside, C., Womack, J.E., Zanotti, M. and Zaragoza, P. (1997). A medium-density genetic linkage map of the bovine genome. Mamm. Gen. 8: 21-28.
- Bishop, M.D., Kappes, S.M., Keele, J.W., Stone, R.T., Sunden, S.L.F., Hawkins, G.A., Toldo, S.S., Fries, R., Grosz, M.D., Yoo, J. and Beattie, C.W. (1994). A genetic linkage map for cattle. Genetics 136: 619-639.
- Brinks, J.S., Clark, R.T. and Rice, F.J. (1961). Estimation of genetic trends in beef cattle. J. Anim. Sci. 20: 903.
- Davis, S.K. and Taylor, J.F. (1995). Mechanisms of Genetic Control of Beef Carcass Merit Traits. Research Project Summary Outline - Interim Report A & M University, Texas, pp. 7.
- Frisch, J.E. (1981). Changes occurring in cattle as a consequence of selection for growth in a stressful environment. J. Agric. Sci. 96: 23-38.
- Kirkpatrick, B.W. (1992). Identification of a conserved microsatellite site in the porcine and bovine insulin-like growth factor-I gene 5' flank. Anim. Genet. 23: 543-548.
- Li, C.C. and Horvitz, D.G. (1953). Some methods of estimating inbreeding coefficient. Am. J. Hum. Genet. 5: 107-117.
- Medrano, J.F. and Aguilar-Cordova, E. (1990). Genotyping of bovine kappa-casein loci following DNA sequence amplification. Biotechnology 8: 144-146.
- Moody, D.E., Pomp, D., Newman, S. and McNeil, M.D. (1994). Characterization of DNA polymorphisms and their association with growth and maternal traits in line 1 Hereford cattle. In: World Congress on Genetics Applied to Livestock Production, 5, Guelph, Canada, August 1994. Proceedings, Guelph Organizing Committee, 1994, 21: 221-224.
- Moore, S.S., Byrne, K., Berger, K.T., Barendse, W., Mccarthy, F., Womack, J.E. and Hetzel, D.J.S. (1994). Characterization of 65 bovine microsatellites. Mamm. Gen. 5: 84-90.
- Ron, M., Yoffe, O., Ezra, E., Medrano, J.F. and Weller, J.I. (1994). Determination of effects of milk protein genotype on production of Israeli Holstein. J. Dairy Sci. 77: 1106-1113.
- Rosa, A.J.M. (1997). Caracterizaçăo da raça Nelore e teste de paternidade por marcadores moleculares. Master's thesis, USP, Săo Paulo, Brazil.
- Schlee, P., Graml, R., Schallenberger, E., Schams, D., Rottmann, O., Olbrich-Bludau, A. and Pirchner, F. (1994). Growth hormone and insuline-like growth factor I concentrations in bulls of various growth hormone genotypes. Theor. Appl. Genet.88: 497-500.
- Snedecor, G.W. and Cochran, W.G. (1967). Statistical Methods. 6th edn. The Iowa State University Press, Ames.
- Swofford, D.L. and Selander, R.B. (1989). Biosys-1. A Computer Program for the Analysis of Allelic Variation in Population Genetics and Biochemical Systematics. Release 1.7, Swofford, D.L. Illinois Natural History Survey, Champaign, IL.
- Vaiman, D., Eggen, A., Mercier, D., Bahri-Darwich, I., Grohs, C., Bruneau, D., Laurent, P., Chaput, B., Oustry, A., Frelat, G., Levéziel, H. and Cribiu, E.P. (1995). A genetic and physical map of bovine chromosome 3. Anim. Genet. 26: 21-25.
- Weir, B.S. (1996). GeneticData Analysis: Methods for Discrete Population Genetic Data 2nd edn. Sinauer Associates, Sunderland, MA.
- Wright, S. (1951). The genetical structure of populations. Ann. Eugen. 15: 323-354.
Publication Dates
-
Publication in this collection
17 Feb 2000 -
Date of issue
Dec 1999
History
-
Received
27 May 1998