Abstract
The grasshopper species Orthoscapheus rufipes and Eujivarus fusiformis were analyzed using several cytogenetic techniques. The karyotype of O. rufipes, described here for the first time, had a diploid number of 2n = 23, whereas E. fusiformis had a karyotype with 2n = 21. The two species showed the same mechanism of sex determination (XO type) but differed in chromosome morphology. Pericentromeric blocks of constitutive heterochromatin (CH) were detected in the chromosome complement of both species. CMA3/DA/DAPI staining revealed CMA3-positive blocks in CH regions in four autosomal bivalents of O. rufipes and in two of E. fusiformis. The location of active NORs differed between the two species, occurring in bivalents M6 and S9 of O. rufipes and M6 and M7 of E. fusiformsi. The rDNA sites revealed by FISH coincided with the number and position of the active NORs detected by AgNO3 staining. The variability in chromosomal markers accounted for the karyotype differentiation observed in the tribe Abracrini.
constitutive heterochromatin; fluorochromes; FISH; Orthoptera; ribosomal DNA
RESEARCH ARTICLE
Comparative cytogenetic analysis of two grasshopper species of the tribe Abracrini (Ommatolampinae, Acrididae)
Marília de França RochaI; Natoniel Franklin de MeloII; Maria José de SouzaIII
ILaboratório de Biodiversidade e Genética de Insetos, Instituto de Ciências Biológicas, Universidade de Pernambuco, Recife, PE, Brazil
IIEmpresa Brasileira de Pesquisa Agropecuária, Petrolina, PE, Brazil
IIIDepartamento de Genética, Centro de Ciências Biológicas, Universidade Federal de Pernambuco, Recife, PE, Brazil
Send correspondence to Send correspondence to: Maria José de Souza Departamento de Genética Centro de Ciências Biológicas Universidade Federal de Pernambuco Av. Prof. Moraes Rego S/N, Cidade Universitária 50732-970 Recife, PE, Brazil E-mail: mjslopes.ufpe@yahoo.com.br
ABSTRACT
The grasshopper species Orthoscapheus rufipes and Eujivarus fusiformis were analyzed using several cytogenetic techniques. The karyotype of O. rufipes, described here for the first time, had a diploid number of 2n = 23, whereas E. fusiformis had a karyotype with 2n = 21. The two species showed the same mechanism of sex determination (XO type) but differed in chromosome morphology. Pericentromeric blocks of constitutive heterochromatin (CH) were detected in the chromosome complement of both species. CMA3/DA/DAPI staining revealed CMA3-positive blocks in CH regions in four autosomal bivalents of O. rufipes and in two of E. fusiformis. The location of active NORs differed between the two species, occurring in bivalents M6 and S9 of O. rufipes and M6 and M7 of E. fusiformsi. The rDNA sites revealed by FISH coincided with the number and position of the active NORs detected by AgNO3 staining. The variability in chromosomal markers accounted for the karyotype differentiation observed in the tribe Abracrini.
Key words: constitutive heterochromatin, fluorochromes, FISH, Orthoptera, ribosomal DNA.
Introduction
The grasshopper subfamily Ommatolampinae (Acrididae) comprises nine tribes and more than 50 genera that have a wide geographic distribution, with most of them being found in North, Central and South America (Amedgnato, 1974; Carbonell, 1977). Although numerous studies have dealt with species of the family Acrididae, the analysis of most Neotropical species, especially of the subfamily Ommatolampinae, has been restricted to conventional staining (Ferreira et al., 1980; Mesa et al., 1982; Mesa and Fontanetti, 1983); at least 20 species of Ommatolampinae have been studied using conventional techniques (Carbonell et al., 1980; Ferreira et al., 1980; Mesa et al., 1982; Mesa and Fontanetti, 1983; Cella and Ferreira, 1991). The use of specific techniques for chromosome identification has been applied only to the species Abracris flavolineata (Cella and Ferreira, 1991). Despite the small number of species studied so far, the data obtained indicate that this subfamily is characterized by a significant number of species (> 40%) with derived karyotypes originating from centric fusions or other rearrangements. These karyotypes are found in species of the genera Pycnosarcus and Lagidacris (17, XO) - Pycnosarcini, Bucephalacris (21, XO) - Dellini, Abracris, Eujivarus and Omalotettix (21, XO), and Jodacris and Siltaces (19, XO) - Abracrini (Carbonell et al., 1980; Ferreira et al., 1980; Mesa et al., 1982; Mesa and Fontanetti, 1983).
Constitutive heterochromatin (CH) accounts for a significant part of the genome in grasshoppers and is characterized by a low gene density and the presence of highly repetitive sequences. Some studies have shown extensive polymorphism in this type of chromatin in grasshoppers, including variation in the location and size of the CH blocks, as well as heterogeneity in these regions. The use of base-specific fluorochromes has contributed to the characterization of CH (King and John, 1980; Santos et al., 1983; John et al., 1985), although for Neotropical grasshoppers the data obtained with such probes are limited to a few species of the families Acrididae and Romaleidae (Souza et al., 1998; Loreto and Souza, 2000; Pereira and Souza, 2000; Souza et al., 2003; Rocha et al., 2004; Loreto et al., 2005; Souza and Melo, 2007). Other techniques, such as fluorescent in situ hybridization (FISH), have identified important differences in the composition of CH that involve variable quantities of repetitive DNA sequences in these regions, including satellite or ribosomal DNA (rDNA) (Rodríguez Iñigo et al., 1993, 1996; Loreto et al., 2008). Since, in contrast to silver nitrate (AgNO3) staining, FISH does not depend on the presence of a transcription product it permits the identification of active or inactive rDNA sequences in the genome (López-León et al., 1999).
In this study, we used the C-banding technique and staining with base-specific fluorochromes (CMA3 and DAPI) to examine the distribution of CH and the proportion of GC and AT base pairs in karyotypes of the grasshoppers Orthoscapheus rufipes and Eujivarus fusiformis; silver nitrate impregnation and FISH were used to determine the position and variability of the nucleolar organizer regions (NORs). The results described here improve our understanding of chromosomal organization in these species and display new light on the chromosomal phylogeny of the Ommatolampinae.
Material and Methods
Specimens of O. rufipes and E. fusiformis were collected in two rainforest areas in Pernambuco State, northeastern Brazil. Fifteen male and nine female specimens of O. rufipes and seven male and two female specimens of E. fusiformis were collected in Gurjaú forest, in the municipality of Cabo (8°17'12" S; 35°2'6" W). Ten male and eight female E. fusiformis specimens were also collected in the Dois Irmãos zoological-botanical garden, in the municipality of Recife (8°3'14" S; 34°52'52" W).
Cytological preparations were obtained from testes and ovarioles by the classic squashing technique. The ovarioles were pretreated with 0.1% colchicine for 6 h prior to preparation. The material was fixed in ethanol:acetic acid (3:1, v/v). Conventional staining was done with 2% lacto-acetic orcein.
C-banding, triple staining with CMA3/DA/DAPI and AgNO3 staining were done as described by Sumner (1972), Schweizer et al. (1983) and Rufas et al. (1987), respectively. FISH was done as described by Moscone et al. (1996) using 18S and 25S rDNA probes from Arabidopsis thaliana (Unfried et al., 1989; Unfried and Gruendler, 1990). The probes were labeled with bio-11-dUTP by nick translation (Life Technologies) and detected using rat antibiotin (Dakopatts M0743, Dako) and TRITC (tetramethyl-rhodamine isothiocyanate)-conjugated anti-antibiotin (Dakopatts R0270, Dako) antibodies. The preparations were counterstained with DAPI (2 μg/mL) and mounted with Vectashield H-1000 (Vector).
For fluorescent in situ hydridization (FISH), images of cells were captured with a Cytovision system coupled to an Olympus BX51 microscope. For the other techniques, the cells were photographed with a Leica microscope. The images were mounted using CorelDraw Graphics Suite 12.
Results
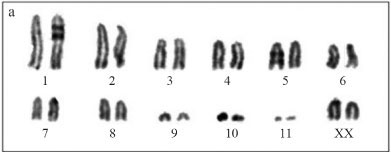
The karyotype of O. rufipes, described here for the first time, consisted of a diploid number of 2n = 23 and an XO sex determination mechanism for males, and 2n = 24 and XX for females. The chromosomes of O. rufipes (Figure 1a-c) were acrotelocentric and were classified according to size into two large pairs (L1-L2), six medium sized pairs (M3-M8) including the X chromosome, and three small pairs (S9-S11). In contrast, E. fusiformis had a diploid number of 2n = 21 and an XO sex determination mechanism. The L1 pair was submetacentric and the other chromosomes were acrotelocentric. There were two large pairs (L1-L2), five medium sized pairs (M3-M7) including the X chromosome, and three small pairs (S8-S10) (Figure 1b-d). In both species, the X chromosome showed variable heteropyknotic behavior during meiotic prophase I (Figure 1c,d).
The CH blocks were located in pericentromeric regions of the chromosomes and varied in size between and within the species studied (Figure 2a,c). In O. rufipes (Figure 2a), the CH blocks were small and located on all chromosomes of the complement, whereas in E. fusiformis (Figure 2c) these blocks were very small and found only on some bivalents. CMA3/DA/DAPI staining revealed CMA3-positive blocks on four bivalents of O. rufipes, including one interstitial block on L2, proximal and telomeric blocks on M5, and pericentromeric blocks on M6 and S9 (Figure 2b). The CMA3-positive blocks of the L2 and M5 bivalents were not detected by C-banding. Eujivarus fusiformis had two medium sized chromosomes (M6 and M7) with CMA3-positive blocks (Figure 2d). In both species, DAPI staining was homogenous throughout all of the chromosomes (data not shown).
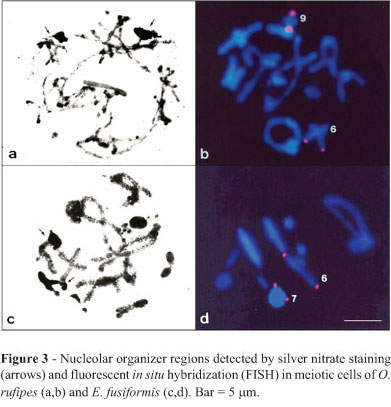
Active NORs were observed during prophase I (pachytene-diplotene) in both species, and were located in bivalents M6 and S9 of O. rufipes and in M6 and M7 of E. fusiformis. At both sites, the NORs coincided with CMA3-positive blocks. Additionally, the rDNA sites detected by FISH coincided with the results of AgNO3 staining in both species (Figure 3a-d). Table 1 summarizes the data obtained with conventional staining, C-banding, base-specific fluorochromes, AgNO3 staining, and FISH for the two species studied.
Discussion
Variations in the karyotypes of grasshoppers have been reported, with the most frequent being chromosomal rearrangements such as inversions, reciprocal translocations and centric fusion/fission, in addition to variability in the pattern of CH distribution and the occurrence of extra chromosomal material. Among the ten Neotropical Acrididae subfamilies studied cytogenetically by Mesa et al. (1982), the Copiocerinae, Melanoplinae and Ommatolampinae were characterized by derived karyotypes (81.8%, 48.4% and 42.9%, respectively) that resulted mainly from centric fusions and inversions.
The karyotypes of 31 species of the subfamily Ommatolampinae are known, with 21 of them belonging to the tribe Abracrini (Carbonell et al., 1980; Ferreira et al., 1980; Mesa et al., 1982; Mesa and Fontanetti, 1983; Cella and Ferreira, 1991; present study). Despite the small number of species studied so far, more than 40% of them show variations in diploid number, a finding that identifies this subfamily as an important group for studying chromosome evolution. Table 2 summarizes the chromosome number, sex mechanism and chromosome morphology of representatives of the tribe Abracrini (Ommatolampinae, Acrididae).
Orthoscapheus rufipes had a basic karyotype (23, XO) consisting of acrotelocentric chromosomes, which was similar to that widely found in the family Acrididae (Mesa et al., 1982; Santos et al., 1983; Bugrov, 1996). On the other hand, the karyotype of E. fusiformis (2n = 21, XO) is considered to be a derived karyotype among acridoid grasshoppers. Of the 21 Abracrini species listed in Table 2, 14 have derived karyotypes. These species belong to the genera Abracris and Eujivarus (21, XO, one pair of metacentric autosomes), Omalotettix (21, XO, pairs 3 and 6 submetacentric, pair 5 subacrocentric, and the remaining chromosomes acrocentric), Jodacris (19, XO, acrocentric chromosomes) and Sitalces (19, XO, two pairs of metacentric autosomes). Among the three species of Abracris studied cytologically, Abracris sp. is the only one with 2n = 21 and XO, with one metacentric pair originating from the centric fusion of two medium sized chromosomes. Abracris dilecta and A. flavolineata have 2n = 23 and XO. However, whereas A. dilecta has acrocentric chromosomes, A. flavolineata has chromosomes with two arms throughout the karyotype complement, in contrast to the basic karyotype of Acridoidea (Mesa et al., 1982; Cella and Ferreira, 1991).
An ancient fusion appears to have been involved in the phylogeny of Eujivarus since four of the five species whose karyotype has been studied show a reduction in the diploid number to 2n = 21 with XO (Ferreira et al., 1980; Mesa et al., 1982). In contrast, more recent fusions and polymorphisms in this type of chromosomal rearrangement have been observed in grasshoppers such as Cornops aquaticum (Mesa et al., 1982), Eyprepocnemis plorans (Arana et al., 1987), Leptysma argentina (Colombo, 1993) and Dichroplus pratensis (Bidau and Martí, 2002).
The CH of O. rufipes and E. fusiformis was preferentially located in pericentromeric regions, in agreement with descriptions for most species of the family Acrididae (King and John, 1980; Santos et al., 1983; Rocha et al., 2004; Souza and Melo, 2007). In Abracris flavolineata, in addition to the pericentromeric regions, CH blocks also occur on the short arms of all chromosomes, except for pair M7 (Cella and Ferreira, 1991). This divergence in the pattern of distribution suggests that rearrangements in CH, such as amplifications or losses, may contribute to karyotype evolution in the Abracrini.
Differences in the base composition of CH (detected by CMA3/DA/DAPI staining) were observed in the species studied. In most chromosomes of both species, there was no predominance of AT or GC base pairs. However, GC-rich CMA3-positive regions were detected in some CH blocks, especially those located in NORs. This pattern has also been reported for other grasshopper species (John et al., 1985; Camacho et al., 1991; Souza et al., 1998; Loreto and Souza, 2000; Pereira and Souza, 2000; Rocha et al., 2004). In some cases, such as in Xyleus angulatus, Phaeoparia megacephala and Cornops frenatum frenatum, CMA3-positive blocks have been detected in all chromosomes of the complement (Souza et al., 1998; Pereira and Souza, 2000; Rocha et al., 2004). On the other hand, in most species, GC-rich CH blocks occur on only some chromosomes of the karyotype (Schweizer et al., 1983; John et al., 1985; Loreto and Souza, 2000; Souza et al., 2003; Rocha et al., 2004; Loreto et al., 2005; Souza and Melo, 2007), including O. rufipes and E. fusiformis.
Three categories of CH were identified in O. rufipes: (1) CH detected by C-banding, (2) CH detected by C-banding and CMA3 staining and restricted to NORs (pairs M6 and S9), and (3) CH detected by CMA3 staining, but not by C-banding, and unrelated to NORs (pairs L2 and M5). According to Sumner (1990), CH is not always detectable by the C-banding technique since its visualization is related to the size of the segments, with blocks less than 106 or 107 base pairs (bp) generally not being detected by this method. The finding that the third type of CH was GC-rich but showed no functional relationship to rDNA sites suggested a different organization for these two regions.
Two rDNA sites preferentially located on medium sized autosomes were observed in O. rufipes and E. fusiformis, a pattern commonly found among representatives of Neotropical Acrididae (Rocha et al., 2004; Souza and Melo, 2007; Loreto et al., 2008). The presence of NORs on bivalent S9, as observed in O. rufipes, has also been described for most Neotropical species studied so far, especially those of the subfamilies Leptysminae (Rocha et al., 2004) and Gomphocerinae (Loreto et al., 2008). According to Loreto et al. (2008), bivalent S9 probably represents the ancestral location of rDNA sites in Neotropical gomphocerine species since this pattern was identified in five species analyzed (Rhammmatocerus brasiliensis, R. brunneri, R. palustris, R. pictus and Amblytropidia sp.). Ribosomal DNA sites on S9 were also observed in five of six leptysmine species studied (Loreto and Souza, 2000; Rocha et al., 2004) and in the ommatolampine O. rufipes. Together, these findings suggest a pattern of ancestrality for Neotropical acridid grasshoppers.
In a broad investigation of the location and expression of ribosomal genes in 49 grasshopper species, Cabrero and Camacho (2008) showed that most Old World representatives have 1-3 rDNA sites and only six species had 5-10 sites. The predominant NOR locations were bivalents 2, 3, 6 and 9 and the X chromosome. In the subfamily Gomphocerinae, signals were detected mainly in bivalents 2, 3 and the X chromosome in species with 2n = 17, whereas in species with 2n = 23 most signals were restricted to bivalent 9. In Oedipodinae, NORs were generally found on bivalents 6 and 9. These data suggest that the location of rDNA sites on chromosome 9 represents an ancestral condition.
NORs located on large chromosomes or in the X chromosome have not yet been described for Brazilian species of the family Acrididae, in contrast to observations for the romaleid species Radacridium nordestinum (L2) and Radacridium mariajoseae (X) (Rocha et al., 1997), Xyleus angulatus (L3, M4 and X) (Souza et al., 1998), and some Old World acridid species (Rufas et al., 1985; Cabrero and Camacho, 1986, 2008).
Many rDNA sites are located in pericentromeric regions of medium and/or small chromosomes (Loreto and Souza, 2000; Rocha et al., 2004), as observed in the present study. However, some NORs occur in proximal regions, as observed in Stenacris xanthochlora and Tucayaca parvula (M8) (Rocha et al., 2004), or in interstitial regions, as described in Schistocerca pallens (M5) and S. flavofasciata (M5 and M6) (Souza and Melo, 2007). In the Old World grasshoppers studied by Cabrero and Camacho (2008), many rDNA sites detected by FISH were found in proximal regions (52.4%), although a significant number was observed in interstitial regions (34.9%) and a minority was distal (12.7%). In addition, about 13% of the 126 rDNA sites detected by FISH were silent.
In conclusion, this study provides the first detailed cytogenetic results for E. fusiformis and O. rufipes. The two species showed significant karyotype differentiation depending on the staining method used. Our findings suggest possible pathways of chromosome evolution in these species. However, additional cytogenetic and molecular analyses of other species of the tribe Abracrini are necessary to improve our understanding of the evolutionary patterns within this group.
Acknowledgments
The authors thank Dr. Carlos S. Carbonell (University of Montevideo, Uruguay), for taxonomic identification of the species studied, Dr. Marcelo Guerra (Department of Botany, UFPE) for providing access to the Laboratory of Plant Cytogenetics and for allowing us to do the FISH experiments, and Francisca Tavares de Lira for technical assistance. We also thank Dr. Neide Santos (Department of Genetics, UFPE) for critically reading the manuscript. This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE), Brazil.
Received: September 11, 2010; Accepted: January 19, 2011.
Associate Editor: Yatiyo Yonenaga-Yassuda
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Amedgnato C (1974) Les genres d'acridiens néotropicaux, leur classification par familles, sous-familles et tribus. Acrida 3:193-204.
- Arana P, Santos JL and Henriques-Gil N (1987) Interference relationships in grasshopper reciprocal translocation heterozygotes. Heredity 59:85-93.
- Bidau CJ and Martí DA (2002) Geographic distribution of Robertsonian fusions in Dichroplus pratensis (Melanoplinae, Acrididae): The central-marginal hypothesis reanalyzed. Cytogenet Genome Res 96:66-74.
- Bugrov AG (1996) Karyotypes of the short-horned orthopteran insects (Orthoptera, Caelifera) from Russia, Kazakhstan, Central Asia, and the Caucasus. Folia Biol (Krakow) 44:15-25.
- Cabrero J and Camacho JPM (1986) Cytogenetics studies in gomphocerine grasshoppers II. Chromosomal location of active nucleolar organizing regions. Can J Genet Cytol 28:540-544.
- Cabrero J and Camacho JPM (2008) Location and expression of ribosomal RNA genes in grasshoppers: Abundance of silent and cryptic loci. Chromosome Res 16:595-607.
- Camacho JPM, Cabrero JM, Viseras E, López-León MD, Navas-Castillo J and Alche JD (1991) G banding in two species of grasshopper and its relationship to C, N, and fluorescence banding techniques. Genome 34:638-643.
- Carbonell CS (1977) Origin, evolution and distribution of the neotropical acridomorph fauna (Orthoptera): A preliminary hypothesis. Rev Soc Ent Argentina 36:153-175.
- Carbonell CS, Mesa A and Conduta VL (1980) Systematics and karyotype of the grasshopper genera Ommalotettix and Abracris Proc Acad Nat Sci Philadelphia 132:279-284.
- Cella DM and Ferreira A (1991) The cytogenetics of Abracris flavolineata (Orthoptera, Caelifera, Ommatolampinae, Abracrini). Rev Bras Genet 14:315-329.
- Colombo PC (1993) Chromosome polymorphisms and natural selection in Leptysma argentina (Orthoptera). II. Gametic phase disequilibrium and differential adult male viability. Heredity 71:295-299.
- Ferreira A, Mesa A and Carbonell CS (1980) The chromosomes of neotropical short-horned grasshoppers of the tribe Abracrini (Orthoptera, Acridoidea, Acrididae, Ommatolampinae). Naturalia 5:89-95.
- John B, King M, Schweizer D and Mendelak M (1985) Equilocality of heterochromatin distribution and heterogeneity in acridid grasshoppers. Chromosoma 91:185-200.
- King M and John B (1980) Regularities and restrictions governing C-band variation in acridoid grasshoppers. Chromosoma 76:123-150.
- López-León MD, Cabrero JM and Camacho JPM (1999) Unusually high amount of inactive ribosomal DNA in the grasshopper Stauroderus scalaris Chromosome Res 7:83-88.
- Loreto V and Souza MJ (2000) Karyotype, constitutive heterochromatin and nucleolar organizer regions (NORs) in Belosacris coccineipes (Acrididae, Leptysminae). Genet Mol Biol 23:575-579.
- Loreto V, Stadtler E, Melo NF and Souza MJ (2005) A comparative cytogenetic analysis between the grasshopper species Chromacris nuptialis and C. speciosa (Romaleidae): Constitutive heterochromatin variability and rDNA sites. Genetica 125:253-260.
- Loreto V, López-León MD, Camacho JPM and Souza MJ (2008) Comparative analysis of rDNA location in five Neotropical gomphocerine grasshopper species. Genetica 132:95-101.
- Mesa A, Ferreira A and Carbonell CS (1982) Cariologia de los Acridoideos neotropicales: Estado actual de su conocimiento y nuevas contribuciones. Annls Soc Ent Fr (N S) 18:507-526.
- Mesa A and Fontanetti CS (1983) Karyotypes of nine Brazilian species of acridids (Orthoptera, Acridoidea). Rev Bras Genet 6:295-305.
- Moscone EA, Matzke MA and Matzke AJM (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105:231-236.
- Pereira LG and Souza MJ (2000) Nature and distribution of constitutive heterochromatin and NOR location in the grasshopper Phaeoparia megacephala (Romaleidae, Orthoptera). Cytobios 103:111-119.
- Rocha MF, Souza MJ and Tashiro T (1997) Karyotype variability in the genus Radacridium (Orthoptera, Romaleidae, Romaleinae). Cytologia 62:53-60.
- Rocha MF, Souza MJ and Moura RC (2004) Karyotype analysis, constitutive heterochromatin and NOR distribution in five grasshopper species of the subfamily Leptysminae (Acrididae). Caryologia 57:107-116.
- Rodríguez Iñigo E, Bella JL and Garcia de La Vega C (1993) Heterochromatin differentiation between two species of the genus Dociostaurus (Orthoptera, Acrididae). Heredity 70:458-465.
- Rodríguez Iñigo E, Fernández-Calvin B, Capel J and Garcia de La Vega C (1996) Equilocality and heterogeneity of constitutive heterochromatin: In situ localization of two families of highly repetitive DNA in Dociostaurus (Orthoptera). Heredity 76:70-76.
- Rufas JS, Esponda P and Gosálvez J (1985) NOR and nucleolus in the spermatogenesis of acridoid grasshoppers. Genetica 66:139-144.
- Rufas JS, Giménez-Abian J, Suja JA and Garcia de La Vega C (1987) Chromosome organization in meiosis revealed by light microscope analysis of silver-stained cores. Genome 29:706-712.
- Santos JL, Arana P and Giraldez R (1983) Chromosome C-banding patterns in Spanish Acridoidea. Genetica 61:65-74.
- Schweizer D, Mendelak M, White MJD and Contreras N (1983) Cytogenetics of the parthenogenetic grasshopper Warramaba virgo and its bisexual relatives. X. Patterns of fluorescent banding. Chromosoma 88:227-236.
- Souza MJ and Melo FN (2007) Chromosome study in Schistocerca (Orthoptera, Acrididae, Cyrtacanthacridinae): Karyotypes and distribution patterns of constitutive heterochromatin and nucleolus organizer regions (NORs). Genet Mol Biol 30:54-59.
- Souza MJ, Rufas JS and Orellana J (1998) Constitutive heterochromatin, NOR location and FISH in the grasshopper Xyleus angulatus (Romaleidae). Caryologia 51:73-80.
- Souza MJ, Haver ORP and Melo FN (2003) Karyotype, C- and fluorescence banding patterns NOR location and FISH in the grasshopper Xestotrachelus robustus (Romaleidae). Caryologia 56:261-267.
- Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304-306.
- Sumner AT (1990) Chromosome Banding. 1st edition. Unwin Hyman Ltd., London, 434 pp.
- Unfried I and Gruendler P (1990) Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana Nucleic Acids Res 18:4011.
- Unfried I, Stocker U and Gruendler P (1989) Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Nucleic Acids Res 17:7513.
Publication Dates
-
Publication in this collection
02 June 2011 -
Date of issue
2011
History
-
Received
11 Sept 2010 -
Accepted
19 Jan 2011