Abstract
Drought stress is the main limiting factor of soybean yield. Currently, genetic engineering has been one important tool in the development of drought-tolerant cultivars. A widely used strategy is the fusion of genes that confer tolerance under the control of the CaMV35S constitutive promoter; however, stress-responsive promoters would constitute the best alternative to the generation of drought-tolerant crops. We characterized the promoter of α-galactosidase soybean (GlymaGAL) gene that was previously identified as highly up-regulated by drought stress. The β-glucuronidase (GUS) activity of Arabidopsis transgenic plants bearing 1000- and 2000-bp fragments of the GlymaGAL promoter fused to the uidA gene was evaluated under air-dried, polyethylene glycol (PEG) and salt stress treatments. After 24 h of air-dried and PEG treatments, the pGAL-2kb led to an increase in GUS expression in leaf and root samples when compared to the control samples. These results were corroborated by qPCR expression analysis of the uidA gene. The pGAL-1kb showed no difference in GUS activity between control and treated samples. The pGAL-2kb promoter was evaluated in transgenic soybean roots, leading to an increase in EGFP expression under air-dried treatment. Our data indicates that pGAL-2kb could be a useful tool in developing drought-tolerant cultivars by driving gene expression.
Keywords:
α-galactosidase gene; salt stress; drought-tolerant soybean; cis-acting element; raffinose degradation pathway
Introduction
Soybean (Glycine max L. Merr) is a valuable commodity due to its utilization in the pharmaceutical industry, biodiesel production and food for humans and animals (Tran and Mochida, 2010Tran L-SP and Mochida K (2010) Functional genomics of soybean for improvement of productivity in adverse conditions. Funct Integr Genomics 10:447-462.). Therefore, the global soybean production was estimated to be 324.2 million metric tons in 2016/2017 (USDA, 2016USDA (2016) World Agricultural Production. Foreign Agricultural Service, Washington, DC, p 5-16.), of which approximately 64% of its production is concentrated in the USA and Brazil. Despite this, the yield and production of soybean has been impacted by the occurrence of drought-stress. Recently, in 2012, the drought in the USA was the most intense since the 1950s, triggering a 20% loss of yield (Zulauf, 2012Zulauf C (2012) 2012 Drought: Yield Loss, Revenue Loss, and Harvest Price Option. Farmadocdaily. University of Illinois, Urbana-Champaign, 5 p.; Rippey, 2015Rippey BR (2015) The U.S. Drought of 2012. Weather and Climate Extremes 10:57-64.). In the same period, the loss of Brazilian soybean production was approximately 13% due to drought stress (Conab, 2012Conab (2012) Acompanhamento da Safra Brasileira de Grãos 2011/12. V: Sexto Levantamento. Conab, Brasília, 35 p.). Moreover, the most severe drought stress period in Brazil occurred between 2003 and 2005, when the loss of soybean was greater than 20% of production (Polizel et al., 2011Polizel AMM, Nakashima K, Yamanaka N, Farias JR, de Oliveira MC, Marin SR, Abdelnoor RV, Marcelino-Guimarães FC, Fuganti R, Rodrigues FA, et al. (2011) Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A: AtDREB1A for the improvement of drought tolerance. Genet Mol Res 4:3641-3656.). Therefore, the development of drought-tolerant soybean cultivars is crucial and should have high priority. Among the strategies available, the development of transgenic crops by the overexpression of drought-tolerant genes has been proven to be successful and can have a significant impact on agricultural production (Silvente et al., 2012Silvente S, Sobolev AP and Lara M (2012) Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS ONE 7:e38554.; Rahman et al., 2016Rahman H, Ramanathan V, Nallathambi J, Duraialagaraja S and Muthurajan R (2016) Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol 16(suppl 1):7-20.).
The constitutive cauliflower mosaic virus 35S RNA promoter (CaMV35S) has been broadly used to control the expression of several genes responsive to drought due to the strong and conspicuous activity presented when introduced in plant genomes (Luo et al., 2013Luo X, Wu J, Li Y, Nan Z, Guo X, Wang Y, Zhang A, Wang Z, Xia G and Tian Y (2013) Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS ONE 8:e54002.; Novák et al., 2013Novák J, Pavlu J, Novák O, Nozková-Hlavácková V, Spundová M, Hlavinka J, Koukalová S, Skalák J, Cerny M and Brzobohaty B (2013) High cytokinin levels induce a hypersensitive-like response in tobacco. Ann Bot 112:41-55.; Bhauso et al., 2014Bhauso TD, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR, Patel K and Rajam MV (2014) Overexpression of bacterial mtlD gene in peanut improves drought tolerance through accumulation of mannitol. Sci World J 2014:1-10.; Withanage et al., 2015Withanage SP, Hossain MA, Kumar MS, Roslan HAB, Abdullah MP, Napis SB and Shukor NAA (2015) Overexpression of Arabidopsis thaliana gibberellic acid 20 oxidase (AtGA20ox) gene enhance the vegetative growth and fiber quality in kenaf (Hibiscus cannabinus L.) plants. Breed Sci 65:177-191.; Du et al., 2016Du H, Shen X, Huang Y, Huang M and Zhang Z (2016) Overexpression of Vitreoscilla hemoglobin increases waterlogging tolerance in Arabidopsis and maize. BMC Plant Biol 16:35.). However, the constitutive overexpression of genes can affect plant development and metabolism (Hsieh et al., 2002Hsieh T-H, Lee J-T, Yang P-T, Chiu L-H, Charng Y-Y, Wang Y-C and Chan M-T (2002) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiology 129:1086-1094.; Homrich et al., 2012Homrich MS, Wiebke-Strohm B, Weber RLM and Bodanese-Zanettini MH (2012) Soybean genetic transformation: A valuable tool for the functional study of genes and the production of agronomically improved plants. Genet Mol Biol 35:998-1010.). Indeed, the control of the expression of drought-tolerant genes by stress-responsive and/or tissue-specific promoters has been used as alternative for the elimination of negative effect of constitutive gene expression driven by the CaMV35S promoter (Chan Ju et al., 2012Chan Ju L, Ha Yeon L, Woong Bom K, Bok-Sim L, Jungeun K, Raza A, Hyun AK, So Young Y, Cheol-Goo H and Suk-Yoon K (2012) Screening of tissue-specific genes and promoters in tomato by comparing genome wide expression profiles of Arabidopsis orthologues. Mol Cells 34:53-59.; Banerjee et al., 2013Banerjee J, Sahoo DK, Dey N, Houtz RL and Maiti IB (2013) An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and tobacco plants. PLoS ONE 8:e79622.; Yan et al., 2015Yan H, Ma L, Wang Z, Lin Z, Su J and Lu B-R (2015) Multiple tissue-specific expression of rice seed-shattering gene SH4 regulated by its promoter pSH4. Rice 8:1-10.). Recently, stress-responsive promoters, such as the PeNAC1 (Wang et al., 2016Wang J-Y, Wang J-P and Yang H-F (2016) Identification and functional characterization of the NAC gene promoter from Populus euphratica. Planta 244:417-427.), ZmGAPP (Hou et al., 2016Hou J, Jiang P, Qi S, Zhang K, He Q, Xu C, Ding Z, Zhang K and Li K (2016) Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS ONE 11:e0154041.), BBX24 (Imtiaz et al., 2015Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J and Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from Chrysanthemum in Arabidopsis. Plant Mol Biol 89:1-19.), and GmNCED1 and GmMYB363P promoters (Li et al., 2014Li H, Fa Wei, Zhang Jie, Wang Nan, Li Qiang and Huan C (2014) Soybean adverse situation induced gene promoter and application thereof. In Google Patents. CN 103820451 A. Issued May 28, 2014.; Zhang et al., 2014Zhang J, Chen S, Li Q, Ma B, Liu Y, Zhang W, Lin Q and He S (2014). Soybean derived drought induced type promoter GmMYB363P and application thereof. In Google patent. CN 104152454 A. Issued Nov.19, 2014.), have been characterized. Among gene promoters previously characterized as drought-responsive, the AtRD29A promoter showed stronger activity during water deficit stress in transgenic plants when compared to the control plants (Yamaguchi-Shinozaki and Shinozaki, 1993Yamaguchi-Shinozaki K and Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331-340.). Therefore, it has been successfully used to drive the expression of drought-resistant genes in different plant species (Kasuga et al., 1999Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K and Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotech 17:287-291.; Polizel et al., 2011Polizel AMM, Nakashima K, Yamanaka N, Farias JR, de Oliveira MC, Marin SR, Abdelnoor RV, Marcelino-Guimarães FC, Fuganti R, Rodrigues FA, et al. (2011) Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A: AtDREB1A for the improvement of drought tolerance. Genet Mol Res 4:3641-3656.; Datta et al., 2012Datta K, Baisakh N, Ganguly M, Krishnan S, Yamaguchi Shinozaki K and Datta SK (2012) Overexpression of Arabidopsis and rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol J 10:579-586.; Saint Pierre et al., 2012Saint Pierre C, Crossa JL, Bonnett D, Yamaguchi-Shinozaki K and Reynolds MP (2012) Phenotyping transgenic wheat for drought resistance. J Exp Bot 63:1799-1808.; Bihmidine et al., 2013Bihmidine S, Lin J, Stone JM, Awada T, Specht JE and Clemente TE (2013) Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237:55-64.; Engels et al., 2013Engels C, Fuganti-Pagliarini R, Marin SRR, Marcelino-Guimarães FC, Oliveira MCN, Kanamori N, Mizoi J, Nakashima K, Yamaguchi-Shinozaki K and Nepomuceno AL (2013) Introduction of the rd29A: AtDREB2A CA gene into soybean (Glycine max L. Merril) and its molecular characterization in leaves and roots during dehydration. Genet Mol Biol 36:556-565.). However, the use of the AtRD29A promoter is restricted by patent protection (Shinozaki et al., 2007Shinozaki K, Seki M and Nanjo T (2007) Environmental stress responsive promoter. In: Google Patent. US 20070006348 A1. Issued Jan 4, 2007.). Moreover, soybean promoters could be an alternative to avoid the use of exotic DNA in soybean transgenic plants. Therefore, the isolation and characterization of new drought-responsive promoters from soybean are crucial and will augment the set of tools available for the development of drought-tolerant cultivars.
In previous studies, we identified a soybean drought stress-responsive gene, the α-galactosidase gene (GlymaGAL) and characterized its expression pattern in sensitive (BR 16) and tolerant (EMBRAPA 48) soybean cultivars under drought stress conditions in the soil or hydroponic systems. GlymaGAL showed high expression levels, especially in the leaves of a drought-tolerant cultivar (EMBRAPA48) submitted to water deprivation (Guimarães-Dias et al., 2012Guimarães-Dias F, Neves-Borges AC, Viana AAB, Mesquita RO, Romano E, de Fátima Grossi-de-Sá M, Nepomuceno AL, Loureiro ME and Alves-Ferreira M (2012) Expression analysis in response to drought stress in soybean: Shedding light on the regulation of metabolic pathway genes. Genet Mol Biol 35:222-232.; Guimaraes-Dias F, 2013, PhD Thesis, Federal University of Rio de Janeiro, Rio de Janeiro). At3g57520 (AtSIP2), the putative Arabidopsis homolog of GlymaGAL, was originally annotated as raffinose synthase (Taji et al., 2002Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K and Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417-426.). However, recently, AtSIP2 was characterized as an alkaline α-galactosidase belonging to the raffinose degradation pathway (Peters et al., 2010Peters S, Egert A, Stieger B and Keller F (2010) Functional identification of Arabidopsis ATSIP2 (At3g57520) as an alkaline α-galactosidase with a substrate specificity for raffinose and an apparent sink-specific expression pattern. Plant Cell Physiol 51:1815-1819.). Studies have shown that the raffinose oligosaccharide family is present in large quantities in legumes and is involved in seed desiccation, cold and drought responses (Sengupta et al., 2015Sengupta S, Mukherjee S, Basak P and Lahiri Majumder A (2015) Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Frontiers Plant Sci 6:656-667.). In our current study, we isolated and characterized in silico the 1004-bp (pGAL-1kb) and 2010-bp (pGAL-2kb) sequences upstream to the start codon of the GlymaGAL. Moreover, we evaluated their activities in the leaves and roots of Arabidopsis transgenic plants and soybean transgenic roots under drought and osmotic stress.
Material and Methods
Cis-element analyses
The genomic sequences of pGAL-1kb and pGAL-2kb, which are upstream of the start codon of the soybean α-galactosidase gene of the soybean genome (GlymaGAL), were obtained using the genome browser tool in the Phytozome database v.9 (Goodstein et al., 2012Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40:D1178-D1186.). The cis-regulatory elements related to the response to water deficit were selected based on information from the literature (Urao et al., 1993Urao T, Yamaguchi-Shinozaki K, Urao S and Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529-1539.; Baker et al., 1994Baker SS, Wilhelm KS and Thomashow MF (1994) The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24:701-713.; Yamaguchi-Shinozaki and Shinozaki, 1994Yamaguchi-Shinozaki K and Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6:251-264.; Iwasaki et al., 1995Iwasaki T, Yamaguchi-Shinozaki K and Shinozaki K (1995) Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet 247:391-398.; Abe et al., 1997Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D and Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859-1868.; Busk and Pagès, 1998Busk PK and Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425-435.; Abe et al., 2003Abe H, Urao T, Ito T, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63-78.; Dubouzet et al., 2003Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751-763.; Narusaka et al., 2003Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal 34:137-148.; Tran et al., 2004Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481-2498.; Behnam et al., 2013Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.) (Table 1). The presence and frequency of these cis-elements in the pGAL-1kb and pGAL-2kb genes were determined by the PLACE database (Plant Cis Program-acting Regulatory) (Higo et al., 1999Higo K, Ugawa Y, Iwamoto M and Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297-300.).
Isolation and cloning of the GlymaGAL promoters of soybean
The promoter sequences of the GlymaGAL, pGAL-1kb and pGAL-2kb genes were amplified using specific primers (Table S1). The genomic DNA used as a template was extracted from a drought-tolerant soybean cultivar (EMBRAPA48) according to a CTAB-based protocol (Doyle and Doyle, 1987Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11-15.). The amplification reactions were performed in a 50 μL final volume, which contained 100 ng of template DNA, 0.3 μM of each primer, 2 mM of MgSO4, 0.3 mM of each dNTP, 1X Pfx amplification buffer and 1 U of high fidelity platinum Pfx DNA polymerase (Thermo Fisher Scientific, Carlsbad, CA, EUA) according to the manufacturer's instructions. The reaction mixtures were submitted to the following cycling steps: 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C or 55 °C (for pGAL-1kb and pGAL-2kb, respectively) for 1 min and extension at 68 °C for 2 min. The thermal profile ended with a final extension at 68 °C for 5 min. The PCR products were analyzed by 1% agarose gel electrophoresis and visualized by UV fluorescence after staining with ethidium bromide. Next, the PCR products were purified using a DNA Clean and Concentrator kit (Zymo Research CA, USA) according to the manufacturer's instructions. The concentration and purity analyses of each purified DNA were evaluated by a NanoDrop™ spectrophotometer ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA).
The purified products were first cloned into a gateway pENTR/D-TOPO entry vector (Life Technologies, Victoria, Australia) and used in the transformation of TOP10 chemically competent Escherichia coli cells according to the manufacturer's instructions, generating pGAL-1kb::pENTR and pGAL-2kb::pENTR. The plasmids of the positive colonies were extracted by a GeneJet Plasmid Miniprep kit (Fermentas, Glen Burnie, MD, USA) according to the manufacturer's instructions, and the concentration and purity of each purified DNA were analyzed by a spectrophotometer. At least three clones were sequenced, and the sequence was compared with the expected promoter sequence in the soybean reference genome (Goodstein et al., 2012Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40:D1178-D1186.).
The pGAL-1kb::pENTR and pGAL-2kb::pENTR constructs were subsequently cloned by recombination into the binary vector Gateway® pKGWFS7 (Invitrogen, Carlsbad, CA), which contained the uidA (encoding the GUS α, β-glucuronidase) and the EGFP (encoding the enhanced green fluorescent) gene sequences, generating the pGAL-1kb::pKGWFS7 and pGAL-2kb::pKGWFS7 clones (with the GUS and EGFP genes driven by pGAL-1kb and pGAL-2kb promoters, respectively). The recombination reactions were performed in a final volume of 8 μL according to the manufacturer's instructions under the following conditions: 1 μL of LR Clonase II (Invitrogen), 150 ng of pGAL-1kb::pENTR or pGAL-2kb::pENTR entry clones, 150 ng of the destination vector pKGWFS7 and TE buffer (1 mM EDTA, 10 mM Tris-HCl). After incubating each mixture for 1 h at 25 °C, 1 μL of proteinase k solution (2 μg/μL) was added, followed by incubation for 10 min at 37 °C. Next, 2 μL of each reaction was used to transform TOP10 chemically competent E. coli cells. Then, positive clones of each construct were confirmed by colony PCR reactions (using promoter-specific forward primers and the EGFP-specific reverse primer) (Table S1). The plasmids of these positive colonies were extracted by the GeneJet Plasmid Miniprep kit (Fermentas) according to the manufacturer's instructions and sequencing (Macrogen, Gasan-dong, South Korea). The concentration and quality analyses of each purified DNA were evaluated by a spectrophotometer.
The AtRD29A (positive control) and CaMV35S (negative control) promoter sequences (Yamaguchi-Shinozaki and Shinozaki, 1993Yamaguchi-Shinozaki K and Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331-340.) were cloned using the same vectors and methods described above to the pGAL constructs, generating the RD29A::pKGWFS7 and 35S::pKGWFS7 clones, respectively (with GUS and EGFP coding sequences submitted to control of the RD29A and 35S promoters, respectively). We also used the DR5::GUS construct as a negative control (with the uidA gene driven by the DR5 promoter) (Chen et al., 2013Chen Y, Yordanov YS, Ma C, Strauss S and Busov VB (2013) DR5 as a reporter system to study auxin response in Populus. Plant Cell Rep 32:453-463.).
Transformation and selection of transgenic plants
The pGAL-1kb::pKGWFS7, pGAL-2kb::pKGWFS7, pRD29A::pKGWFS7 and pDR5::GUS constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation, which was subsequently transferred into Arabidopsis thaliana ecotype Columbia (wild-type) by the floral dip method (Clough and Bent, 1998Clough SJ and Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735-743.). The respective transformed plants, pGAL-1kb::GUS, pGAL-2kb::GUS, pRD29A::GUS and pDR5::GUS were grown in a pot containing substrate, vermiculite and perlite (2: 1: 0.5) at a controlled temperature of 22 °C ± 2 under a 16-h light/8-h dark photoperiod with a light intensity of 100 μmol m−2.s −1 and 60% relative humidity.
Transgenic seeds with a single T-DNA were selected by segregation rates on one-half MS medium (Murashige and Skoog, 1962Murashige T and Skoog F (1962) A revised medium for a rapid growth and bioassays with tobacco tissues cultures. Plant Physiol 15:473-479.) agar plates containing 50 μg/mL kanamycin and maintained under the same conditions as mentioned above. The T1, T2 and T3 plantlets were transferred to soil and maintained under the same conditions until the seeds were collected. Then, three T3 homozygous transgenic lines expressing each construct were employed for abiotic stress treatments.
The pGAL-2kb::pKGWFS7 and 35S::pKGWFS7 constructs were introduced into Agrobacterium rhizogenes K599 by electroporation, which was subsequently used to transform roots of the tolerant soybean cultivar (Embrapa 48) using the syringe method (Kuma et al., 2015Kuma KM, Lopes-Caitar VS, Romero CCT, Silva SMH, Kuwahara MK, Carvalho MCCG, Abdelnoor RV, Dias WP and Marcelino-Guimarães FC (2015) A high efficient protocol for soybean root transformation by Agrobacterium rhizogenes and most stable reference genes for RT-qPCR analysis. Plant Cell Rep 34:1987-2000.). After seven days of co-cultivation, the plants were transferred into a selective medium of 100 μg/mL of cefotaxime and 100 μg/mL of spectinomycin, maintained under a 16-h light/8-h dark photoperiod and cycled at 25 °C ± 2 for 10 days. Transformation was confirmed through a visual inspection of EGFP expression using a fluorescence stereomicroscope (Leica M205 FA). Non-fluorescent roots were not excised to avoid injury. Finally, three transgenic plants for each construct (pGAL-2kb::GUS and 35S::GUS) were employed for abiotic stress treatments.
Abiotic stress treatments
The activities of the pGAL-2kb, pGAL-1kb, pRD29A and pDR5 promoters in the transgenic Arabidopsis plants under water privation stress were evaluated before and after salt stress, air-dried and polyethylene glycol (PEG) assays. In all experiments, the seeds harvested from pGAL-1kb::GUS (lines L1, L2, L3), pGAL-2kb::GUS (lines L1, L2, L3), RD29::GUS and DR5::GUS transgenic plants were initially surface sterilized and maintained in 4 °C for 4 days to break dormancy. Then, approximately 100 seeds for each line/treatment were germinated on one half-strength MS medium 1.2% agar in plates (150 mm diameter), which were positioned vertically and cultivated until 15 days old. They were grown under a continuous light photoperiod with a light intensity of 100 μmol m−2 s −1, cycled at 22 °C ± 2 and 60% relative humidity and then submitted to four different treatments.
In the drought treatment by air drying (air-dried), a set of plates harboring 20 seedlings of each Arabidopsis transgenic line remained open for 6, 12 or 24 h (Rodrigues et al., 2012Rodrigues FA, Marcolino-Gomes J, de Fátima Corrêa Carvalho J, do Nascimento LC, Neumaier N, Farias JRB, Carazzolle MF, Marcelino FC and Nepomuceno AL (2012) Subtractive libraries for prospecting differentially expressed genes in the soybean under water deficit. Genet Mol Biol 35:304-314.; Nobres et al., 2016Nobres P, Patreze CM, Waltenberg FP, Correa MF, Tavano ECDR, Mendes BMJ and Alves-Ferreira M (2016) Characterization of the promoter of the homeobox gene CaHB12 in Coffea arabica. Trop Plant Biol 9:50-62.). Meanwhile, a set of plates with control plants remained closed. For the soybean transgenic roots submitted to the air-dried treatment, a set of Magenta vessel GA-7 (Sigma) harboring 15 soybean plants remained open for 24 h, while a set of Magenta vessels with control plants remained closed.
In the PEG treatment, 15 seedlings of each Arabidopsis transgenic line were transferred to plates containing PEG 8000 (Verslues et al., 2006Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J and Zhu J-K (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523-539.). In this case, after the seeds germinated on one half-strength MS medium agar until 15 days old, the seedlings were transferred to plates (150 mm diameter) with one half-strength MS medium agar supplement (700 g PEG 8000 diluted in 1 L of water), submitting the seedlings to a water potential (Ψ) = −1.7 MPa. Meanwhile, control plants were maintained in a PEG-free solution. The PEG-infused plates were incubated overnight (approximately for 15 hours) before the plants were transferred.
Subsequently, the salt-stress treatments with 20 mL of 200 mM NaCl were added to cultivation medium harboring 20 seedlings of each transgenic line (Soussi et al., 1998Soussi M, Ocaña A and Lluch C (1998) Effects of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J Exp Bot 49:1329-1337.), which were submitted to this condition for 6, 12 or 24 h. Control plants were maintained in a NaCl-free solution.
In all treatments, control plants were collected at the end of the experiment to rule out external changes or influences in the results. After the stress treatments, qualitative and quantitative GUS analyses were performed. All treatments were conducted with three independent biological replicates.
Histochemical GUS assay of Arabidopsis transgenic plants
The histochemical GUS assay was performed after stress application in the transgenic seedlings for 6, 12 or 24 h in the air-dried treatment, PEG treatment or salt treatments. The histochemical GUS assay was performed following the methods of Jefferson (1989Jefferson RA (1989) The GUS reporter gene system. Nature 342:837-838.) to assess the promoter activity in roots and leaves of the treated seedlings. The seedlings were observed under a stereomicroscope (Leica S8 APO) with a magnification of 10X and photographed by a Leica EC3 camera, and the image was adjusted in high resolution using LASEZ software version 3.0 (Leica). The histochemical GUS assay was performed with three independent biological replicates and three plants for each line.
Total RNA isolation and transcript level analysis
To validate the results obtained by histochemical GUS assay and to quantitatively analyze the pGAL-2kb activity, qPCR assays were performed using root and leaf RNA samples of three lineages (pGAL-2kb::GUS (L1, L2 and L3), pRD29A::GUS (positive control) and pDR5::GUS (negative control)) submitted to the air-dried, PEG or salt stress conditions at 24 h and the non-stressed condition (control plants). The leaf RNA samples of each line/treatment (a total of five plants for each line/treatment) were extracted with Trizol Reagent (Invitrogen), followed by TURBO DNase enzyme (Ambion, Thermo Fisher) treatment according to the manufacturer's instructions. Meanwhile, the root RNA samples each line/treatment (a total of 10 plants for each line/treatment) were extracted using the RNeasy Plant Mini Kit (Qiagen, Inc., Valencia, CA, USA) followed by DNase treatment, as indicated by the manufacturer's protocol. The total RNA concentration and purity were determined using a spectrophotometer, while the RNA integrity was tested by electrophoretic separation in a 1% agarose gel. Three independent biological samples were collected for the relative expression studies.
The cDNA synthesis reactions with leaf and root RNA samples were initially performed using the SuperScript III Reverse Transcriptase enzyme (Invitrogen) following the manufacturer's instructions. The resulting cDNAs were used for qPCR assays run in triplicate. For the qPCR reactions, the uidA (GUS) gene-specific primers were used (Table S1).
The qPCRs were carried out in optical 96-well plates in a 7500 Fast Real-Time PCR detection system (Applied Biosystems) following the manufacturer's instructions. The amplification reactions were performed in a 20 μL final volume containing 10 μL cDNA (1:50); SYBER® Green 1X (Thermo Fisher Scientific); 0.4 μM of each primer (forward and reverse) (Síntese biotecnologia); 0.025 mM dNTP; PCR buffer (-Mg) 1X; 3 mM MgCl2; 0.25 U Platinum Taq DNA Polymerase (Thermo Fisher Scientific) and 0.4 μL ROX reference dye (Thermo Fisher Scientific).
The reaction mixtures were incubated at 94 °C for 5 min, followed by 40 cycles of 94 °C for 15 s, 60 °C for 10 s, 72 °C for 15 s and 60 °C for 35 s. Subsequently, a melting curve analysis was run from 30 °C to 100 °C for 1 min.
The melting curve and gel electrophoresis analyses of the amplification products confirmed that the primers amplified only a single product of expected size (data not shown). The primer set efficiencies and the Ct cutoff cycles (cycle thresholds) were estimated for each experimental set by Online real-time PCR Miner software (Zhao and Fernald, 2005Zhao S and Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comp Biol 12:1047-1064.), and these values were converted into normalized relative quantities (NRQs) by the program QBASE version 1.3.5 (Hellemans et al., 2007Hellemans J, Mortier G, De Paepe A, Speleman F and Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19.1-R19.14.). To determine the most stable combination of the reference genes At4g34270, At4g38070 and At5g12240 (Czechowski et al., 2005Czechowski T, Stitt M, Altmann T, Udvardi MK and Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5-17.) we used NormFinder software (Andersen et al., 2004Andersen CL, Jensen JL and Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245-5250.). As the At4g34270 and At4g38070 genes exhibited a stable expression pattern under abiotic stress, they were used as housekeeping genes for the normalization of uidA expresion. The quantitative expression data were analyzed statically by Student's t-test and variance analysis (ANOVA) methods using Assistat v 7.7 software (Silva and Azevedo, 2002Silva FAS and Azevedo CAV (2002) Versão do programa computacional Assistat para o sistema operacional Windows. Rev Bras Prod Agroindustr 4:71-78.).
Results
In silico analysis of the frequency of water deficit response cis-elements
In silico analysis of the 2-kb fragment of the GlymaGAL promoter allowed us to identify 13 cis-acting elements previously associated with the water deficit response (Table 1) (Urao et al., 1993Urao T, Yamaguchi-Shinozaki K, Urao S and Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529-1539.; Baker et al., 1994Baker SS, Wilhelm KS and Thomashow MF (1994) The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24:701-713.; Yamaguchi-Shinozaki and Shinozaki, 1994Yamaguchi-Shinozaki K and Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6:251-264.; Iwasaki et al., 1995Iwasaki T, Yamaguchi-Shinozaki K and Shinozaki K (1995) Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet 247:391-398.; Abe et al., 1997Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D and Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859-1868.; Busk and Pagès, 1998Busk PK and Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425-435.; Abe et al., 2003Abe H, Urao T, Ito T, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63-78.; Dubouzet et al., 2003Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751-763.; Narusaka et al., 2003Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal 34:137-148.; Tran et al., 2004Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481-2498.; Behnam et al., 2013Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.). The presence and frequency of these cis-elements upstream of the start codon of GlymaGAL revealed that the pGAL-1kb and pGAL-2kb sequences have several putative water deficit response cis-acting elements (Figure 1). Among the 13 different cis-acting elements identified as associated with the drought response, only the ABREATRD22 motif was not found in the GlymaGAL promoter. However, the frequency of cis-elements was distinct over each promoter fragment (Table 1, Figure 1). The DRE, MYCATERD1, ABRE and GBOX motifs were found exclusively in pGAL-2kb (Table 1).
Genomic DNA sequence and different segments with different cis-elements in the drought-stress response of the promoter of the GlymaGAL gene from soybean. The immediate upstream nucleotide of the start codon of GlymaGAL is designated as position 1. The sequence represents a single-stranded DNA.
Activity profile of the pGAL-1kb and pGAL-2kb promoters in transgenic Arabidopsis under water stress conditions
Three plants of each transgenic lineage were evaluated for the respective promoter expression level by histochemical GUS assay and photographed in bright field microscopy after 6, 12 and 24 h of the air-dried, PEG and salt stress treatments. Plants bearing the pRD29A::GUS and pDR5::GUS constructs were used as positive and negative controls, respectively (Figures 2-5).
pGAL-1kb::GUS histochemical assay. Three transgenic lineages, pGAL-1kb::GUS (L1, L2 and L3), pRD29::GUS positive control and pDR5::GUS negative control, were submitted to control and 6, 12 and 24 h of the air-dried, salt stress and PEG conditions. The order of the sample photos is as follows: root, cotyledonal leaf, young leaf and totally expanded leaf. The data shown are representative of three independent lines (n = 3). Scale bars = 2 mm.
Histochemical analysis and expression profile analyses of the GUS reporter gene under control of the pGAL-2kb promoter. The order of the sample photos is as follows: root, cotyledonal leaf, young leaf and totally expanded leaf. A) The leaves and roots of plants grown under normal conditions and air-dried treatment (6, 12 and 24 h) were compared to the pRD29::GUS and pDR5::GUS plants under normal conditions and treated (6, 12 and 24 h). The data shown are representative of three independent lines (n = 3). B and C). The expression levels of uidA mRNA in leaves and roots under control (non-treated) and air-dried treatment. Values are means ± SD (n = 3). The relative expression values, represented on the y-axis, were obtained by qPCR experiments and calculated using the 2−ΔΔCt method. The At4g34270 and At4g38070 genes were used as endogenous controls to normalize data. Asterisks indicate significant differences of samples under air-dried treatment and non-treatment (Student's t-test * p < 0.05 and ** p < 0.01). Total RNA was extracted from the leaves and roots of three independent T3 lines of 2-week-old transgenic plants after 24 h of drought (air-dried) treatment. Scale bars = 2 mm.
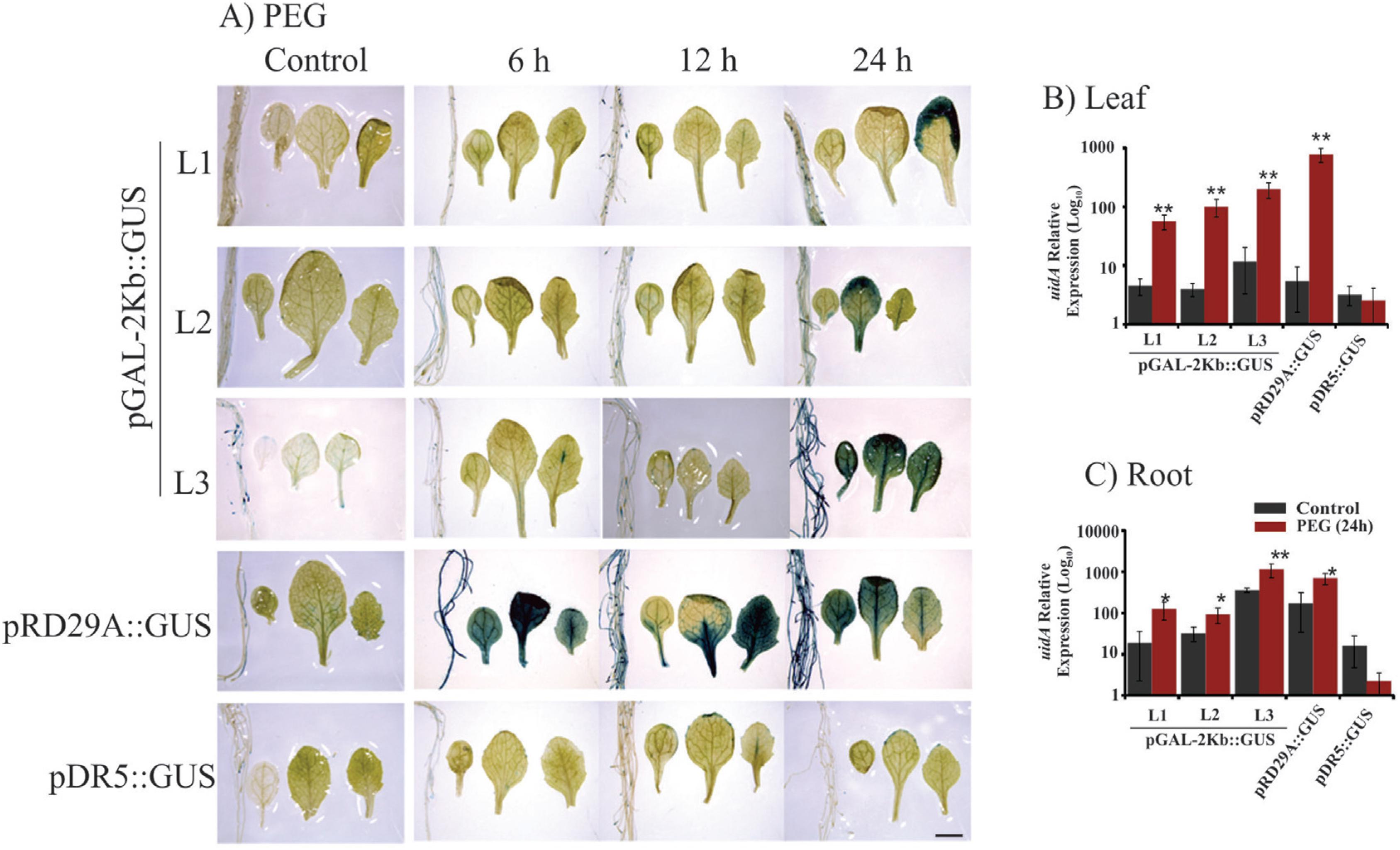
Histochemical analysis and expression profile analyses of the GUS reporter gene under control of the pGAL-2kb promoter. The order of the sample photos is as follows: root, cotyledonal leaf, young leaf and totally expanded leaf. A) The leaves and roots of plants grown under normal conditions and PEG treatment (6, 12 and 24 h) were compared to the pRD29::GUS and pDR5::GUS plants under normal conditions and treated conditions (6, 12 and 24 h). The data shown are representative of three independent lines (n = 3). B and C) The expression levels of uidA mRNA in leaves and roots under PEG treatment and no treatment. Values are means ± SD (n = 3). The relative expression values, represented on the y-axis, were obtained by qPCR experiments and calculated using the 2−ΔΔCt method. The At4g34270 and At4g38070 genes were used as endogenous controls to normalize data. Asterisks indicate significant differences of samples under PEG treatment and no treatment (Student's t-test * p < 0.05 and ** p < 0.01). Total RNA was extracted from the leaves and roots of three independent T3 lines of 2-week-old transgenic plants after 24 h of PEG treatment. Scale bars = 2 mm.
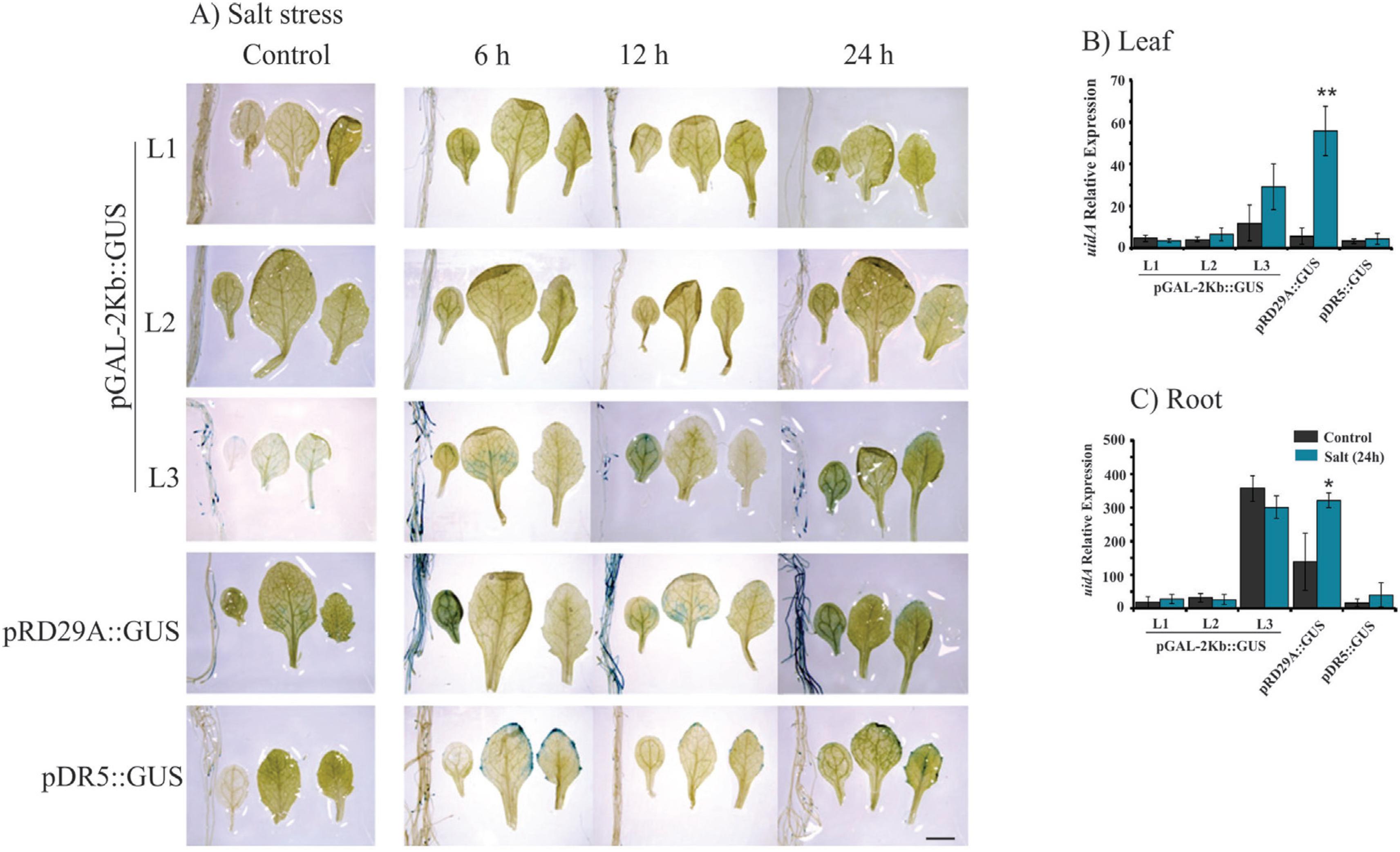
Histochemical analysis and expression profile analyses of the GUS reporter gene under control of the pGAL-2kb promoter. The order of the sample photos is as follows: root, cotyledonal leaf, young leaf and totally expanded leaf. A) The leaves and roots of plants grown under normal conditions and salt treatment (6, 12 and 24 h) were compared to the pRD29::GUS and pDR5::GUS plants under normal conditions and treated conditions (6, 12 and 24 h). The data shown are representative of three independent lines (n = 3). B and C) The expression levels of uidA mRNA in leaf and roots samples under control (non-treated) and salt treatment. Values are means ± SD (n = 3). The relative expression values, represented on the y-axis, were obtained by qPCR experiments and calculated using the 2−ΔΔCt method. The At4g34270 and At4g38070 genes were used as endogenous controls to normalize data. Asterisks indicate significant differences of samples under salt treatment and no treatment (Student's t-test * p < 0.05 and ** p < 0.01). Total RNA was extracted from the leaves and roots of three independent T3 lines of 2-week-old transgenic plants after 24 h of salt treatment. Scale bars = 2 mm.
Plants bearing the pGAL-1kb::GUS construct did not show any GUS activity when submitted to stress treatments (Figure 2). However, the pGAL-2kb::GUS transgenic plants submitted to the air-dried treatments for 24 h showed a strong GUS activity in roots and leaves. However, no signal was observed in these plants at earlier time points (6 and 12 h) (Figure 3A). PEG treatment of pGAL-2kb::GUS transgenic plants triggered the same temporal expression pattern as the air-dried treatment; GUS activity was observed after 24 h (Figure 4A). qPCR analysis of pGAL-2kb transgenic plants substantiated the histochemical assays, showing that the uidA gene was significantly up-regulated (Student's t-test * p < 0.05 and ** p < 0.01) in the leaves and roots of transgenic lines when submitted to the air-dried (Figure 3B, C) and PEG treatments after 24 h (Figure 4B, C). In contrast, no signal was observed in pGAL-2kb::GUS transgenic plants under salt stress in leaves and roots (Figure 5A). Again, this result was confirmed by qPCR (Figure 5B, C).
Activity profile of the pGAL-2kb soybean promoter in soybean transgenic roots under drought stress conditions
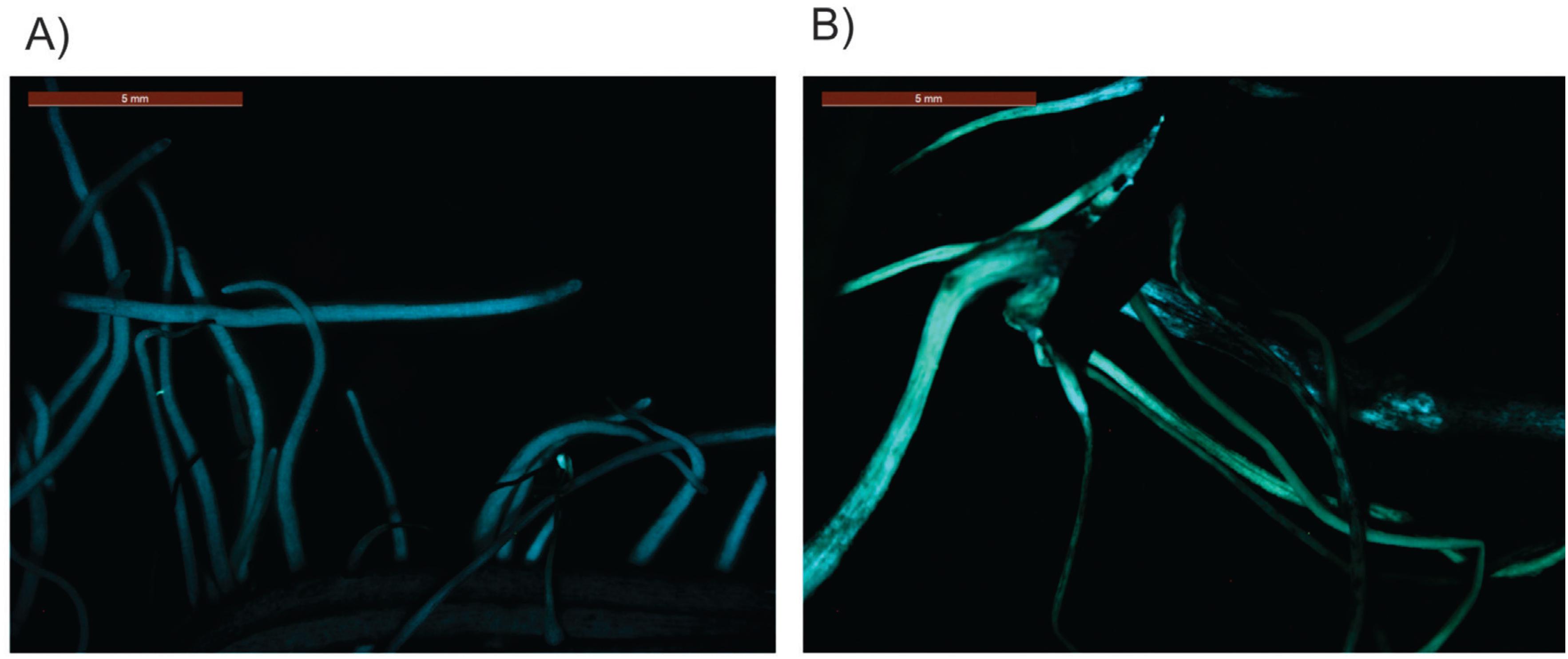
To investigate whether the pGAL::2kb promoter is also active in soybean plants, we performed a soybean root transformation according to Kuma et al. (2015)Kuma KM, Lopes-Caitar VS, Romero CCT, Silva SMH, Kuwahara MK, Carvalho MCCG, Abdelnoor RV, Dias WP and Marcelino-Guimarães FC (2015) A high efficient protocol for soybean root transformation by Agrobacterium rhizogenes and most stable reference genes for RT-qPCR analysis. Plant Cell Rep 34:1987-2000.. Transformed roots submitted to the air-dried treatment showed a substantial increase in the EGFP signal when compared to untransformed roots under the same stress condition (Figure 6).
Expression EGFP in A. rhizogenes transformed soybean roots submitted to 24 h of air- dried treatment. (A) 35S::GUS (B) pGAL-2kb::GUS.
Discussion
In this study, we evaluated the activity of pGAL-1kb and pGAL-2kb promoter sequences of the soybean α-galactosidase gene in Arabidopsis and soybean transgenic plants under drought and salt stress. The pGAL-2kb promoter had high activity in the roots of Arabidopsis and soybean transgenic plants submitted to water deficit by the air-dried treatment (Figure 3 and 6). Similar results were also observed in Arabidopsis leaves submitted to the air-dried and PEG treatments (Figure 3 and 4). However, the pGAL-2kb promoter sequence did not show a significant change in the activity of Arabidopsis transgenic plants under salt stress conditions (Figure 5). These results showed that the pGAL-2kb promoter sequence is able to drive inducible expression primarily by drought stress.
Our results showed that the pGAL-1kb sequence lacks activity under the tested conditions, which is likely due to the absence of specific cis-elements involved in controlling the dehydration stress response (Figure 2). Other authors have also shown the importance of distal promoter regions in stress-associated responses (Behnam et al., 2013Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.; Imtiaz et al., 2015Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J and Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from Chrysanthemum in Arabidopsis. Plant Mol Biol 89:1-19.). For example, the 3.0-kb AtNCED3 promoter sequence was able to drive uidA expression in response to dehydration, while the 1.5-kb AtNCED3 promoter region was not functional (Tan et al., 2003Tan B-C, Joseph LM, Deng W-T, Liu L, Li Q-B, Cline K and McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35:44-56.; Behnam et al., 2013Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.). Imtiaz et al. (2015)Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J and Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from Chrysanthemum in Arabidopsis. Plant Mol Biol 89:1-19. showed that the 2.7-kb CmBBX24 promoter sequence of Chrysanthemum has activity in Arabidopsis transgenic plants under drought and salt stress, but the deletion of fragments from the promoter's distal end led to a lower promoter activity under drought stress. When compared to the pGAL-1kb promoter sequence, pGAL-2kb has four additional cis-motifs previously associated with the response to drought stress, including one ABRE, one MYCATERD1, one GBOX and one DRE (Table 1). Previous results have shown that the presence of these cis-elements has been associated with the response to dehydration and/or abiotic stresses (Abe et al., 1997Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D and Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859-1868.; Narusaka et al., 2003Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal 34:137-148.; Lata and Prasad, 2011Lata C and Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62:4731-4748.; Behnam et al., 2013Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.). However, only experimental data can validate the role of these cis-elements in the GlymaGAL promoter.
Another possible explanation for the lack of activity of the pGAL-1kb sequence is the frequency and the spatial organization of its cis-elements. Furthermore, the promoter activity analysis of pGAL-1kb was carried out in a heterologous system, which may also explain the negative result. Soybean and A. thaliana evolutionarily split 90 million years ago, so cis-elements and transcription factors and promoter organization is likely to differ between these species and to interfere in the regulation of pGAL-1kb (Shoemaker et al., 1996Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, et al. (1996) Genome duplication in soybean (Glycine subgenus Soja). Genetics 144:329-338.; Mühlhausen et al., 2013Mühlhausen A, Lenser T, Mummenhoff K and Theißen G (2013) Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J 73:824-835.).
The use of the drought stress-responsive promoter originally isolated from the soybean genome may have advantages as a biotechnological tool to improve drought stress tolerance in soybean cultivars. Its use may minimize the unexpected promoter activity caused by cryptic cis-elements introduced with the use of foreign DNA in soybean transgenic cultivars. In addition to pGAL-2kb, two other soybean promoters, GmNCED1 and GmMYB363, which present activity in response to abiotic stresses, were recently characterized and patented (Li et al., 2014Li H, Fa Wei, Zhang Jie, Wang Nan, Li Qiang and Huan C (2014) Soybean adverse situation induced gene promoter and application thereof. In Google Patents. CN 103820451 A. Issued May 28, 2014.; Zhang et al., 2014Zhang J, Chen S, Li Q, Ma B, Liu Y, Zhang W, Lin Q and He S (2014). Soybean derived drought induced type promoter GmMYB363P and application thereof. In Google patent. CN 104152454 A. Issued Nov.19, 2014.).
The activity of the GmNCED1 promoter was significantly induced by salt stress in the roots of tobacco transgenic plants (Li et al., 2014Li H, Fa Wei, Zhang Jie, Wang Nan, Li Qiang and Huan C (2014) Soybean adverse situation induced gene promoter and application thereof. In Google Patents. CN 103820451 A. Issued May 28, 2014.). The GmMYB363 promoter activity was induced in soybean transgenic roots under PEG 6000 treatment (Zhang et al., 2014Zhang J, Chen S, Li Q, Ma B, Liu Y, Zhang W, Lin Q and He S (2014). Soybean derived drought induced type promoter GmMYB363P and application thereof. In Google patent. CN 104152454 A. Issued Nov.19, 2014.). Compared to pGmNCED1 and pGmMYB363, the soybean pGAL-2kb promoter is active in roots as well as in leaves during drought stress. The promoter activity in both organs may represent an advantage to provide wide and effective protection under water deficit when driving the expression of drought-tolerant genes in transgenic plants. Nevertheless, pGmMYB363 and pGmNCED1 have shorter sequences than the pGAL-2kb promoter (1,384 and 1,253 bp, respectively), which is an important characteristic. However, we identified a high number of cis-elements in the pGAL-2kb promoter between 1000 and 1500 bp upstream of the start codon. Furthermore, we did not find cis-elements in the distal region between 1,500 and 2,000 bp. Therefore, it is important to evaluate the 1.5-kb sequence of the pGAL promoter to improve its use as a biotech tool. In addition, other constructs containing pGAL-2kb deletions may also be active as drought-responsive, as efficient as the entire sequence. Imtiaz et al. (2015)Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J and Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from Chrysanthemum in Arabidopsis. Plant Mol Biol 89:1-19. showed that the deletion of the 1,162-bp fragment between −2,552 (full promoter) and −1,390 significantly reduced promoter activity in leaves and roots, while no significant decrease in promoter activity was observed with further deletions from −1,390 to −780, −780 to 600 and −600 to −480. Thus, further analyses will aid in better characterizing the use of the pGAL-2kb promoter as a new drought stress-responsive promoter. Here we showed that the full-length pGAL-2kb sequence is a potential candidate for use in genetic engineering to induce a response to drought stress in soybean.
Acknowledgments
We are grateful to EMBRAPA-CENARGEN and EMBRAPA-Soja (Brazilian Agricultural Research Corporation) for supplying RNA and soybean seeds. This work was supported by CNPq (National Council for Scientific and Technological Development), CAPES (Coordination for the Improvement of Higher Education Personnel/Ministry of Education) and FAPERJ (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Grant E-26/110.847/2010) and an award from CNPq (Grant 307376/2013-3) to MAF.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D and Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859-1868.
- Abe H, Urao T, Ito T, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63-78.
- Andersen CL, Jensen JL and Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245-5250.
- Baker SS, Wilhelm KS and Thomashow MF (1994) The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24:701-713.
- Banerjee J, Sahoo DK, Dey N, Houtz RL and Maiti IB (2013) An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and tobacco plants. PLoS ONE 8:e79622.
- Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M and Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20:315-324.
- Bhauso TD, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR, Patel K and Rajam MV (2014) Overexpression of bacterial mtlD gene in peanut improves drought tolerance through accumulation of mannitol. Sci World J 2014:1-10.
- Bihmidine S, Lin J, Stone JM, Awada T, Specht JE and Clemente TE (2013) Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237:55-64.
- Busk PK and Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425-435.
- Chan Ju L, Ha Yeon L, Woong Bom K, Bok-Sim L, Jungeun K, Raza A, Hyun AK, So Young Y, Cheol-Goo H and Suk-Yoon K (2012) Screening of tissue-specific genes and promoters in tomato by comparing genome wide expression profiles of Arabidopsis orthologues. Mol Cells 34:53-59.
- Chen Y, Yordanov YS, Ma C, Strauss S and Busov VB (2013) DR5 as a reporter system to study auxin response in Populus. Plant Cell Rep 32:453-463.
- Clough SJ and Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J 16:735-743.
- Conab (2012) Acompanhamento da Safra Brasileira de Grãos 2011/12. V: Sexto Levantamento. Conab, Brasília, 35 p.
- Czechowski T, Stitt M, Altmann T, Udvardi MK and Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5-17.
- Datta K, Baisakh N, Ganguly M, Krishnan S, Yamaguchi Shinozaki K and Datta SK (2012) Overexpression of Arabidopsis and rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol J 10:579-586.
- Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11-15.
- Du H, Shen X, Huang Y, Huang M and Zhang Z (2016) Overexpression of Vitreoscilla hemoglobin increases waterlogging tolerance in Arabidopsis and maize. BMC Plant Biol 16:35.
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751-763.
- Engels C, Fuganti-Pagliarini R, Marin SRR, Marcelino-Guimarães FC, Oliveira MCN, Kanamori N, Mizoi J, Nakashima K, Yamaguchi-Shinozaki K and Nepomuceno AL (2013) Introduction of the rd29A: AtDREB2A CA gene into soybean (Glycine max L. Merril) and its molecular characterization in leaves and roots during dehydration. Genet Mol Biol 36:556-565.
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40:D1178-D1186.
- Guimarães-Dias F, Neves-Borges AC, Viana AAB, Mesquita RO, Romano E, de Fátima Grossi-de-Sá M, Nepomuceno AL, Loureiro ME and Alves-Ferreira M (2012) Expression analysis in response to drought stress in soybean: Shedding light on the regulation of metabolic pathway genes. Genet Mol Biol 35:222-232.
- Hellemans J, Mortier G, De Paepe A, Speleman F and Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19.1-R19.14.
- Higo K, Ugawa Y, Iwamoto M and Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297-300.
- Homrich MS, Wiebke-Strohm B, Weber RLM and Bodanese-Zanettini MH (2012) Soybean genetic transformation: A valuable tool for the functional study of genes and the production of agronomically improved plants. Genet Mol Biol 35:998-1010.
- Hou J, Jiang P, Qi S, Zhang K, He Q, Xu C, Ding Z, Zhang K and Li K (2016) Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS ONE 11:e0154041.
- Hsieh T-H, Lee J-T, Yang P-T, Chiu L-H, Charng Y-Y, Wang Y-C and Chan M-T (2002) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiology 129:1086-1094.
- Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J and Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from Chrysanthemum in Arabidopsis. Plant Mol Biol 89:1-19.
- Iwasaki T, Yamaguchi-Shinozaki K and Shinozaki K (1995) Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet 247:391-398.
- Jefferson RA (1989) The GUS reporter gene system. Nature 342:837-838.
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K and Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotech 17:287-291.
- Kuma KM, Lopes-Caitar VS, Romero CCT, Silva SMH, Kuwahara MK, Carvalho MCCG, Abdelnoor RV, Dias WP and Marcelino-Guimarães FC (2015) A high efficient protocol for soybean root transformation by Agrobacterium rhizogenes and most stable reference genes for RT-qPCR analysis. Plant Cell Rep 34:1987-2000.
- Lata C and Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62:4731-4748.
- Li H, Fa Wei, Zhang Jie, Wang Nan, Li Qiang and Huan C (2014) Soybean adverse situation induced gene promoter and application thereof. In Google Patents. CN 103820451 A. Issued May 28, 2014.
- Luo X, Wu J, Li Y, Nan Z, Guo X, Wang Y, Zhang A, Wang Z, Xia G and Tian Y (2013) Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PLoS ONE 8:e54002.
- Mühlhausen A, Lenser T, Mummenhoff K and Theißen G (2013) Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J 73:824-835.
- Murashige T and Skoog F (1962) A revised medium for a rapid growth and bioassays with tobacco tissues cultures. Plant Physiol 15:473-479.
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K and Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal 34:137-148.
- Nobres P, Patreze CM, Waltenberg FP, Correa MF, Tavano ECDR, Mendes BMJ and Alves-Ferreira M (2016) Characterization of the promoter of the homeobox gene CaHB12 in Coffea arabica Trop Plant Biol 9:50-62.
- Novák J, Pavlu J, Novák O, Nozková-Hlavácková V, Spundová M, Hlavinka J, Koukalová S, Skalák J, Cerny M and Brzobohaty B (2013) High cytokinin levels induce a hypersensitive-like response in tobacco. Ann Bot 112:41-55.
- Peters S, Egert A, Stieger B and Keller F (2010) Functional identification of Arabidopsis ATSIP2 (At3g57520) as an alkaline α-galactosidase with a substrate specificity for raffinose and an apparent sink-specific expression pattern. Plant Cell Physiol 51:1815-1819.
- Polizel AMM, Nakashima K, Yamanaka N, Farias JR, de Oliveira MC, Marin SR, Abdelnoor RV, Marcelino-Guimarães FC, Fuganti R, Rodrigues FA, et al. (2011) Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A: AtDREB1A for the improvement of drought tolerance. Genet Mol Res 4:3641-3656.
- Rahman H, Ramanathan V, Nallathambi J, Duraialagaraja S and Muthurajan R (2016) Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol 16(suppl 1):7-20.
- Rippey BR (2015) The U.S. Drought of 2012. Weather and Climate Extremes 10:57-64.
- Rodrigues FA, Marcolino-Gomes J, de Fátima Corrêa Carvalho J, do Nascimento LC, Neumaier N, Farias JRB, Carazzolle MF, Marcelino FC and Nepomuceno AL (2012) Subtractive libraries for prospecting differentially expressed genes in the soybean under water deficit. Genet Mol Biol 35:304-314.
- Saint Pierre C, Crossa JL, Bonnett D, Yamaguchi-Shinozaki K and Reynolds MP (2012) Phenotyping transgenic wheat for drought resistance. J Exp Bot 63:1799-1808.
- Sengupta S, Mukherjee S, Basak P and Lahiri Majumder A (2015) Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Frontiers Plant Sci 6:656-667.
- Shinozaki K, Seki M and Nanjo T (2007) Environmental stress responsive promoter. In: Google Patent. US 20070006348 A1. Issued Jan 4, 2007.
- Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, et al. (1996) Genome duplication in soybean (Glycine subgenus Soja). Genetics 144:329-338.
- Silva FAS and Azevedo CAV (2002) Versão do programa computacional Assistat para o sistema operacional Windows. Rev Bras Prod Agroindustr 4:71-78.
- Silvente S, Sobolev AP and Lara M (2012) Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS ONE 7:e38554.
- Soussi M, Ocaña A and Lluch C (1998) Effects of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J Exp Bot 49:1329-1337.
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K and Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana Plant J 29:417-426.
- Tan B-C, Joseph LM, Deng W-T, Liu L, Li Q-B, Cline K and McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35:44-56.
- Tran L-SP and Mochida K (2010) Functional genomics of soybean for improvement of productivity in adverse conditions. Funct Integr Genomics 10:447-462.
- Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K and Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481-2498.
- Urao T, Yamaguchi-Shinozaki K, Urao S and Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529-1539.
- USDA (2016) World Agricultural Production. Foreign Agricultural Service, Washington, DC, p 5-16.
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J and Zhu J-K (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523-539.
- Wang J-Y, Wang J-P and Yang H-F (2016) Identification and functional characterization of the NAC gene promoter from Populus euphratica Planta 244:417-427.
- Withanage SP, Hossain MA, Kumar MS, Roslan HAB, Abdullah MP, Napis SB and Shukor NAA (2015) Overexpression of Arabidopsis thaliana gibberellic acid 20 oxidase (AtGA20ox) gene enhance the vegetative growth and fiber quality in kenaf (Hibiscus cannabinus L.) plants. Breed Sci 65:177-191.
- Yamaguchi-Shinozaki K and Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331-340.
- Yamaguchi-Shinozaki K and Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6:251-264.
- Yan H, Ma L, Wang Z, Lin Z, Su J and Lu B-R (2015) Multiple tissue-specific expression of rice seed-shattering gene SH4 regulated by its promoter pSH4. Rice 8:1-10.
- Zhang J, Chen S, Li Q, Ma B, Liu Y, Zhang W, Lin Q and He S (2014). Soybean derived drought induced type promoter GmMYB363P and application thereof. In Google patent. CN 104152454 A. Issued Nov.19, 2014.
- Zhao S and Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comp Biol 12:1047-1064.
- Zulauf C (2012) 2012 Drought: Yield Loss, Revenue Loss, and Harvest Price Option. Farmadocdaily. University of Illinois, Urbana-Champaign, 5 p.
Internet Resources
- NetPrimer Software, www.premierbiosoft.com/netprimer (January, 2013).
» www.premierbiosoft.com/netprimer - real-time PCR Miner software, miner.ewindup.info (March, 2015).
» miner.ewindup.info - Phytozome portal version 9, phytozome.jgi.doe.gov (January, 2013).
» phytozome.jgi.doe.gov - PLACE database, www.dna.affrc.go.jp/PLACE/ (January, 2013).
» www.dna.affrc.go.jp/PLACE/
-
Associate Editor: Marcia Pinheiro Margis
Supplementary material
The following online material is available for this article:
Publication Dates
-
Publication in this collection
27 Mar 2017 -
Date of issue
2017
History
-
Received
04 Mar 2016 -
Accepted
21 Nov 2016