Abstract
Somatic embryos of the commercial soybean (Glycine max) cultivar IAS5 were co-transformed using particle bombardment with a synthetic form of the Bacillus thuringiensis delta-endotoxin crystal protein gene cry1Ac, the beta-glucuronidase reporter gene gusA and the hygromycin resistance gene hpt. Hygromycin-resistant tissues were proliferated individually to give rise to nine sets of clones corresponding to independent transformation events. The co-bombardment resulted in a co-transformation efficiency of 44%. Many histodifferentiated embryos and 30 well-developed plants were obtained. Twenty of these plants flowered and fourteen set seeds. The integration and expression of the cry1Ac, gusA and hpt transgenes into the genomes of a sample of transformed embryos and all T0, T1, T2 and T3 plants were confirmed by Gus activity, PCR, Southern and western blot, and ELISA techniques. Two T0 plants out of the seven co-transformed plants produced seeds and were analyzed for patterns of integration and inheritance until the T3 generation. Bioassays indicated that the transgenic plants were highly toxic to the velvetbean caterpillar Anticarsia gemmatalis, thus offering a potential for effective insect resistance in soybean.
Anticarsia gemmatalis; cry1Ac gene; IAS5 cultivar; insect resistance; transgenic soybean
PLANT GENETICS
RESEARCH ARTICLE
Resistance to Anticarsia gemmatalis Hübner (Lepidoptera, Noctuidae) in transgenic soybean (Glycine max (L.) Merrill Fabales, Fabaceae) cultivar IAS5 expressing a modified Cry1Ac endotoxin
Milena Schenkel HomrichI; Luciane Maria Pereira PassagliaI; Jorge Fernando PereiraII; Paulo Fernando BertagnolliII; Giancarlo PasqualiIII; Mohsin Abbas ZaidiIV; Illimar AltosaarIV; Maria Helena Bodanese-ZanettiniI
IDepartamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil
IICentro Nacional de Pesquisa de Trigo, EMBRAPA, Passo Fundo, RS, Brazil
IIIDepartamento de Biologia Molecular e Biotecnologia, Instituto de Biociências, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil
IVBiochemistry, Microbiology and Immunology Department, University of Ottawa, Ottawa Ontario, Canada
Send correspondence to Send correspondence to: Maria Helena Bodanese-Zanettini Departamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul Caixa Postal 15053 91501-970, Porto Alegre, RS, Brazil E-mail: maria.zanettini@ufrgs.br
ABSTRACT
Somatic embryos of the commercial soybean (Glycine max) cultivar IAS5 were co-transformed using particle bombardment with a synthetic form of the Bacillus thuringiensis delta-endotoxin crystal protein gene cry1Ac, the b-glucuronidase reporter gene gusA and the hygromycin resistance gene hpt. Hygromycin-resistant tissues were proliferated individually to give rise to nine sets of clones corresponding to independent transformation events. The co-bombardment resulted in a co-transformation efficiency of 44%. Many histodifferentiated embryos and 30 well-developed plants were obtained. Twenty of these plants flowered and fourteen set seeds. The integration and expression of the cry1Ac, gusA and hpt transgenes into the genomes of a sample of transformed embryos and all T0, T1, T2 and T3 plants were confirmed by Gus activity, PCR, Southern and western blot, and ELISA techniques. Two T0 plants out of the seven co-transformed plants produced seeds and were analyzed for patterns of integration and inheritance until the T3 generation. Bioassays indicated that the transgenic plants were highly toxic to the velvetbean caterpillar Anticarsia gemmatalis, thus offering a potential for effective insect resistance in soybean.
Key words:Anticarsia gemmatalis, cry1Ac gene, IAS5 cultivar, insect resistance, transgenic soybean.
Introduction
Soybean (Glycine max (L.) Merrill Fabales, Fabaceae) is one of the most important sources of edible oil and protein, resulting in special interest in the genetic improvement of this crop. The velvetbean caterpillar (Anticarsia gemmatalis Hübner) is a major soybean pest, mainly occurring in soybean growing regions of North and South America (Panizzi and Corrêa-Ferreira, 1997; Macrae et al., 2005), causes extremely high levels of defoliation when infestation is heavy and can severely damage axillary meristems, with a single caterpillar being able to consume up to 110 cm2 of soybean foliage (Walker et al., 2000). However, various authors have reported that A. gemmatalis can be controlled by the delta-endotoxin (d-endotoxin) produced by some strains of Bacillus thuringiensis (Stewart et al.,1996; Walker et al., 2000; Macrae et al., 2005).
Various DNA delivery methods and plant tissues have been used in developing transgenic soybean plants. Such techniques include particle bombardment of shoot meristems (McCabe et al., 1988; Christou et al., 1989), embryogenic suspension cultures (Finer and McMullen, 1991; Stewart et al., 1996) and Agrobacterium tumefaciens-mediated T-DNA delivery into cotyledonary nodes derived from five-day to seven-day old seedlings (Donaldson and Simmonds, 2000; Olhoft et al., 2003), mature seeds (Paz et al., 2006), immature zygotic cotyledons (Yan et al., 2000), somatic embryos derived from immature cotyledons (Parrott et al., 1989) or embryogenic suspension cultures (Trick and Finer, 1998).
A simple procedure for the establishment and proliferation of somatic embryos from immature cotyledons, more rapid and less labor-intensive than embryogenic suspension cultures, was developed by Santarém et al. (1997), using a semi-solid medium described by Wright et al. (1991). Using this procedure transgenic soybean plants have been generated from the soybean cultivars Jack (Santarém and Finer, 1999) and IAS5 (Droste et al., 2002) expressing the b-glucuronidase (Gus, EC 3.2.1.31) reporter gene gusA. The transgenic plants obtained in both of these studies were fertile, indicating that this strategy is a promising tool for the recovery of transgenic soybean plants.
Earlier studies have indicated that soybean regeneration is genotype-specific with differences in their responses to in vitro culture and transformation. There is no efficient transformation system for a wide range of soybean cultivars (Hofmann et al., 2004; Ko et al., 2004), which explains why very few reports on genetic transformation of commercial soybean cultivars are available. According to Meurer et al. (2001) there are two major routes to improve embryogenic culture-based soybean regeneration and transformation protocols with the goal of increasing the recovery of transgenic fertile lines. One option is to screen large numbers of new soybean cultivars and genotypes for embryogenic potential, while another option is to use existing protocols and cultivars, such as the Jack cultivar, coupled with traditional breeding programs for introgressing transgenic traits into other genotypes. The second option was followed by Stewart et al. (1996) to introduce insect resistance traits into soybean and Yan et al. (2000) for engineering soybean protein modification.
The Brazilian soybean cultivar IAS5 has commonly been used in genetic improvement programs and recommended by the Brazilian Agricultural Ministry (Ministério da Agricultura, Pecuária e Abastecimento, MAPA) for commercial growing in the Brazilian states of Goiás, Minas Gerais, Paraná, São Paulo and Rio Grande do Sul (MAPA, 2007). This cultivar has shown good reliable response in embryogenic systems (Santos et al., 1997; Droste et al., 2002).
The study described in this paper was aimed at developing transgenic soybean with resistance to A. gemmatalis larvae. Somatic embryos of the IAS5 cultivar growing on semi-solid medium were transformed with a synthetic Bacillus thuringiensis d-endotoxin crystal protein cry1Ac gene by particle bombardment and the cry1Ac transgene insertion pattern in the transgenic plants analyzed in the primary transformants. Transmission and expression of the transgenes were also characterized in the T1, T2 and T3 generations. Insect feeding assays indicated that the transgenic plants were highly toxic to A. gemmatalis larvae.
Materials and Methods
Plant material and plasmids used for soybean transformation
In the experiments described in this paper we started with seeds of Glycine max cultivar IAS5 supplied by Embrapa Soja, Londrina, PR, Brazil. Pods containing immature seeds 3-5 mm in length were harvested from field-grown plants. Somatic embryogenesis from immature cotyledons was induced, proliferated and maintained as described by Droste et al. (2002).
The two plant transformation vector plasmids used were: pGusHyg, a pUC18 derivative carrying gusA and the hygromycin resistance gene hpt, both driven by the cauliflower mosaic virus (CaMV) 35S promoter with a nopaline synthase gene (nos) terminator; and pGEM4Z-Cry1Ac, a pGEM4Z derivative containing the truncated synthetic cry1Ac gene under control of the cauliflower mosaic virus (CaMV) 35S promoter with a nos terminator. The cry1Ac used in this study was synthesized at the University of Ottawa using the recursive polymerase chain reaction (PCR) approach (Prodromou and Pearl, 1992). The original bacterial cry1Ac has a G+C content of 37% while the synthetic version was designed with a G+C content of 47.7% for expression in dicotyledonous plants, with the overall modifications resulting in higher Cry1Ac expression levels in transgenic plants (Sardana et al., 1996).
Transformation and regeneration of transgenic plants
Embryogenic tissue from immature cotyledons of soybean cultivar IAS5 was transformed by particle bombardment using the particle inflow gun (PIG; Finer et al., 1992) as described by Droste et al. (2002). Briefly, 20 embryogenic clusters, equivalent to about 70 mg of tissue, were placed in a petri dish containing D20 medium (Wright et al., 1991) and bombarded once with M10 tungsten particles (Dupont, USA) coated with a 4:1 molar ratio of pGEM4Z-Cry1Ac (4 µg µL-1) to pGusHyg (1 µg µL-1) plasmid DNA. This procedure was repeated to produce 15 replicate plates. The bombarded tissue was cultured for 14 days on D20 medium containing 12.5 mg L-1 of the selective agent hygromycin-B, after which the tissue was cultured for three months on the same medium containing 25 mg L-1 hygromycin-B. For the establishment and proliferation of embryogenic tissues after selection pieces of green tissue were subcultured every 14 days for 56 days in plates containing fresh D20 medium without antibiotics.
Three months after bombardment, hygromycin-resistant embryogenic soybean tissues were visually selected, counted and separately cultured for the establishment and proliferation of lines corresponding to putative independent transformation events. Out of 60 independent pieces of hygromycin-resistant tissues we established nine proliferative lines. Lines 18 and 43 subsequently being shown to consist of plants resulting from two different transformation events so that these lines were sub-divided into a and b plants. Different plants from the same line were categorized as families, with, for example 18a1 being plant 1 from line 18a. To stimulate histodifferentiation, clusters of hygromycin-resistant embryos were transferred to plates containing MSM6 maturation medium (Finer and McMullen, 1991) for 60 days.
The histodifferentiated somatic embryos were transferred to MS0 conversion medium, containing Murashige and Skoog (MS) salts solution (Murashige and Skoog, 1962), B5 vitamins solution (Gamborg et al., 1968), 3% (w/v) sucrose, 0.3% (w/v) Phytagel (Sigma, USA), pH 6.4. After a further 20-30 days the germinated embryos were individually transferred from the petri plates to 100 mL flasks containing 25 mL of the same medium to continue regeneration of the plantlets.
After about 30-40 days in the flasks the regenerated plantlets were transferred to vermiculite contained in plastic cups covered with polyvinyl chloride film in which the remained for a further 30 days during which time they were gradually exposed to ambient levels of relative humidity (RH » 50%). After that, plants were transferred to pots containing 1 kg of soil and maintained at 26 °C ± 3 °C (RH » 80%) in a growth chamber under a 14 h photoperiod and a light intensity of 13,500 lux provided by 60 W daylight type fluorescent lamps (Osram, Brazil). The plants remained in these pots until physiological maturity (» 150 days). These plants were the primary transgenic regenerants (T0), i.e., transgenic plants recovered from the explant originally subjected to particle bombardment. Thirty well-developed plantlets were transplanted ex vitro, of which 20 reached maturity and flowered and 14 set seeds. All embryos/plants derived from an independent piece of hygromycin-resistant tissue were noted as being clonal embryos/plants.
For progeny analysis the T1 seeds, equivalent to the F1 seeds for non-transgenic plants, obtained from self-pollination of T0 plants were planted in pots containing 8 kg of soil fertilized with an equivalent of 500 kg/ha of 0-25-25 (N-P-K) and 6 t/ha lime. The seeds were sown in December 2004 and harvested in April 2005, in a greenhouse, under natural light at 25 ± 5 °C, relative humidity of about 50% ± 10%, and a photoperiod varying from 14 h in December to 12 h in April. Plants were grown until physiological seed maturity. The T1 plants were saved to produce the T2 generation which was in turn grown to maturity under the same conditions as their parents and saved to produce the T3 generation. The chi-square (c2) test was used to confirm the expected Mendelian segregation patterns of 3:1 and 2:1 (transgenic: non-transgenic plants). Some T3 plants were reciprocally crossed to non-transgenic plants. Ten plants of the 18a1 family were used as the pollen donor and 10 as the pollen recipient. Twenty-five plants of 18a4 family were crossed as pollen recipient. Ten flowers were pollinated per plant.
We also investigated the number of homozygous plants positive for both Cry1Ac and GusA. A T3 transformant plant from the 18a4 family that served as the pollen recipient in the backcross set four seeds which gave raise to four plants positive for both Cry1Ac and GusA, indicating that this T3 transformant plant was homozygous. To check this we tested 20 seeds obtained from self-pollination of the T3 transformant plant, which were germinated on wet filter paper to produce seedlings which we analyzed for Gus activity and cry1Ac expression. We also used this methodology, and the same number of selfed seeds for each plant, to investigate the possible homozygous nature of 253 out of the 309 T3 plants positive for both Cry1Ac and GusA.
PCR analysis and Southern blot hybridization of digested genomic DNA
We used the cetyltrimethylammonium bromide (CTAB) procedure (Doyle and Doyle, 1987), with some modifications, to extract genomic DNA from 72 of the histodifferentiated soybean embryos and leaf tissues of all of the 20 soybean plants which reached maturity. The extracted DNA samples were individually assayed for the presence of cry1Ac, gusA and hpt. The PCR primers used to detect cry1Ac were cryFOR (5GGGGATCCATGGATA ACAATCCGAAC3) and cryREV (5CAGTCGACATT CAGCCTCGAGTGTTG3), which amplify a 1845 bp region of cry1Ac. The reaction mixture (25 µL) consisted of 200 µM dNTPs, 1 unit of Taq DNA polymerase (Invitrogen), 1X Reaction buffer with 2 mM MgCl2, 100 nM of each primer and 100 ng of each DNA sample. Amplifications were carried out by pre-cycling at 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, 52 °C for 1 min and 72 °C for 2 min, with a final extension step of 5 min at 72 °C. The PCR primers used to detect gusA were gusAFOR (5GGTGGGAAAGCGCGTTACAAG3) and gusAREV (5GGATTCCGGCATAGTTAAAGG3) amplifying 622 bp, while to detect hpt we used hptFOR (5GC GATTGCTGATCCCCATGTGTAT3) and hptREV (5G GTTTCCACTATCGGCGAGTACTT3) which amplify 512 bp. The reaction mixtures for both genes were as described above while the amplification conditions for both these genes consisted of pre-cycling at 94 °C for 5 min, followed by 30 cycles of 94 °C for 45 s, 52 °C for 45 s and 72 °C for 45 s, with a final extension of 2 min at 72 °C. All amplifications reactions were carried out in a PCR Express Thermal Cycler (Thermo Hybaid, UK). After amplification the products were separated by electrophoreses on 1% (w/v) agarose gel and transferred overnight onto Hybond N+ membranes (GE Healthcare) for the Southern blot procedure following standard protocols (Sambrook and Russel, 2001). Probe labeling, hybridization, stringency washes and detection were carried out as specified by the ECL kit (GE Healthcare). The DNA blotting was probed with a 1.8 kb PCR fragment containing the cry1Ac coding sequence purified from agarose gel using the GFX kit (GE Healthcare). Hybridizing bands were detected by exposure to Kodak X-OMAT autoradiography films for 2 h.
The Southern blot hybridization of digested genomic DNA was carried out using 20 µg of total genomic DNA from putative transgenic and non-transgenic control plants digested overnight at 37 °C with the restriction enzymes BamHI, KpnI and SalI (Promega, USA). Digested genomic DNA of each plant was separated by electrophoresis on 0.8% (w/v) agarose gel and transferred from the gel to a Hybond N+ nylon membrane. Probe labeling, hybridization, stringency, washes and detection were carried out as specified above.
Gus histochemical assay and protein expression analysis
Leaf disks or seedlings were assayed for Gus activity using the improved histochemical staining protocol (Jefferson, 1987).
For protein expression analysis 0.2 g of fresh leaf tissue was excised from T0 plants and homogenized in 500 mL of extraction buffer [containing 50 mM of 1 M Tris-HCl (pH 6.8), 0.2% (w/v) polyvinylpyrrolidone (PVP-40, Sigma) and 1% (v/v) b-mercaptoethanol (Sigma)]. Samples were stirred for 30 min at 4 °C and then clarified by centrifugation at 10,000 g. Protein concentration was determined by the Bradford method (Bradford, 1976) with bovine serum albumin as protein standard. For each plant, 50 µg of crude protein extract was subjected to 10% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. The presence of the Cry1Ac protein was detected using polyclonal antibody specific for Cry1Ac d-endotoxin from Bacillus thuringiensis (kindly supplied by Dr. Lidia Mariana Fiuza, UNISINOS, RS, Brazil). The protein bands were visualized using the ECL Western Blot Detection and Analysis System (GE Healthcare). Detection of Cry1Ac protein produced by T1, T2 and T3 plants was monitored by a double-sandwich enzyme-linked immunosorbent assay (ELISA) procedure using the Trait Check Bt 1Ac cotton leaf/seed kits (Strategic Diagnostics Inc., USA).
Insect bioassays
Insecticidal activity of T2 transgenic plants toward A. gemmatalis larvae was evaluated using a detached leaf-feeding assay. The plants were heterozygous for cry1Ac as determined by segregation analysis and were Cry1Ac-positive as ascertained by double-sandwich ELISA. Leaflets from large trifoliate leaves were placed in 100 mm x 20 mm petri dishes containing a 90 mm Whatman n. 1 filter paper (Whatman International, UK) saturated with distilled water to maintain high humidity. The amount of leaf tissue was kept as uniform as possible from one dish to another. Leaf samples were infested with 20 neonate A. gemmatalis larvae per plate. We used 10 transgenic plants from the 18a1 family and 30 from the 18a4 family with four replicate plates per plant, were included in the bioassay. Three non-transgenic IAS5 plants of the same age as the transgenic plants, with four replicate plates per plant, were used as controls. After 24 h, the percent foliage consumption was estimated. Remaining leaf remnants were removed and replaced by a 1 cm3 of a solid artificial diet (Greene et al., 1976). Leaf consumption percentages were converted to scores as follows: 0 = no consumption, 1 = less than 50%; 2 = more than 50%; and 3 = 100% consumption. The number of dead larvae and alive larvae was determined 24 h, 48 h, 72 h and 96 h after the start of the assay.
Results and Discussion
Transformation and regeneration of transgenic plants
From the 60 independent pieces of hygromycin-resistant tissue we obtained nine proliferative lines from which 613 histodifferentiated somatic embryos were transferred to conversion medium. In the later stages of the experiment, 30 well-developed plantlets were transplanted ex vitro, 20 of which reached maturity and flowered and 14 of these set seeds. All embryos/plants derived from an independent piece of hygromycin-resistant tissue were noted as being clonal embryos or plants.
Transgene integration and expression
We used PCR to screen 72 histodifferentiated embryos and the 20 plants which reached maturity for the presence of cry1Ac, gusA and hpt. Two of the 9 lines (22%) produced no PCR products for any of the genes tested and were considered "escapes" and discarded. The molecular analysis also showed that one piece of hygromycin-resistant tissue could contain two independent transformation events, base on which lines 18 and 43 were subdivided (a and b).
The molecular characterization of transformed lines is presented in Table 1. All 64 embryos and 11 plants of the nine independent lines presented the expected 622 bp gusA fragment and the 512 bp hpt fragment. The presence of the expected 1845 bp cry1Ac fragment was observed in 23 embryos and seven plants derived from four lines (18a, 26, 41 and 43a). Simultaneous occurrence of gusA, hpt and cry1Ac genes in these four lines allowed the co-transformation efficiency to be calculated (4/9 = 44%). The efficiency of our transformation rate is similar to that previously reported for soybean, since in situations in which two genes on independent plasmids have been introduced by particle bombardment the co-transformation rates have been shown to vary from 18% to 50% (Christou and Swain, 1990; Li et al., 2004).
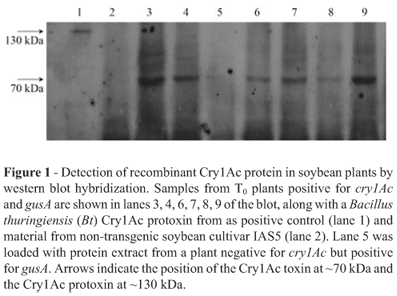
Western blot hybridization was used to evaluate the expression of cry1Ac gene at the protein level. When the proteins were separated electrophoretically on SDS gel, a band of »70 kDa corresponding to the Cry1Ac toxin was detected in plants which were positive for cry1Ac and gusA (Figure 1) but no antibody reactive protein was detected in plants which were negative for cry1Ac and positive for gusA or in non-transgenic plants.
Transgene segregation
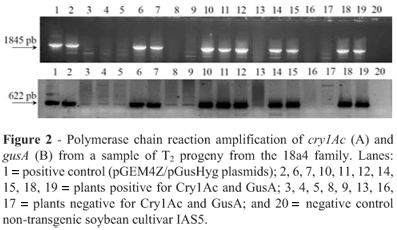
We obtained seven co-transformed T0 plants containing the cry1Ac, gusA and hpt genes but only plants 18a1 and 18a4, derived from the same 18a line (Table 1), produced seeds and were analyzed for patterns of integration and inheritance until the T3 generation. Expression of the cry1Ac gene in the T1 and T2 plants was first determined using the double-sandwich antibody procedure and Gus expression by histochemically assaying discs from young leaves. To validate the results of the preliminary tests, the presence of transgenes were confirmed by PCR analysis in all T1 and T2 plants positive for Cry1Ac and GusA (Figure 2), although PCR was also performed on plants which were negative for Cry1Ac and GusA and the absence of bands suggested that no transgenes could be detected in these plants. The correlation between the presence of transgene DNA and its expression was perfect.
Based on the 48 T1 plants evaluated, it appeared that cry1Ac plus gusA and hpt were linked at one integration site in the initial T0 plants (Table 2). The T1 progenies of both T0 plants segregated fewer plants positive for cry1Ac and gusA than the 3:1 transgenic to non-transgenic progeny predicted by Mendelian principles for a single dominant locus. The T2 and T3 families continued to segregate in an unusual manner, with a large deficiency of transgenic plants (Table 3). Moreover, the segregation ratios indicated that T2 plants were uniformly heterozygous for the transformed traits.
Transgenes are generally expected to behave as dominant genes and segregate in a 3:1 ratio for transgenic to non-transgenic progeny when the plant is self-pollinated, because the transgene locus is considered to be hemizygous in the primary (T0) transformant (Campbell et al., 2000). However, transgenic loci introduced into higher plant species frequently display unpredictable patterns of inheritance and expression, which has occurred at a frequency between 10% and 50% for some transgenic lines (Yin et al., 2004). Unusual segregation ratios could result from a number of factors, including inactivation of transgene expression, insertion leading to a lethal mutation and poor transgene transmission to the progeny. The inactivation of expression is frequently observed when transgenes are present in multiple copies and is responsible for causing abnormal segregation (Yin et al., 2004). However, our PCR amplifications performed on T1 and T2 plants phenotypically negative for Cry1Ac and GusA confirmed the absence of transgene-DNA in these plants so the inactivation of transgene expression cannot be the reason for the non-Mendelian segregation observed in our study.
The integration of foreign DNA into a plant genome can produce an insertion mutation in an essential gene. A lethal mutation reflected by the lack of homozygotes can lead to a 2:1 segregation for transgenic and non-transgenic traits in the progeny of a transgenic plant. Limanton-Grevet and Jullien (2001) attributed the 2:1 segregation observed in a T2 progeny of a transformed Asparagus officinalis line to the absence of homozygotes, but since our results did not fit even a 2:1 segregation pattern (Tables 2 and 3) it appears that other factors could be acting. However, non-Mendelian segregation of transgenes could be due to failure of transmittance from one of the gametes. Christou et al. (1989) credited a 1:1 segregation ratio observed in progeny derived from selfing a transgenic soybean plant to the failure to pass a transgene to the next generation through pollen. Feldmann et al. (1997) reported that reciprocal backcrosses to non-transgenic Arabidopsis plants showed unequal gametic transmission of the kanamycin-resistance (KanR) trait in seven lines for up to six successive generations, two of the lines failing to transmit KanR through the ovule and the extreme KanR deficiency of seedlings of the other five lines being primarily due to failure to transmit the trait through the pollen.
We investigated gametic transgene transmission by crossing several of our transgenic soybean plants with non-transgenic soybean plants. The segregation of the gusA and cry1Ac genes in the F1 backcross (BCF1) was determined histochemically for Gus and by double-sandwich ELISA for Cry1Ac. Our results showed transgene transmission through male and female gametes, but at a substantially reduced rate (Table 4). In the 18a1 family, transmission of the transgenes through the pollen was reduced to 60% to 75% of that expected for a heterozygote while transmission through the ovule dropped to 33% to 49%. Family 18a4 displayed a similar transmission rate (30% to 47%) through the ovule, no crosses being made in which the transformant served as the pollen donor. The low number of crosses in which the transformant served as the pollen donor did not allow us to conclude unequivocally that the transmission through pollen was affected less severely than transmission through the ovule.
Homozygous plants positive for both Cry1Ac and GusA
A 18a4 family T3 transformant that served as the female parent in the backcross set four seeds which resulted in four plants positive for both Cry1Ac and GusA, thus indicating that this T3 transformant plant was homozygous. To check this we tested 20 seeds obtained from self-pollination of that plant which we germinated to produce seedlings which showed Gus activity and expressed Cry1Ac, confirming that the plant was homozygous. Based on this fact, we decided to use the same methodology, and the same number (20) of selfed seeds for each plant, to investigate the possible homozygous nature of another 253 out of the 309 T3 transgenic plants which were positive for both Cry1Ac and GusA. Besides the plant already mentioned, our analyses detected a further 12 homozygous T3 plants positive for both Cry1Ac and GusA. These experiments excluded the possibility of a lethal insertional mutation as a reason for the observed non-Mendelian segregation, because such an occurance would have been reflected by a lack of homozygotes. On the other hand, we assumed that the difficulty in obtaining homozygous plants in the T2 generation could be accounted for the poor transmission of transgenes through pollen and, more importantly, female gametes.
Southern analysis of transformed plants
Genomic DNA of T1, T2 and T3 plants positive for Cry1Ac and GusA was digested with KpnI, an enzyme that cuts plasmid pGEM4Z only once, SalI that does not cleave the transformation plasmid and BamHI which releases the 2.1 kb cry1Ac cassette gene coding region and nos terminator. A representative Southern blot of seven T1 plants (derived from 18a1 and 18a4 T0 plants) positive for Cry1Ac and GusA is shown in Figure 3. The detection of a 2.1 kb band in the DNA digested with BamHI in all plants indicated the presence of at least one intact copy of cry1Ac. The presence of bands larger than 2.1 kb is evidence for rearrangements of the transgene DNA. Analysis of digests with KpnI revealed three cry1Ac fragments in all plants. Analysis with SalI confirmed the presence of three copies for that gene (data not shown). Southern blot was also performed on plants negative for Cry1Ac and GusA in order to determine if transgenes were present but the absence of any hybridization signal indicated that these plants probably had no inserted transgene. All the transgenic plants of the T1, T2 and T3 progenies analyzed showed the same hybridization pattern as the two T0 parental plants (data not shown) and consequently the same copy number. These results indicate that all copies of the gene are inherited as a unit and that the original transgene integration pattern observed in the primary regenerated plants was stably passed on to all progeny plants.
Insecticidal activity
To confirm that the Cry1Ac protein produced in the transgenic plants was functional isolated leaves from transgenic plants and non-transgenic control plants were infested with neonate larvae of A. gemmatalis. All control leaves were completely defoliated after 24 h (Figure 4a) but consumption was significantly reduced in transgenic leaves (Table 5; Figure 4b) and, in comparison to larvae fed on control leaves, larvae fed on transgenic leaves showed browning and severe growth retardation (Figure 4c). In these experiments any remaining leaf tissue was remove 24 h after the start of feeding and replaced with a normal laboratory diet. Within 48 h larvae initially fed on transgenic leaves stopped feeding and most were dead, with only two or less (depending on the replicate) surviving for 96 h. However, 19 out of 20 larvae reared on control leaves survived for 96 h. The mortality of A. gemmatalis affected by expression of cry1Ac in transgenic leaves of plants from the 18a1 and the 18a4 families was similar. This data could be accounted for the fact that both families were derived from the same transformation event. Soybean cultivars containing B. thuringiensis (Bt) toxin gents (Bt soybeans) have not yet been commercialized, although experimental lines have been developed. Parrott et al. (1994) reported that the expression of a native cry1Ab gene prevented the feeding and growth of A. gemmatalis larvae. It has been reported that a transgenic line of the soybean cultivar Jack expressing high levels of synthetic cry1Ac killed all A. gemmatalis larvae and significantly reduced the survival and feeding of the soybean looper (Pseudoplusia includens) and the corn earworm (Helicoverpa zea) in laboratory bioassays (Stewart et al., 1996) and in artificially infested field cages (Walker et al., 2000). More recently, Macrae et al. (2005) evaluated transgenic soybean lines (based on cultivar A5547) expressing synthetic cry1A for resistance against several lepidopteran pests in screenhouse and conventional field trials carried out in the United States and Argentina and found that the Bt lines exhibited virtually complete efficacy against all the pests evaluated.
Intralocus recombination
Co-transformation is one of the best strategies to obtain marker-free transgenic plants since it is based on the principle that a proportion of transformed plants carrying the selectable marker gene will also have integrated the transgene of interest at a second, unlinked, insertion site and the genes can subsequently be removed from such plants by genetic segregation (Ebinuma et al., 2001; Bettany et al., 2002; Park et al., 2004). We used this co-transformation strategy to introduce cry1Ac into soybean with the expectation of obtaining marker-free transgenic plants. However, as the co-transformed cry1Ac, gusA and hpt integrated at a single locus they segregated together. A high incidence of linkage has been demonstrated when using biolistic-mediated co-transformation (Miki and McHugh, 2004) but, nevertheless, we still obtained three transgenic plants negative for Cry1Ac but positive for GusA (Table 3; Figure 5), perhaps because interchromosomal recombination split up the transgenes. It is possible that the transgenic locus contains interspersed genomic DNA within it (Kohli et al., 2003). Intriguingly, up to now, no plants containing the cry1Ac gene alone have been obtained.
In this paper we have outlined the success development of A. gemmatalis-resistant transgenic soybean (cultivar IAS5) plants containing synthetic cry1Ac. Although the segregation was non-Mendelian in the first generations our data clearly demonstrated that the transgenes were stably transmitted and expressed in progenies. Homozygous transgenic plants were obtained in the T3 generation and the agronomic performance and the response of these plants toward field populations of A. gemmatalis are under analysis. Further studies involving combined molecular and cytogenetic analysis can now be performed to determine the transgenic locus organization.
Acknowledgments
We would like to thank the following people for their important contributions to this study: Dr. Flávio Moscardi (EMBRAPA Soja, Londrina, PR), Dr. José Roberto Salvadori (EMBRAPA Trigo, RS), Dr. Luiz Carlos Federizzi (Departamento de Plantas de Lavoura, UFRGS), Dr. Márcia Pinheiro Margis (Departamento de Genética, UFRGS) and Silvia N. Cordeiro Richter (technical assistance). This work was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS, Brazil), Centro do Agronegócio-Casa Rural (Brazil), The Rockfeller Foundation (USA) and Natural Sciences and Engineering Research Council (NSERC, Canada). Milena S. Homrich acknowledges CNPq for her Ph.D. fellowship. The authors thank an anonymous reviewer for valuable suggestions.
Internet Resources
Received: July 10, 2007; Accepted: November 8, 2007.
Associate Editor: Everaldo Gonçalves de Barros
- Bettany AJE, Dalton SJ, Timms E, Dhanoa MS and Morri P (2002) Effect of selectable gene to reporter gene ratio on the frequency of co- transformation and co-expression of uidA and hpt transgenes in protoplast-derived plants of tall fescue. Plant Cell Tissue Organ Cult 68:177-186.
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248-254.
- Campbell BT, Baenziger PS, Mitra A, Sato S and Clemente T (2000) Inheritance of multiple transgenes in wheat. Crop Sci 40:1133-1141.
- Christou P, Swain WF, Yang NS and McCabe DE (1989) Inheritance and expression of foreign genes in transgenic soybean plants. Genetics 88:7500-7504.
- Christou P and Swain WF (1990) Co-transformation frequencies of foreign genes in soybean cell cultures. Theor Appl Genet 79:337-341.
- Donaldson PA and Simmonds DH (2000) Susceptibility to Agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell Rep 19:478-484.
- Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissues. Phytochem Bull 19:11-15.
- Droste A, Pasquali G and Bodanese-Zanettini MH (2002) Transgenic fertile plants of soybean [Glycine max (L.) Merrill] obtained from bombarded embryogenic tissue. Euphytica 127:367-376.
- Ebinuma H, Sugita K, Matsunaga E, Endo S, Yamada K and Komamine A (2001) Systems for the removal of a selection marker and their combination with a positive marker. Plant Cell Rep 20:383-392.
- Feldmann KA, Coury DA and Christianson ML (1997) Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147:1411-22.
- Finer JJ and McMullen MD (1991) Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell Dev Biol-Plant 27:175-182.
- Finer JJ, Vain P, Jones MW and McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:323-328.
- Gamborg OL, Miller RA and Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151-158.
- Greene GL, Leppla NC and Dickerson WA (1976) Velvet bean caterpillar: A rearing procedure and artificial medium. J Econ Entomol 69:487-488.
- Hofmann N, Nelson RL and Korban SS (2004) Influence of media components and pH on somatic embryo induction in three genotypes of soybean. Plant Cell Tissue Organ Cult 77:157-163.
- Jefferson RA (1987) Assaying chimeric genes in plants: The Gus gene fusion system. Plant Mol Biol Rep 5:387-405.
- Ko TS, Nelson RL and Korban SS (2004) Screening multiple soybean cultivars (MG 00 to MG VIII) for somatic embryogenesis following Agrobacterium-mediated transformation of immature cotyledons. Crop Sci 44:1825-1831.
- Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E and Christou P (2003) Transgene integration, organization and interaction in plants. Plant Mol Biol 52:247-258.
- Li HY, Zhu YM, Chen Q, Conner RL, Ding XD, Li J and Zhang BB (2004) Production of transgenic soybean plants with two anti-fungal protein genes via Agrobacterium and particle bombardment. Biol Plant 48:367-374.
- Limanton-Grevet A and Jullien M (2001) Agrobacterium-mediated transformation of Asparagus officinalis L.: Molecular and genetic analysis of transgenic plants. Mol Breed 7:141-150.
- Macrae TC, Baur ME, Boethel DJ, Fitzpatrick BJ, Gao AG, Gamundi JC, Harrison LA, Kabuye VT, Mcpherson RM, Miklos JA et al. (2005) Laboratory and field evaluations of transgenic soybean exhibiting high-dose expression of a synthetic Bacillus thuringiensis cry1A gene for control of Lepidoptera. J Econ Entomol 98:577-587.
- McCabe DE, Swain WF, Martinell BJ and Christou P (1988) Stable transformation of soybean (Glycine max) by particle acceleration. Bio/Technol 6:923-926.
- Meurer CA, Dinkins RD, Redmond CT, McAllister KP, Tucker DT, Walker DR, Parrott WA, Trick HN, Essic JS, Frantz HM et al. (2001) Embryogenic response of multiple soybean [Glycine max (L.) Merr.] cultivars across three locations. In Vitro Cell Dev Biol-Plant 37:62-67.
- Miki B and McHugh S (2004) Selectable marker genes in transgenic plants: Applications, alternatives and biosafety. J Biotechnol 107:193-232.
- Murashige T and Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473-497.
- Olhoft PM, Flagel LE, Donovan CM and Somers DA (2003) Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216:723-735.
- Panizzi AR and Corrêa-Ferreira BS (1997) Dynamics in the insect fauna adaptation to soybean in the tropics. Trends Entomol 1:71-88.
- Park J, Lee YK, Kang BK and Chung WI (2004) Co-transformation using a negative selectable marker gene for the production of selectable marker gene-free transgenic plants. Theor Appl Genet 109:1562-1567.
- Parrott WA, Hoffman LM, Hildebrand DF, Williams EG and Collins GB (1989) Recovery of primary transformants of soybean. Plant Cell Rep 7:615-617.
- Parrott WA, All JN, Adang MJ, Bailey MA, Boerma HR and Stewart CN (1994) Recovery and evaluation of soybean (Glycine max [L.] Merr.) plants transgenic for a Bacillus thuringiensis var. Kurstaki insecticidal gene. In Vitro Cell Dev Biol-Plant 30:144-149.
- Paz MM, Martinez JC, Kalvig AB, Fonger TM and Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25:206-213.
- Prodromou C and Pearl LH (1992) Recursive PCR - A novel technique for total gene synthesis. Protein Eng 5:827-829.
- Sambrook J and Russel DW (2001) Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, New York.
- Santarém ER, Pelissier B and Finer JJ (1997) Effect of explant orientation, pH, solidifying agent and wounding on initiation of soybean somatic embryos. In Vitro Cell Dev Biol-Plant 33:13-19.
- Santarém ER and Finer JJ (1999) Transformation of soybean [Glycine Max (L.)Merrill] using proliferative embryogenic tissue maintained on semi-solid medium. In Vitro Cell Dev Biol-Plant 35:451-455.
- Santos KGB, Mundstock E and Bodanese-Zanettini MH (1997) Genotype-specific normalization of soybean somatic embryogenesis through the use of an ethylene inhibitor. Plant Cell Rep. 16:859-864.
- Sardana R, Dukiandjiev S, Giband M, Cheng X, Cowan K, Sauder C and Altosaar I (1996) Construction and rapid testing of synthetic and modified toxin gene sequences CryIA (b & c) by expression in maize endosperm culture. Plant Cell Rep 15:677-681.
- Stewart CN, Adang MJ, All JN, Boerma HR, Cardineau G, Tucker D and Parrott WA (1996) Genetic transformation, recovery, and characterization of fertile soybean transgenic for a synthetic Bacillus thuringiensis cry1Ac gene. Plant Physiol 112:121-129.
- Trick HN and Finer JJ (1998) Sonication-assisted Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill] embryogenic suspension culture tissue. Plant Cell Rep 17:482-488.
- Walker DR, All JN, McPherson RM, Boerma HR and Parrott WA (2000) Field evaluation of soybean engineered with a synthetic cry1Ac transgene for resistance to corn earworm, soybean looper, velvetbean caterpillar (Lepidoptera, Noctuidae), and lesser cornstalk borer (Lepidoptera, Pyralidae). J Econ Entomol 93:613-622.
- Wright MS, Launis KL, Novitzky R, Duesiing JH and Harms CT (1991) A simple method for the recovery of multiple fertile plants from individual somatic embryos of soybean [Glycine max (L.) Merrill]. In Vitro Cell Dev Biol-Plant 27:153-157.
- Yan B, Reddy MSS, Collins GB and Dinkins RD (2000) Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merrill.] using immature zygotic cotyledon explants. Plant Cell Rep 19:1090-1097.
- Yin Z, Plader W and Malepszy S (2004) Transgene inheritance in plants. J Appl Genet 45:127-144.
- MAPA, Ministério da Agricultura, Pecuária e Abastecimento, http://www.agricultura.gov.br/pls/portal/url/ITEM/ 37AA904436961F63E040A8C07502416D (October 15, 2007).
» link
Publication Dates
-
Publication in this collection
24 June 2008 -
Date of issue
2008
History
-
Received
10 July 2007 -
Accepted
08 Nov 2007