Abstract
Epistasis (gene-gene interaction) is a ubiquitous component of the genetic architecture of complex traits such as susceptibility to common human diseases. Given the strong negative correlation between circulating adiponectin and resistin levels, the potential intermolecular epistatic interactions between ADIPOQ (SNP+45T > G, SNP+276G > T, SNP+639T > C and SNP+1212A > G) and RETN (SNP-420C > G and SNP+299G > A) gene polymorphisms in the genetic risk underlying type 2 diabetes (T2DM) and metabolic syndrome (MS) were assessed. The potential mutual influence of the ADIPOQ and RETN genes on their adipokine levels was also examined. The rare homozygous genotype (risk alleles) of SNP-420C > G at the RETN locus tended to be co-inherited together with the common homozygous genotypes (protective alleles) of SNP+639T > C and SNP+1212A > G at the ADIPOQ locus. Despite the close structural relationship between the ADIPOQ and RETN genes, there was no evidence of an intermolecular epistatic interaction between these genes. There was also no reciprocal effect of the ADIPOQ and RETN genes on their adipokine levels, i.e., ADIPOQ did not affect resistin levels nor did RETN affect adiponectin levels. The possible influence of the ADIPOQ gene on RETN expression warrants further investigation.
adipokine; ADIPOQ; epistasis; interaction; RETN
Lack of evidence for intermolecular epistatic interactions between adiponectin and resistin gene polymorphisms in Malaysian male subjects
Cia-Hin Lau; Sekaran Muniandy
Department of Molecular Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia
Send correspondence to Send correspondence to: Cia-Hin Lau Department of Molecular Medicine, Faculty of Medicine, University of Malaya 50603 Kuala Lumpur, Malaysia. E-mail: lauciahin_4275@yahoo.com
ABSTRACT
Epistasis (gene-gene interaction) is a ubiquitous component of the genetic architecture of complex traits such as susceptibility to common human diseases. Given the strong negative correlation between circulating adiponectin and resistin levels, the potential intermolecular epistatic interactions between ADIPOQ (SNP+45T > G, SNP+276G > T, SNP+639T > C and SNP+1212A > G) and RETN (SNP-420C > G and SNP+299G > A) gene polymorphisms in the genetic risk underlying type 2 diabetes (T2DM) and metabolic syndrome (MS) were assessed. The potential mutual influence of the ADIPOQ and RETN genes on their adipokine levels was also examined. The rare homozygous genotype (risk alleles) of SNP-420C > G at the RETN locus tended to be co-inherited together with the common homozygous genotypes (protective alleles) of SNP+639T > C and SNP+1212A > G at the ADIPOQ locus. Despite the close structural relationship between the ADIPOQ and RETN genes, there was no evidence of an intermolecular epistatic interaction between these genes. There was also no reciprocal effect of the ADIPOQ and RETN genes on their adipokine levels, i.e., ADIPOQ did not affect resistin levels nor did RETN affect adiponectin levels. The possible influence of the ADIPOQ gene on RETN expression warrants further investigation.
Key words: adipokine, ADIPOQ, epistasis, interaction, RETN.
Adiponectin and resistin are important hormones in the regulation of insulin sensitivity (insulin secretion and signaling pathway), energy homeostasis (glycogen synthesis and glycogenolysis), glucose metabolism (gluconeogenesis, glucose uptake and use) and lipid metabolism (fatty acid synthesis and oxidation) (Lau and Muniandy, 2011a). We have recently shown that the mutual regulation of adiponectin and resistin is an important determinant of metabolic disturbances, cardiometabolic function and global metabolic status (Lau and Muniandy, 2011a). Moreover, the relative proportion of adiponectin-to-resistin levels reflect metabolic homeostasis and metabolic disorders much better than adiponectin and resistin levels alone, with a significant inverse correlation between the levels of these two hormones (Lau and Muniandy, 2011a). The similar domain architecture of these adipokines, despite their diametrically opposed physiological effects, suggests a common regulatory mechanism in metabolic homeostasis (Patel et al., 2004). Single nucleotide polymorphisms (SNPs) have been identified in the ADIPOQ (SNP+45T > G, SNP+276G > T, SNP+639T > C and SNP+1212A > G) and RETN (SNP-420C > G and SNP+299G > A) genes. These SNPs of the ADIPOQ (Zhao and Zhao, 2011) and RETN (Wen et al., 2010) genes are the most widely studied for their etiological links to the risk of type 2 diabetes mellitus (T2DM) and metabolic syndrome (MS). In general, T2DM and MS do not follow a Mendelian pattern of inheritance.
There is increasing evidence that gene-gene interactions (epistasis) play a far more important role in an individual's susceptibility to these complex diseases than single polymorphisms on their own since one gene can enhance or hinder the expression of another gene (Moore, 2003). Moreover, there is a growing awareness that the failure to replicate single-locus association studies for common diseases may reflect an underlying genetic architecture in which interactions between genes are the norm rather than the exception (Lehner, 2011). Epistasis occurs when the developmental or physiological effects of a genetic difference at one locus depend on the genotype at another locus, perhaps on a different chromosome (Desai et al., 2007). This dependence is often a consequence of an underlying functional relationship between the two genes (Lehner, 2011). Since epistasis is a ubiquitous component of the genetic architecture of complex diseases, analysis of the combined effects of the ADIPOQ and RETN genes may provide more information about the risk of T2DM and MS than the analysis of a single gene.
Given the strong negative correlation between circulating adiponectin and resistin levels, in this work we examined the potential intermolecular epistatic interactions between ADIPOQ and RETN gene polymorphisms in the genetic risk underlying T2DM and MS. The potential mutual influence of the ADIPOQ and RETN genes on their adipokine levels was also examined. The results of this study provide a better understanding of the intermolecular interactions between ADIPOQ and RETN gene polymorphisms that underlie the pathogenesis of T2DM and MS. Such an analysis of genetic interactions can reveal implicit structural and functional relationships between the ADIPOQ and RETN genes and may provide useful insights for developing therapeutic approaches to treat T2DM and MS.
All subjects were native Malaysian males 40-70 years old from three major ethnic groups (Malay, Chinese and Indian). This study was approved by the University of Malaya Medical Centre (UMMC) Ethics Committee (Ethical Approval Letter no. 612.17). Informed consent was obtained from each participant after thorough explanation of the study. Each subject received a detailed questionnaire about his personal and family history of disease and demographic data. Eight hundred and nine subjects were enrolled in a case-control study in which there were 208 controls, 174 individuals with MS but no T2DM, 171 individuals with T2DM but no MS and 256 individuals with T2DM and MS. MS was defined according to the International Diabetes Federation 2005 diagnostic criteria (Alberti et al., 2005). The sampling method used and the characteristics of this study have been described elsewhere (Lau and Muniandy, 2011a,b).
The subjects were screened for metabolic parameters that included serum fasting levels of adiponectin, resistin, insulin, total cholesterol, LDL cholesterol, HDL cholesterol, triglycerides, plasma glucose and whole blood HbA1C. The anthropometric parameters that were measured or calculated included blood pressure, body mass index (BMI), waist circumference and waist-to-hip ratio (WHR). β-Cell function (HOMA%β index), insulin sensitivity (HOMA%S index) and insulin resistance (HOMA-IR index) were calculated with HOMA calculator software version 2.2.2 (Diabetes Trials Unit, Oxford University). The clinical characteristics and demographic parameters of the subjects have been reported elsewhere (Lau and Muniandy, 2011a).
Genomic DNA was extracted from whole blood using a Wizard Genomic DNA purification kit (Promega Corporation, Madison, WI, USA). SNPs were genotyped using Custom TaqMan® SNP Genotyping assay kits (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's standard protocol. Allelic discrimination analysis was done with a StepOnePlus real-time PCR system version 2.0 (Applied Biosystems). All of the SNPs passed the pre-defined threshold for genotyping call rate. Only subjects with a high genotyping call rate (quality value > 98%) were included in the study. Primers and PCR conditions were designed to amplify the DNA region of each SNP marker. Thirty samples were randomly selected for forward and reverse DNA sequencing (AITBiotech Pte Ltd., Singapore) to verify the genotyping results for each SNP marker. Details of the genotyping and DNA sequencing procedures are described elsewhere (Lau and Muniandy, 2011b).
Dimension reduction correspondence analysis to examine the structural relationships between two SNPs in low-dimensional space was done using the PASW Statistics 18 Program (SPSS Inc, Chicago, IL, USA). Quantitative traits analysis for alleles was done using the HAPSTAT program, version 3.0 (Chapel Hill, NC, USA). In the additive model, an empirical p value was generated after Bonferroni correction with adjusted known confounding factors. The potential nonlinear interactions between ADIPOQ and RETN gene polymorphisms were analyzed with the Generalized Multifactor Dimensionality Reduction (GMDR) Beta program, version 0.7 (University of Virginia, Charlottesville, VA, USA). GMDR analysis included a combined cross-validation/permutation testing procedure that prevented model over-fitting and minimized false-positive results generated by multiple comparisons. Ethnicity was stratified before analysis. All p values were two-tailed, and p values < 0.05 were considered statistically significant.
SNP-420C > G of the RETN gene showed a strong structural relationship with SNP+639T > C and SNP+1212A > G of the ADIPOQ gene (Figure 1). The heterozygous genotype of SNP-420C > G tended to be co-inherited together with the heterozygous genotypes of SNP+639T > C (Figure 1C) and SNP+1212A > G (Figure 1D). SNP+299G > A of the RETN gene shared a strong structural relationship with SNP+639T > C of the ADIPOQ gene (Figure 2). The common homozygous genotypes of SNP+299G > A and SNP+639T > C tended to be co-inherited (Figure 2C). Interestingly, the rare homozygous genotype of SNP-420C > G was generally co-inherited with the common homozygous genotypes of SNP+639T < C (Figure 1C) and SNP+1212A < G (Figure 1D). Furthermore, the rare homozygous genotypes of these SNPs at the ADIPOQ and RETN loci were unlikely to be co-inherited (Figures 1 and 2), which suggested a possible antagonistic relationship between the ADIPOQ and RETN genes.
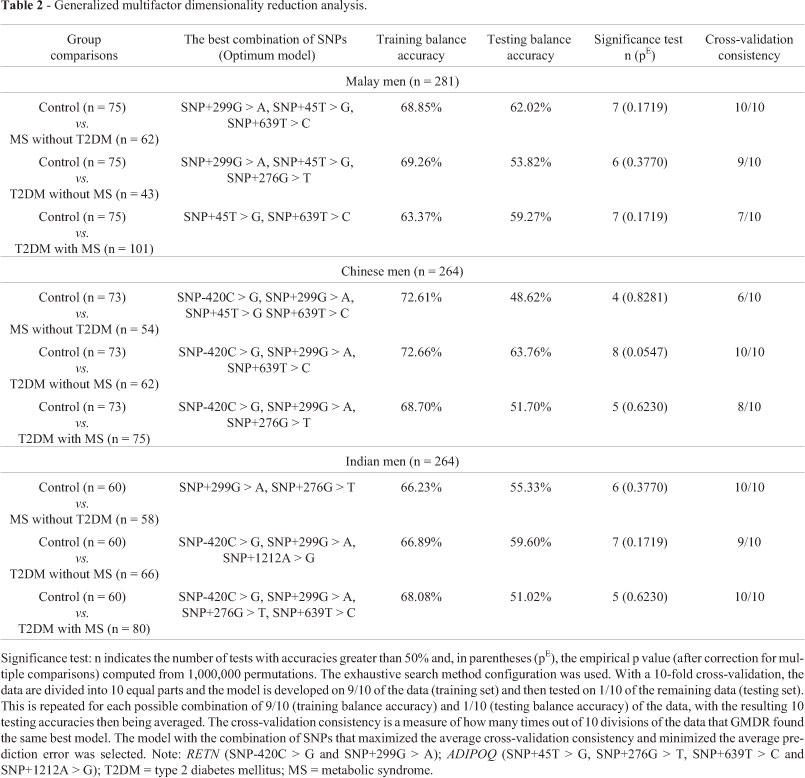
Despite the above relationships, there were no interactions between ADIPOQ (SNP+45T > G, SNP+276G > T, SNP+639T > C and SNP+1212A > G) and RETN (SNP-420C > G and SNP+299G > A) gene polymorphisms and their adipokine levels (Table 1). GMDR analysis showed that none of these SNP combinations was associated with the risk of T2DM and MS among Malaysian Malay, Chinese and Indian men (Table 2). Despite the close structural relationship between the ADIPOQ and RETN genes (Figures 1 and 2), the functional relationship between these genes was unclear (Tables 1 and 2). The extent to which the structural relationship observed here was related to gene transcriptional activity (adipokine levels) and/or metabolic disturbances (risk of T2DM and MS) remains to be determined.
Genome-based linkage and association studies have shown marked variations in the circulating levels of adiponectin (Henneman et al., 2010) and resistin (Menzaghi et al., 2006). We have previously reported a strong association between RETN gene polymorphisms and resistin levels, but only a marginal association between ADIPOQ gene polymorphisms and adiponectin levels (Lau and Muniandy, 2011b). Recent comprehensive linkage disequilibrium mapping studies have shown that ADIPOQ (Heid et al., 2010) and RETN (Asano et al., 2010) gene polymorphisms, including the SNPs examined here, account for the high variablity in the levels of their respective adipokines. Nevertheless, the genetic epistasis of adiponectin and resistin remain largely unexplored.
The results described here show that the ADIPOQ and RETN genes did not have a reciprocal effect on their adipokine levels, i.e., ADIPOQ did not affect resistin levels nor did RETN affect adiponectin levels (Table 1). To our knowledge, an association between RETN gene polymorphisms and adiponectin levels has been reported in only one study in which the G-allele of SNP-420C > G at the RETN locus was associated with lower circulating adiponectin levels in Japanese patients (Miyamoto et al., 2009). The present study is the first to examine the association between ADIPOQ gene polymorphisms and resistin levels. Although not statistically significant, the C-allele of SNP+639T > C and the G-allele of SNP+1212A > G at the ADIPOQ locus possibly provided a small contribution to the elevated serum resistin levels in individuals with these polymorphisms (Table 1). Recent large scale genome-wide association studies have suggested that these rare alleles are also associated with an increased risk of hypoadiponectinemia (Ling et al., 2009; Heid et al., 2010). Since the ADIPOQ and RETN genes encode functionally-related proteins, a deleterious mutation in the ADIPOQ gene could indirectly interfere with RETN expression (probably through adipokine interaction or metabolic pathway rather than via genetic regulation). In agreement with this conclusion, genes with similar molecular functions normally have similar epistatic interactions. Thus, if in a particular condition two metabolic pathways can produce the same metabolite, then inactivation of either pathway alone will have little effect on fitness (Lehner, 2011).
Paradoxically, the rare homozygous genotype (risk alleles) of SNP-420C > G at the RETN locus tended to be co-inherited with the common homozygous genotypes (protective alleles) of SNP+639T > C (Figure 1C) and SNP+1212A > G (Figure 1D) at the ADIPOQ locus. The T-allele of SNP+639T > C and the A-allele of SNP+1212A > G have a protective effect against T2DM and MS (Heid et al., 2010; Siitonen et al., 2011), whereas the G-allele of SNP-420C > G triggers these conditions (Ochi et al., 2007; Miyamoto et al., 2009). Recombination between SNPs in these genes may prevent disease-causing alleles of these SNPs from being co-inherited. Indeed, some studies have reported a prevalence of antagonistic epistasis, i.e., the tendency of multiple mutations to have a mitigating/alleviating effect rather than a reinforcing/aggravating effect (Desai et al., 2007). Recombination and mutations provide the basis for less synergistic or more antagonistic epistasis (Desai et al., 2007). The results described here provide no conclusive evidence for a link between the structural similarity and/or functional similarity (synergistic or antagonistic epistasis) between these two genes. Indeed, the extent to which the ADIPOQ gene interferes with RETN expression remains to be determined.
Although ADIPOQ and RETN are located on different chromosomes, the intermolecular epistatic interaction between these genes is possible (Charalampos and Richard, 2006). Epistatic interactions between mutations in different genes may be caused by different underlying molecular mechanisms (Lehner, 2011). If two genes share a common function, then the loss of one gene could be compensated for by continued activity of the second gene (Lehner, 2011). Since adiponectin and resistin have opposite physiological effects (Lau and Muniandy, 2011a), polymorphisms in ADIPOQ and RETN are likely to affect disease outcome by acting antagonistically during natural selection. Despite the structural similarity between ADIPOQ and RETN (Figures 1 and 2), there was no evidence of an intermolecular epistatic interaction between these genes (Table 2). The ADIPOQ and RETN genes may not, by themselves, exhibit a significant association with the risk of T2DM and MS because their combined effect may be relatively small and dependent on genotypes at other loci; the latter may compensate for variation in the loci under study and may differ across populations. Indeed, recent large-scale systematic reviews and meta-analyses have shown marked heterogeneity among SNPs associated with the ADIPOQ (Zhao and Zhao, 2011) and RETN (Wen et al., 2010) loci.
The findings of this study relate to Malaysian men and may not be applicable to other groups because of genetic differences. Fine-mapping studies are needed since our investigation did not cover upstream and downstream variations in the ADIPOQ and RETN genes. Comprehensive linkage disequilibrium mapping and haplotype-based data mining would allow a detailed analysis of haplotype-haplotype interactions and more insight into the genetic risk factors associated with T2DM and MS. In addition to the ADIPOQ and RETN genes, other genes involved in the susceptibility to T2DM and MS should be included when modelling high-order gene-gene interactions. The availability of more powerful computer-based approaches will allow large scale genome-proteome interaction-based analyses to map epistatic interactions on a wider scale. In this context, understanding the role of "gene-environment" interactions is crucial for the characterization of low-penetrance genes. Further studies with larger sample sizes than that used here are required in order to reach more reliable conclusions.
Acknowledgments
This work was supported by University of Malaya postgraduate research grants (grant nos. PS102-2009A and PS201-2010A), a short-term research university grant (grant no. FS232-2008C) and an e-science fund grant (grant no. 12-02-03-2044). We thank all those who participated in this project, including the nurses of the University of Malaya Medical Centre (UMMC), particulary Madam Farahwahidah who helped with blood sample collection. We also thank members of our laboratory who provided support and helpful discussions throughout this work.
Internet Resources
Received: May 19, 2011; Accepted: August 4, 2011.
Associate Editor: Mara Hutz
License information: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Alberti KG, Zimmet P and Shaw J (2005) The metabolic syndrome - A new worldwide definition. Lancet 366:1059-1062.
- Asano H, Izawa H, Nagata K, Nakatochi M, Kobayashi M, Hirashiki A, Shintani S, Nishizawa T, Tanimura D, Naruse K, et al. (2010) Plasma resistin concentration determined by common variants in the resistin gene and associated with metabolic traits in an aged Japanese population. Diabetologia 53:234-246.
- Charalampos GS and Richard AF (2006) Managing associations between different chromosomes. Science 312:207-208.
- Desai MM, Weissman D and Feldman MW (2007) Evolution can favor antagonistic epistasis. Genetics 177:1001-1010.
- Heid IM, Henneman P, Hicks A, Coassin S, Winkler T, Aulchenko YS, Fuchsberger C, Song K, Hivert MF, Waterworth DM, et al. (2010) Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: Results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 208:412-420.
- Henneman P, Aulchenko YS, Frants RR, Zorkoltseva IV, Zillikens MC, Frolich M, Oostra BA, van Dijk KW and van Duijn CM (2010) Genetic architecture of plasma adiponectin overlaps with the genetics of metabolic syndrome-related traits. Diabetes Care 33:908-913.
- Lau CH and Muniandy S (2011a) Novel adiponectin-resistin (AR) and insulin resistance (IR-AR) indexes are useful integrated diagnostic biomarkers for insulin resistance, type 2 diabetes and metabolic syndrome: A case control study. Cardiovasc Diabetol 10:8.
- Lau CH and Muniandy S (2011b) Adiponectin and resistin gene polymorphisms in association with their respective adipokine levels. Ann Hum Genet 75:370-382.
- Lehner B (2011) Molecular mechanisms of epistasis within and between genes. Trends Genet 27:323-331.
- Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ, Kesaniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP, et al. (2009) Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: The GEMS study. Obesity 17:737-744.
- Menzaghi C, Coco A, Salvemini L, Thompson R, De Cosmo S, Doria A and Trischitta V (2006) Heritability of serum resistin and its genetic correlation with insulin resistance-related features in nondiabetic Caucasians. J Clin Endocrinol Metab 91:2792-2795.
- Miyamoto Y, Morisaki H, Kokubo Y, Yamanaka I, Tomoike H, Okayama A, Yoshimasa Y and Morisaki T (2009) Resistin gene variations are associated with the metabolic syndrome in Japanese men. Obes Res Clin Pract 3:65-74.
- Moore JH (2003) The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered 56:73-82.
- Ochi M, Osawa H, Hirota Y, Hara K, Tabara Y, Tokuyama Y, Shimizu I, Kanatsuka A, Fujii Y, Ohashi J, et al. (2007) Frequency of the G/G genotype of resistin single nucleotide polymorphism at -420 appears to be increased in younger-onset type 2 diabetes. Diabetes 56:2834-2838.
- Patel SD, Rajala MW, Rossetti L, Scherer PE and Shapiro L (2004) Disulfide-dependent multimeric assembly of resistin family hormones. Science 304:1154-1158.
- Siitonen N, Pulkkinen L, Lindstrom J, Kolehmainen M, Eriksson JG, Venojarvi M, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Tuomilehto J and Uusitupa M (2011) Association of ADIPOQ gene variants with body weight, type 2 diabetes and serum adiponectin concentrations: The Finnish Diabetes Prevention Study. BMC Med Genet 12:5.
- Wen Y, Lu P and Dai L (2010) Association between resistin gene -420 C/G polymorphism and the risk of type 2 diabetes mellitus: A meta-analysis. Acta Diabetol, DOI: 10.1007/s00592-010-0247-8.
- Zhao T and Zhao J (2011) Genetic effects of adiponectin on blood lipids and blood pressure. Clin Endocrinol (Oxf) 74:214-222.
- HOMA calculator software ver. 2.2.2, Diabetes Trials Unit, Oxford University, http://www.dtu.ox.ac.uk/homa (April 25, 2011).
- HAPSTAT program, ver. 3.0, http://www.bios.unc.edu/~lin/hapstat (April 3, 2011).
- Generalized Multifactor Dimensionality Reduction (GMDR) Beta program, ver. 0.7, http://sourceforge.net/projects/gmdr (May 18, 2011).
Publication Dates
-
Publication in this collection
15 Dec 2011 -
Date of issue
2012
History
-
Received
19 May 2011 -
Accepted
04 Aug 2011