Abstract
Entomopathogenic fungi can produce a series of chitinases, some of which function synergistically with proteases and other hydrolytic enzymes to degrade the insect cuticle. In the present study, the chitinase gene Ifu-chit2 from Isaria fumosorosea was investigated. The Ifu-chit2 gene is 1,435-bp long, interrupted by three short introns, and encodes a predicted protein of 423 amino acids with a 22 residue signal peptide. The predicted Ifu-Chit2 protein is highly homologous to Beauveria bassiana chitinase Bbchit2 and belongs to the glycohydrolase family 18. Ifu-Chit2 was expressed in Escherichia coli to verify chitinase activity, and the recombinant enzyme exhibited activity with a colloidal chitin substrate. Furthermore, the expression profiles of Ifu-chit2 were analyzed at different induction times under in vivo conditions. Quantitative real-time PCR analysis revealed that Ifu-chit2 expression peaked at two days post-induction. The expression of chitinase Ifu-chit2 in vivo suggests that the chitinase may play a role in the early stage of pathogenesis.
Isaria fumosorosea; chitinase; prokaryotic expression; quantitative real-time PCR
Introduction
Entomopathogenic fungi are widely distributed throughout the fungal kingdom and are an important group of microorganisms that have been used as biological controls against insect pests in many agroecosystems (Hajek and Delalibera Jr, 2010Hajek AE and Delalibera Jr I (2010) Fungal pathogens as classical biological control agents against arthropods. Biocontrol 55:147–158.; Mishra et al., 2013aMishra S, Kumar P and Malik A (2013a) Preparation, characterization, and insecticidal activity evaluation of three different formulations of Beauveria bassiana against Musca domestica. Parasitol Res 112:3485–3495.,bMishra S, Kumar P and Malik A (2013b) Effect of process parameters on the enzyme activity of a novel Beauveria bassiana isolates. Int J Curr Microbiol Appl Sci 2:49–56.). Isaria fumosorosea is a cosmopolitan entomopathogenic fungus parasitizing diverse insect species, including the important agricultural pests diamondback moth and whitefly. I. fumosorosea is being used increasingly as a biological control agent for several insect pests (Ali et al., 2010aAli S, Huang Z and Ren S (2010a) Production of cuticle degrading enzymes by Isaria fumosorosea and their evaluation as a biocontrol agent against diamondback moth. J Pest Sci 83:361–370.,bAli S, Wu J, Huang Z and Ren SX (2010b) Production and regulation of extracellular chitinase from the entomopathogenic fungus Isaria fumosorosea. Biocontrol Sci Technol 20:723–738.).
Chitinases (EC.3.2.1.14) catalyze the hydrolysis of chitin, and these enzymes have diverse functions ranging from nutritional roles in bacteria and archaea, defensive roles in plants, developmental roles in insects and morphogenetic, nutritional and invasive functions in fungi (Li, 2006Li DC (2006) Review of fungal chitinases. Mycopathologia 161:345–360.; Hartl et al., 2012Hartl L, Zach S and Seidl-Seiboth V (2012) Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93:533–543.). During fungal penetration through the host cuticle, entomopathogenic fungi produce hydrolytic enzymes such as proteases, chitinases and lipases, that degrade the insect cuticle and can initiate the infection process (Charnley and St Leger, 1991Charnley AK and St Leger RJ (1991) The role of cuticle-degrading enzymes in fungal pathogenesis in insects. In: Cole GT and Hoch HC (eds) The Fungal Spore Disease Initiation in Plants and Animals. Plenum Press, New York, pp 267–286.; Charnley, 2003Charnley AK (2003) Fungal pathogens of insects: Cuticle degrading enzymes and toxins. Adv Bot Res 40:241–321.). Entomopathogenic fungal chitinases are attractive candidates for the biological control of insect pests in agroforestry (Zhu et al., 2008Zhu YP, Pan JR, Qiu JZ and Guan X (2008) Isolation and characterization of a chitinase gene from entomopathogenic fungus Verticillium lecanii. Braz J Microbiol 39:314–320.).
Chitinases are grouped into glycoside hydrolase families 18 (GH18) and 19 (GH19) by amino acid sequence similarity of their catalytic domains (Henrissat, 1991Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316.). Enzymes in these families differ in their primary sequence, three-dimensional (3D) structure, and molecular mechanisms of catalysis (Henrissat and Bairoch, 1996Henrissat B and Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696.; Henrissat and Davies, 1997Henrissat B and Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7:637–644.). GH family18 are evolutionarily diverse and represent an ancient chitinase family widely distributed in a variety of organisms including bacteria, fungi, animals, and some plant species, while family 19 chitinases are found only in higher plants and some Gram-positive bacteria such as Streptomyces griseus (Ohno et al., 1996Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M and Watanabe T (1996) A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol 178:5065–5070.; Lee et al., 2009Lee YG, Chung KC, Wi SG, Lee JC and Bae HJ (2009) Purification and properties of a chitinase from Penicillium sp. LYG 0704. Protein Express Purif 65:244–250.; Ahmed et al., 2012Ahmed NU, Park JI, Seo MS, Kumar TS, Lee IH, Park BS and Nou IS (2012) Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica. Mol Biol Rep 39:3649–3657.). Filamentous fungi have many different chitinases belonging to GH family 18 (Li, 2006Li DC (2006) Review of fungal chitinases. Mycopathologia 161:345–360.), and all GH family 18 proteins present a common α/β TIM-barrel fold and include a DXXDXDXE sequence motif (Bokma et al., 2002Bokma E, Rozeboom HJ, Sibbald M, Dijkstra BW and Beintema JJ (2002) Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur J Biochem 269:893–901.).
Several chitinases have been characterized from Metarhizium anisopliae and Beauveria bassiana in vivo (Fang et al., 2005Fang WG, Leng B, Xiao YH, Jin K, Ma JC, Fan YH, Feng J, Yang XY, Zhang YJ and Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370.; Bhanu Prakash et al., 2012Bhanu Prakash GV, Padmaja V, Jami SK and Kirti PB (2012) Expression of chitinase genes of Metarhizium anisopliae isolates in lepidopteran pests and on synthetic media. J Basic Microbiol 52:628–635.). The Chit1 chitinase from M. anisopliae was shown to have little or no effect on virulence, while overproduction of chitinase Bbchit1 in B. bassiana did enhance the biocontrol activity of the fungus against aphids (Screen et al., 2001Screen SE, Hu G and St Leger RJ (2001) Transformants of Metarhizium anisopliae sf. anisopliae overexpressing chitinase from Metarhizium anisopliae sf. acridum show early induction of native chitinase but are not altered in pathogenicity to Manduca sexta. J Invertebr Pathol 78:260–266.; Fang et al., 2005Fang WG, Leng B, Xiao YH, Jin K, Ma JC, Fan YH, Feng J, Yang XY, Zhang YJ and Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370.). Chitinase chi2 of M. anisopliae has been reported to be responsible for virulence in a bioassay of transgenic isolates with the chitinase gene overexpressed and silenced (Boldo et al., 2009Boldo JT, Junges A, do Amaral KB, Staats CC, Vainstein MH and Schrank A (2009) Endochitinase CHI2 of the biocontrol fungus Metarhizium anisopliae affects its virulence toward the cotton stainer bug Dysdercus peruvianus. Curr Genet 55:551–560.).
Currently, I. fumosorosea produces chitinases that are effective in hydrolyzing and destroying the cuticle of various insects (Ali et al., 2010aAli S, Huang Z and Ren S (2010a) Production of cuticle degrading enzymes by Isaria fumosorosea and their evaluation as a biocontrol agent against diamondback moth. J Pest Sci 83:361–370.,bAli S, Wu J, Huang Z and Ren SX (2010b) Production and regulation of extracellular chitinase from the entomopathogenic fungus Isaria fumosorosea. Biocontrol Sci Technol 20:723–738.). The I. fumosorosea chitinase gene Ifu-chit1 was isolated by our group previously and transgenic B. bassiana overexpressing this gene showed significantly improved virulence against the moth Dendrolimus punctatus compared with the wild-type strain (Tang et al., 2009Tang Q, Zhang YP, Xie L, Song LH, Li ZZ and Huang B (2009) Generation of transgenic Beauveria bassiana strains with chitinase gene from Isaria fumosorosea and its increased virulence against Dendrolimus punctatus (Lepidoptera, Lasiocampidae). Acta Entomol Sin 52:755–762.). Whether there are more chitinase genes in I. fumosorosea is currently unknown. In this study, we identified another chitinase gene from I. fumosorosea (Ifu-chit2) and characterized it using bioinformatics, Rapid Amplification of cDNA Ends (RACE) and heterologous expression in Escherichia coli. Furthermore, the expression profiles of Ifu-chit2 under in vivo conditions were investigated.
Materials and Methods
Fungal strains, host insects and growth conditions
The fungal strain used in this study was I. fumosorosea strain RCEF3304 (isolated from Diptera pupa from Jiangxi province, China), obtained from the Anhui Provincial Key Laboratory of Microbial Pest Control, Hefei, China. Mycelia were harvested from chitinase induction medium containing ground chitin (1%, w/v) in basal salts medium according to Wang’s procedure (Wang et al., 2013Wang ZX, Meng HM, Zhuang ZL, Chen MJ, Xie L and Huang B (2013) Molecular cloning of a novel subtilisin-like protease (Pr1A) gene from the biocontrol fungus Isaria farinosa. Appl Entomol Zool 48:477–487.).
For in vivo experiments, larvae of the greater wax moth (Galleria mellonella) obtained from Ruiqing Bait (Jiangsu, China) were used as host insects for fungal development. A conidial suspension (100 μL at 108 conidia/mL) was evenly spread onto potato dextrose agar (PDA) and grown for 12 days at 25 °C. Fifty insects were rolled on the conidiating culture, individually placed in sterilised plastic vials and incubated at 25 °C. Different stages of infection were monitored over a pathogenesis period of 4 days without feeding. At each of the observed infection stages, 5–7 insects were ground to a fine powder in liquid nitrogen for total RNA extraction.
Cloning and sequencing of the full length cDNA and genomic DNA of Ifu-chit2
Total RNA was extracted from ground, frozen mycelia using TRizol reagent (Invitrogen, CA, USA) and quantified using a spectrophotometer. 1 μg of RNA was reverse transcribed using a PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, China). Reverse-transcribed cDNA was used as template for PCR amplification with degenerate PCR primers designed based on conserved domains of fungal chitinases to amplify the conserved fragment (Table 1). PCR was carried out following manufacturer’s instructions. Based on the acquired core region sequence, two pairs of gene-specific primers were designed for 5′- and 3′-RACE PCR to amplify the 5′- and 3′- ends of Ifu-chit2, respectively (Table 1). The 5′ and 3′ UTR regions were amplified using a SMART RACE cDNA Amplification Kit (Clontech, USA), and products were cloned and sequenced.
DNA was extracted using a slight modification of the CTAB method and resuspended in pre-warmed sterile deionized water. The DNA sequence of Ifu-chit2 gene was amplified with primers designed based on the Ifu-chit2 chitinase cDNA using I. fumosorosea genomic DNA as template.
Bioinformatics analysis of Ifu-Chit2
Protein parameters were calculated using Protparam at Expasy (Gasteiger et al., 2005Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD and Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker HM (ed) The Proteomics Protocols handbook. Humana Press, Totowa, pp 571–607.), and signal peptide prediction was carried out using the SignalP 4.0 server (Petersen et al., 2011Petersen TN, Brunak S, von Heijne G and Nielsen H (2011) SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786.). Sequences of homologs from other species were obtained using the BLASTP tool, and homologous sequences were used for multiple sequence alignment and generation of a phylogenetic tree by applying the neighbor-joining (NJ) methods in MEGA version 4.1 with ClustalW. Confidence limits were estimated from 1000 bootstrapping replicates.
Expression and purification of recombinant Ifu-chit2 protein
Primers 5-CCGGAATTCATGCTGGGTTTCCTCA GGAAAT-3 and 5-ATAAGAATGCGGCCGCCTA GTGGTGATGGTGATGGTG AGCCATGCTATTCCT GATATTG-3 (restriction enzyme sites are underlined; the 6His-tag sequence is in bold) were designed for PCR amplification based on the encoding region of the Ifu-chit2 gene. The PCR product was cloned into the pET-28a (+) vector (Novagen), resulting in recombinant expression vector pET-28a-Ifu-chit2. This was transformed into E. coli strain BL21 (DE3) chemically competent cells (TransGen, China) and grown at 37 °C in Luria-Bertani (LB) medium containing 50 μg/mL kanamycin. Ifu-Chit2 expression was induced by addition of isopropyl β-D-thiogalactoside (IPTG) to a final concentration of 0.5 mM and cultivation was continued for an additional 6 h at 28°C. Cells were collected at 1–6 h by centrifugation (4,000 rpm for 10 min), analyzed by 12% SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Cells induced for 5 h were harvested and loaded onto a His-Bind Niagarose column (Cwbio, China) and the target protein was purified according to the manufacturer’s instructions.
Western blot analysis
For Western blot analysis, 12% SDS-PAGE was carried out to separate proteins prior to transfer onto a nitro-cellulose membrane (Pall) at 100 mA for 1 h using a Semi-Dry Trans-Blot Cell (Bio-Rad). Non-specific protein-protein interactions were blocked using 5% non-fat dry milk in TBST buffer (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween-20) for 1 h. Membranes were incubated overnight at 4 °C with mouse anti-His antibody (Abmart, Shanghai) diluted 1:5,000 in blocking buffer, and washed three times for 5 min each time in TBST buffer. The second anti-body, AP-conjugated goat anti-mouse IgG (Promega) was diluted 1:7,500 in TBST buffer, incubated with the membrane for 1 h, and washed three times for 5 min each time in TBST buffer, followed by visualization with Western blue stabilized substrate for alkaline phosphatase (Promega).
Chitinase activity assay
Chitinase activity was measured as described previously with modifications (Mauch et al., 1984Mauch F, Hadwiger LA and Boller T (1984) Ethylene: Symptom, not signal for the induction of chitinase and β-1, 3-glucanase in pea pods by pathogens and elicitors. Plant Physiol 76:607–611.). Incubations consisted of 350 μL 100 mM sodium acetate buffer (pH 6.5), 60 μg of suitably diluted enzyme and 200 μL of 1% colloidal chitin. After 1 h incubation at 37 °C, reactions were stopped by centrifugation at 10,000 rpm for 5 min. Supernatants (300 μL) were boiled with 100 μL of potassium tetraborate buffer for 3 min, and 2.5 mL of DMAB re-agent (10% (w/v) 4-(dimethyl amino) benzaldehyde in glacial acetic acid: 11.5 M HCl (87.5:12.5, v/v) was added to the reactions and then incubated at 37 °C for 20 min. The change in absorbance at 420 nm of the supernatants (suitably diluted enzyme boiled for 30 min as a control) was recorded using a Spectra Max M2 (Molecular Devices, USA). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the release of 1 μmol of GlcNAc per mL in 1 h.
Quantitative real-time PCR analysis of Ifu-chit2 gene expression under in vivo conditions
For Ifu-chit2 expression studies, infected insects were collected for real-time PCR after 1d (1 day after inoculation), 2d (2 days after inoculation), 3d (melanization of the infected insect) and 4d (death insect), respectively. Total RNA was isolated from harvested infected insects using Trizol (Invitrogen, USA). Expression of Ifu-chit2 was quantified using a quantitative real-time PCR assay performed in a 7500 Real-Time PCR System (Applied Bio-systems, USA), using the SYBR Green kit (TaKaRa, China) following the manufacturer’s instructions. Specific primers for Ifu-chit2 and translation elongation factor (TEF) were designed for real-time PCR amplification (Table 1). Synthesis of cDNA and quantitative real-time PCR was performed in triplicate for each gene. The expression levels of Ifu-chit2 in vivo at 1 day after induction were given a value of one, and relative expression levels were calculated using the formula 2-ΔΔCT with TEF as the internal control for each sample (Livak and Schmittgen, 2001Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408.).
Results
Cloning and sequence analysis of Ifu-chit2
Degenerate PCR primers corresponding to conserved domains of entomopathogenic fungal chitinases were designed as described above, and a 766-bp fragment was amplified using primers chit2-F1 and chit2-R1. A BLAST search of the sequenced PCR product revealed high homology to chitinase genes from Cordyceps confragosa (72%), Isaria farinosa (72%), Aphanocladium album (71%) and B. bassiana (70%), which indicated that the fragment was a partial sequence of the chitinase Ifu-chit2 from I. fumosorosea.
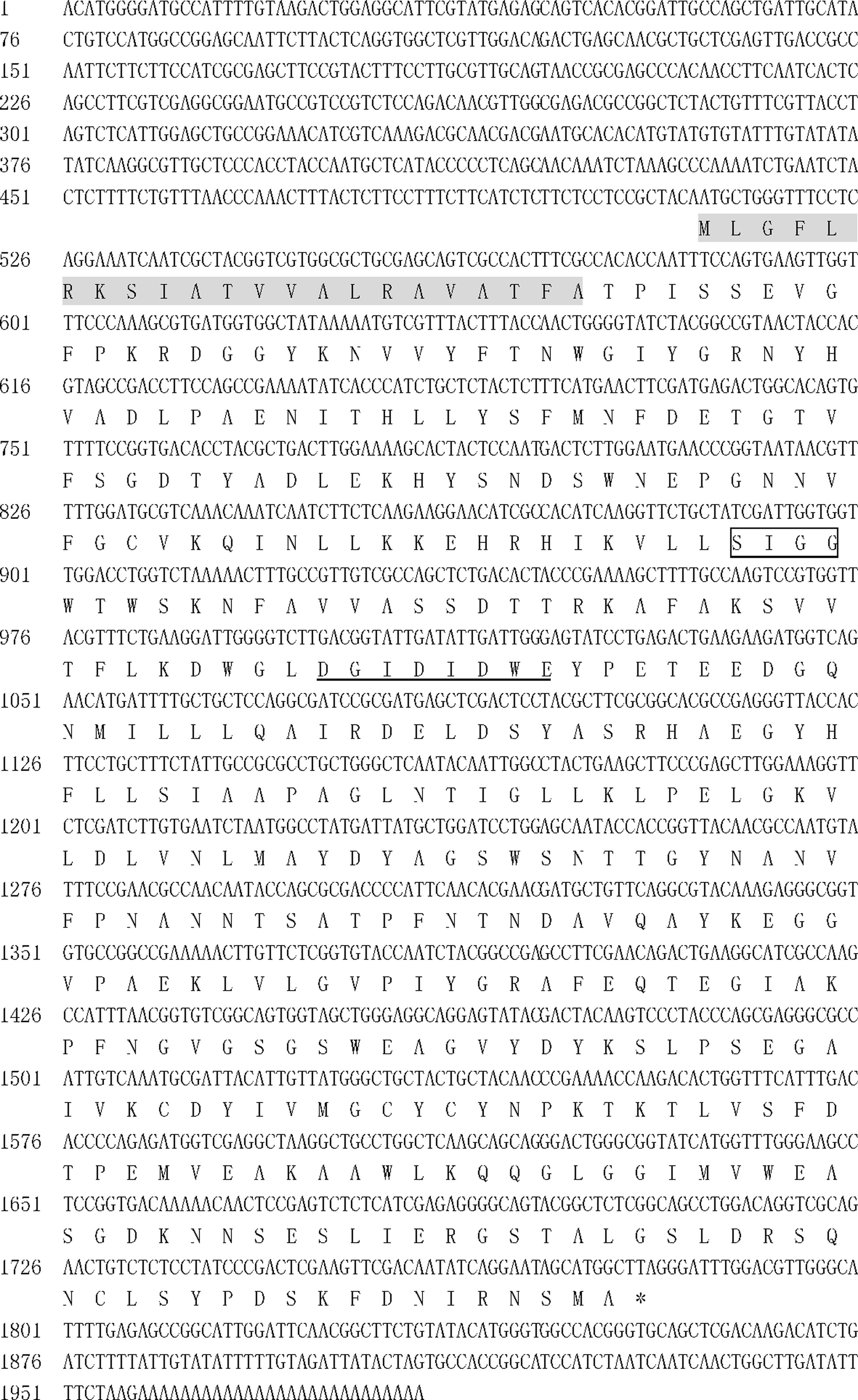
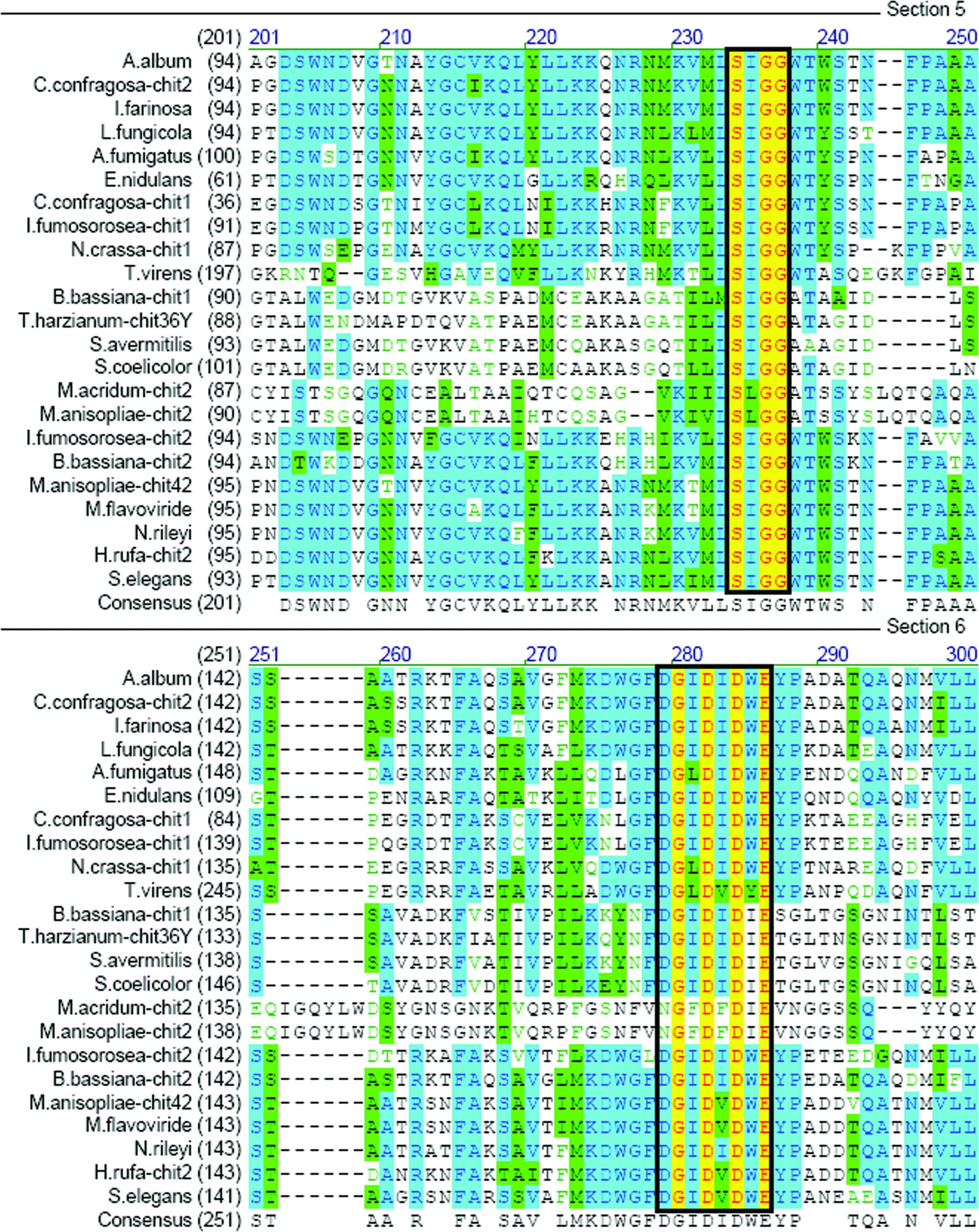
The 5′ and 3′ ends of the partial sequence were extended from gene-specific primers using the RACE approach. The 5′-UTR, ORF, and 3′-UTR were 510 bp, 1272 bp, and 202 bp, respectively (Figure 1). The deduced protein was 423 aa with a predicted molecular mass of 46.57 kDa (Figure 1), a predicted isoelectric point (pI) of 5.22, and a 22-aa signal peptide at the N-terminus (shaded, Figure 1). Comparison of the predicted Ifu-Chit2 chitinase with fungal orthologs revealed two highly conserved regions of the catalytic domain (SIGG and DGIDIDWE) that correspond to a substrate-binding site and catalytic residues, respectively. This confirmed that Ifu-Chit2 was a member of glycosyl hydrolase family 18 (Figure 2). A phylogenetic tree showed that Ifu-Chit2 is more closely related to B. bassiana chit2 than to chitinases of other species (Figure 3).
Full-length cDNA and deduced amino acid sequence of Ifu-chit2 from Isaria fumosorosea. Numbers to the left indicate the nucleotide and amino acid positions. The conserved sequences SIGG and DGIDIDWE are boxed and underlined, respectively. An asterisk indicates the end of the protein sequence, and the signal peptide is shaded.
Multiple sequence alignment of the core region of the catalytic domains of Ifu-Chit2 and related chitinases. Identical amino acids are high-lighted in yellow, and similar residues are shown in green or light-green. The consensus (average) sequence is shown below the aligned sequences. Highly conserved motifs SxGG and DxxDxDxE are boxed. GenBank accession numbers for are listed in parentheses: A. album CHIT (CAA45468), C. confragosa CHIT2 (AAV98692), I. farinosa CHIT (ABD64606), Lecanicillium fungicola CHIT (AAP45631), Aspergillus fumigatus CHIT (AAO61686), Emericella nidulans CHIT (BAA35140), C. confragosa CHIT1 (AAX56960), I. fumosorosea CHIT1 (FJ377733), Neurospora crassa CHIT1 (EAA36073), Trichoderma virens CHIT (AAL84697), B. bassiana CHIT1 (AY145440), T. harzianum CHIT36Y (AAL01372), Streptomyces avermitilis CHIT (NP_826813), S. coelicolor CHIT (NP_626743), Metarhizium acridum CHIT2 (AJ293217), M. anisopliae CHIT2 (AAY34347), B. bassiana CHIT2 (AY147011), M. anisopliae CHIT42 (AAB81998), M. flavoviride CHIT (CAB44709), Nomuraea rileyi CHIT (AY264288), Hypocrea rufa CHIT (AAF19617), Stachybotrys elegans CHIT (AAM70478).
Phylogenetic analysis of chitinase sequences from different pathogenic fungi. The phylogenetic tree was created by the neighbor-joining method in Mega version 4.1 using default settings based on the multiple sequence alignment. A bootstrap value is attached to each branch. GenBank accession numbers are referenced in the legend of Figure 2.
Moreover, a knowledge-based protein model tool from the SWISS-MODEL program was used to predict the 3D structure of the Ifu-Chit2 protein based on the tertiary structures of related chitinases in the database. The model exhibited the expected α/β-barrel consisting of eight α-helices and eight parallel β-strands that alternate along the peptide backbone, which is consistent with Coccidioides immitis CiX1 and Aspergillus fumigatus ChiB1 (Figure 4) (Li, 2006Li DC (2006) Review of fungal chitinases. Mycopathologia 161:345–360.).
Predicted structure of Isaria fumosorosea Ifu-Chit2. The top view of the TIM barrel structure of the superimposed model is shown. Red letters indicate the conserved sequences SIGG and DGIDIDWE.
The genomic sequence of Ifu-chit2 was 1435 bp due to the presence of three introns at 121–176, 277–331 and 380–431 (data not shown). The exon/intron splice sites (5′GT-AG3′) were representative of gap junctions reported for other fungi chitinase genes isolated from C. confragosa (ABD77096), A. album (X64104) and Aspergillus nidulans (D87063).
Expression of recombinant Ifu-Chit2 in E. coli
The Ifu-chit2 encoding sequences were successfully inserted into pET-28a (+) vectors and transformed into E. coli BL21 (DE3), and SDS-PAGE analysis confirmed that the Ifu-Chit2 protein was expressed following induction. A protein with a molecular mass of approximately 50 kDa was clearly visible, which was slightly larger than the predicted size (Figure 5). The maximum expression was achieved 5 h after induction at 28 °C. Western blot analysis with anti-His antibody confirmed that the overexpressed protein included a His-tag (Figure 5), and the protein was purified using a His-Band Ni-agarose column (Figure.5). Purified recombinant Ifu-Chit2 exhibited an enzyme activity of 32.7 U/mL, which was much higher than that of Vlchit1 protein from Verticillium lecanii (Zhu et al., 2008Zhu YP, Pan JR, Qiu JZ and Guan X (2008) Isolation and characterization of a chitinase gene from entomopathogenic fungus Verticillium lecanii. Braz J Microbiol 39:314–320.). The result indicated that Ifu-Chit2 was successfully expressed in active forms at a higher level.
SDS-PAGE and Western blot analysis of the expression of recombinant Ifu-Chit2 in E. coli. Lane M: pre-stained protein marker; Lane 1: total protein of cells containing empty vector pET-28a with IPTG induction; Lane 2: total protein of cells containing expression vector pET-28a-Ifu-chit2 without IPTG induction; Lanes 3–8: total protein of cells containing expression vector pET-28a-Ifu-chit2 induced by IPTG at 28 °C for 1, 2, 3, 4, 5 and 6 h, respectively; Lane 9: purified recombinant Ifu-Chit2 (50 kDa). Lane 10: total protein of cells containing empty vector pET-28a with IPTG induction; Lane 11: total protein of cells containing expression vector pET-28a-Ifu-chit2 without IPTG induction; Lane 12: total protein of cells containing expression vector pET-28a-Ifu-chit2 induced by IPTG at 28 °C for 4 h; Lane13: Purified recombinant Ifu-Chit2.
Expression profiles of Ifu-chit2 under in vivo conditions
The expression profiles of Ifu-chit2 under in vivo conditions were investigated using quantitative real-time PCR (Figure 6). A single product-specific melting curve was obtained using for Ifu-chit2, indicating that the primers amplified efficiently and were specific for the gene of interest. Ifu-chit2 gene was found to be expressed constitutively during all stages of insect infection examined (1–4 days). The data normalized to endogenous reference gene were presented as the fold-change in gene expression during different stages of infection and relative to the levels of expression observed at 1 day after infection. Ifu-chit2 expression levels peaked at 2 days after infection (Figure 6). Neither chitinase gene was expressed in uninoculated insects.
Expression analysis of Ifu-chit2 at different times under in vivo conditions. mRNA transcripts were measured at 1, 2, 3 and 4 days after inoculation using qRT-PCR. Relative expression levels were calculated using the comparative ΔΔCt method. The constitutively expressed translation elongation factor gene served as endogenous control, and expression levels are shown as relative to those at 1 day after inoculation, which were given a value of 1. Data are expressed as the mean ± SE (standard error) of three independent experiments. Student’s t-test was used to determine the statistical significance of differences between groups. Differences are considered significant for p < 0.05.
Discussion
Like most fungal pathogens, I. fumosorosea may use a combination of chitinases, proteases and lipases to penetrate the insect cuticle and access the host hemocoel, and extracellular chitinases may therefore be important for virulence. In the present study, we successfully isolated a gene from I. fumosorosea that encodes for a chitinase (Ifu-Chit2). Ifu-Chit2 was successfully expressed in E. coli BL21 (DE3) with the product of recombinant protein Ifu-Chit2 (50 kDa).
Most chitinase genes cloned and characterized to date have been classified into two families of glycosyl hydrolases (family 18 and 19), based on amino acid sequence similarity (Henrissat, 1991Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316.; Henrissat and Bairoch, 1993Henrissat B and Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788.). Our results indicated that Ifu-Chit2 belonged to family 18, as it included the characteristic substrate binding and catalytic motifs (SXGG and DXXDXDXE) (Lu, et al., 2005Lu ZX, Laroche A and Huang HC (2005) Isolation and characterization of chitinases from Verticillium lecanii. Can J Microbiol 51:1045–1055.; Baratto, et al., 2006Baratto CM, Dutra V, Boldo JT, Leiria LB, Vainstein MH and Schrank A (2006) Isolation, characterization, and transcriptional analysis of the chitinase chi2 gene (DQ011663) from the biocontrol fungus Metarhizium anisopliae var. anisopliae. Curr Microbiol 53:217–221.). Residues that are essential for chitinase activity, particularly Asp164, Asp167, Asp169 and Glu171, were found to be present in the Ifu-Chit2 catalytic domain, indicating a similar catalytic activity and structure to previously characterized enzymes of this family (Liu, et al., 2008Liu Z, Yang Q, Hu S, Zhang J and Ma J (2008) Cloning and characterization of a novel chitinase gene (chi46) from Chaetomium globosum and identification of its biological activity. Appl Microbiol Biotechnol 80:241–252.). The chitin-binding domain (ChBD) plays an important role in permitting chitinases to bind specifically to insoluble chitin (Limón, et al., 2004Limón MC, Chacón MR, Mejías R, DelgadoJarana J, Rincón AM, Codón AC and Benítez T (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl Microbiol Biotechnol 64:675–685.). Most bacterial chitinases in family 18 are characterized by the presence of a signal peptide, a catalytic domain, and a ChBD (Wu, et al., 2001Wu ML, Chuang YC, Chen JP, Chen CS and Chang MC (2001) Identification and characterization of the three chitin-binding domains within the multidomain chitinase chi92 from Aeromonas hydrophila JP101. Appl Environ Microbiol 67:5100–5106.). However, Ifu-Chit2 lacks the ChBD domain, as observed previously for some fungal chitinases (Li, 2006Li DC (2006) Review of fungal chitinases. Mycopathologia 161:345–360.). Ifu-Chit2 does include a signal sequence at the N-terminus, suggesting that it is secreted. Moreover, teleomorph of I. fumosorosea and B. bassiana belong to the genus Cordyceps sensu suto according to phylogenetic studies (Sung et al., 2007Sung GH, Hywel-Jones NL, Sung JM, Luangsa-Aard JJ, Shrestha B and Spataforal JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol 57:5–59.). Interestingly, to some extent, the relationship between Ifu-Chit2 and Bb-chit2 in the phylogenetic tree was consistent with the phylogenetic relationship between I. fumosorosea and B. bassiana.
It has been reported that the efficient up-regulation of chitinase expression during insect infection may be responsible for virulence in various fungi (Hartl et al., 2012Hartl L, Zach S and Seidl-Seiboth V (2012) Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93:533–543.). Recently, Bhanu Prakash et al. (2012)Bhanu Prakash GV, Padmaja V, Jami SK and Kirti PB (2012) Expression of chitinase genes of Metarhizium anisopliae isolates in lepidopteran pests and on synthetic media. J Basic Microbiol 52:628–635. reported that the four M. anisopliae chitinase genes chi, chi 1, chi 2 and chi 3 isolated from the insect hosts Spodoptera litura and Helicoverpa armigera during six developmental stages of the pathogen, were up-regulated in S. litura during mycosed and conidiated condition, whereas in H. armigera they were only expressed 48 h after incubation (Bhanu Prakash et al., 2012Bhanu Prakash GV, Padmaja V, Jami SK and Kirti PB (2012) Expression of chitinase genes of Metarhizium anisopliae isolates in lepidopteran pests and on synthetic media. J Basic Microbiol 52:628–635.). In the present study, most insects were dead four days after induction. Quantitative real-time PCR showed that Ifu-chit2 was up-regulated at two days after inoculation and then decreased over the duration of the experiment. We speculate that the expression of Ifu-chit2 acts against G. mellonella in the early stage of the infection process in order to solubilize the host cuticle.
Many filamentous fungi have been found to produce more than one kind of chitinase. Research has demonstrated that these chitinases have a mutually synergistic and complementary effect among them. Previous reports indicated that both the chitinase CHI2 and CHIT30 of M. anisopliae analyzed by molecular genetic tools play important roles for pathogenicity in the infection process (Boldo et al., 2009Boldo JT, Junges A, do Amaral KB, Staats CC, Vainstein MH and Schrank A (2009) Endochitinase CHI2 of the biocontrol fungus Metarhizium anisopliae affects its virulence toward the cotton stainer bug Dysdercus peruvianus. Curr Genet 55:551–560.; Staats et al., 2013Staats CC, Kmetzsch L, Lubeck I, Junges A, Vainstein MH and Schrank A (2013) Metarhizium anisopliae chitinase CHIT30 is involved in heat-shock stress and contributes to virulence against Dysdercus peruvianus. Fungal Biol 117:137–144.). It has been reported previously that Trichoderma harzianum chitinases CHIT33 and CHIT37 are able to enhance CHIT42 activity of degrading phytopathogenic cell walls (de la Cruz et al., 1992de la Cruz J, Hidalgo-Gallego A, Lora JM, Benitez T, Pintor-Toro JA and Llobell A (1992) Isolation and characterization of three chitinases from Trichoderma havzianum. Eur J Biochem 206:859–867.).
Although little is known about the roles of chitinases in the infection process of entomopathogenic fungi, chitinases are thought to be key enzymes during the early stages of the infection process. Furthermore, the other hydrolytic enzymes like proteases, lipase and chitosanase secreted during the infection process might compensate for the penetration functions. Taken together, our data demonstrate that expression of Ifu-chit2 is part of a series of responses of I. fumosorosea triggered by the presence of insect host, and may play a role in the early stage of pathogenesis. However, further overexpression and gene knockout experiments for Ifu-chit2 gene are needed to determine its exact roles in pathogenicity.
In conclusion, we characterized the I. fumosorosea chitinase gene Ifu-chit2 and expressed the recombinant protein in E. coli. Expression profiles of Ifu-chit2 under in vivo conditions were also determined. Whether there are any other chitinases in I. fumosorosea needs to be studied further.
Acknowledgments
This work was supported by the Special Fund for Forestry Scientific Research in the Public Interest (Grant No. 201204506), the Special Fund for Agro-scientific Research in the Public Interest (201003079) and the National Natural Science Foundation of China (Grant No. 31272096, No. 31471821 and No. 31201568).
-
Associate Editor: Célia Maria de Almeida Soares
References
- Ahmed NU, Park JI, Seo MS, Kumar TS, Lee IH, Park BS and Nou IS (2012) Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica Mol Biol Rep 39:3649–3657.
- Ali S, Huang Z and Ren S (2010a) Production of cuticle degrading enzymes by Isaria fumosorosea and their evaluation as a biocontrol agent against diamondback moth. J Pest Sci 83:361–370.
- Ali S, Wu J, Huang Z and Ren SX (2010b) Production and regulation of extracellular chitinase from the entomopathogenic fungus Isaria fumosorosea Biocontrol Sci Technol 20:723–738.
- Baratto CM, Dutra V, Boldo JT, Leiria LB, Vainstein MH and Schrank A (2006) Isolation, characterization, and transcriptional analysis of the chitinase chi2 gene (DQ011663) from the biocontrol fungus Metarhizium anisopliae var. anisopliae Curr Microbiol 53:217–221.
- Bhanu Prakash GV, Padmaja V, Jami SK and Kirti PB (2012) Expression of chitinase genes of Metarhizium anisopliae isolates in lepidopteran pests and on synthetic media. J Basic Microbiol 52:628–635.
- Bokma E, Rozeboom HJ, Sibbald M, Dijkstra BW and Beintema JJ (2002) Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis Eur J Biochem 269:893–901.
- Boldo JT, Junges A, do Amaral KB, Staats CC, Vainstein MH and Schrank A (2009) Endochitinase CHI2 of the biocontrol fungus Metarhizium anisopliae affects its virulence toward the cotton stainer bug Dysdercus peruvianus. Curr Genet 55:551–560.
- Charnley AK (2003) Fungal pathogens of insects: Cuticle degrading enzymes and toxins. Adv Bot Res 40:241–321.
- Charnley AK and St Leger RJ (1991) The role of cuticle-degrading enzymes in fungal pathogenesis in insects. In: Cole GT and Hoch HC (eds) The Fungal Spore Disease Initiation in Plants and Animals. Plenum Press, New York, pp 267–286.
- de la Cruz J, Hidalgo-Gallego A, Lora JM, Benitez T, Pintor-Toro JA and Llobell A (1992) Isolation and characterization of three chitinases from Trichoderma havzianum Eur J Biochem 206:859–867.
- Fang WG, Leng B, Xiao YH, Jin K, Ma JC, Fan YH, Feng J, Yang XY, Zhang YJ and Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370.
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD and Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker HM (ed) The Proteomics Protocols handbook. Humana Press, Totowa, pp 571–607.
- Hajek AE and Delalibera Jr I (2010) Fungal pathogens as classical biological control agents against arthropods. Biocontrol 55:147–158.
- Hartl L, Zach S and Seidl-Seiboth V (2012) Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93:533–543.
- Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316.
- Henrissat B and Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788.
- Henrissat B and Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696.
- Henrissat B and Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7:637–644.
- Lee YG, Chung KC, Wi SG, Lee JC and Bae HJ (2009) Purification and properties of a chitinase from Penicillium sp. LYG 0704. Protein Express Purif 65:244–250.
- Li DC (2006) Review of fungal chitinases. Mycopathologia 161:345–360.
- Limón MC, Chacón MR, Mejías R, DelgadoJarana J, Rincón AM, Codón AC and Benítez T (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl Microbiol Biotechnol 64:675–685.
- Liu Z, Yang Q, Hu S, Zhang J and Ma J (2008) Cloning and characterization of a novel chitinase gene (chi46) from Chaetomium globosum and identification of its biological activity. Appl Microbiol Biotechnol 80:241–252.
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408.
- Lu ZX, Laroche A and Huang HC (2005) Isolation and characterization of chitinases from Verticillium lecanii Can J Microbiol 51:1045–1055.
- Mauch F, Hadwiger LA and Boller T (1984) Ethylene: Symptom, not signal for the induction of chitinase and β-1, 3-glucanase in pea pods by pathogens and elicitors. Plant Physiol 76:607–611.
- Mishra S, Kumar P and Malik A (2013a) Preparation, characterization, and insecticidal activity evaluation of three different formulations of Beauveria bassiana against Musca domestica Parasitol Res 112:3485–3495.
- Mishra S, Kumar P and Malik A (2013b) Effect of process parameters on the enzyme activity of a novel Beauveria bassiana isolates. Int J Curr Microbiol Appl Sci 2:49–56.
- Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M and Watanabe T (1996) A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol 178:5065–5070.
- Petersen TN, Brunak S, von Heijne G and Nielsen H (2011) SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786.
- Screen SE, Hu G and St Leger RJ (2001) Transformants of Metarhizium anisopliae sf. anisopliae overexpressing chitinase from Metarhizium anisopliae sf. acridum show early induction of native chitinase but are not altered in pathogenicity to Manduca sexta J Invertebr Pathol 78:260–266.
- Staats CC, Kmetzsch L, Lubeck I, Junges A, Vainstein MH and Schrank A (2013) Metarhizium anisopliae chitinase CHIT30 is involved in heat-shock stress and contributes to virulence against Dysdercus peruvianus Fungal Biol 117:137–144.
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-Aard JJ, Shrestha B and Spataforal JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol 57:5–59.
- Tang Q, Zhang YP, Xie L, Song LH, Li ZZ and Huang B (2009) Generation of transgenic Beauveria bassiana strains with chitinase gene from Isaria fumosorosea and its increased virulence against Dendrolimus punctatus (Lepidoptera, Lasiocampidae). Acta Entomol Sin 52:755–762.
- Wang ZX, Meng HM, Zhuang ZL, Chen MJ, Xie L and Huang B (2013) Molecular cloning of a novel subtilisin-like protease (Pr1A) gene from the biocontrol fungus Isaria farinosa Appl Entomol Zool 48:477–487.
- Wu ML, Chuang YC, Chen JP, Chen CS and Chang MC (2001) Identification and characterization of the three chitin-binding domains within the multidomain chitinase chi92 from Aeromonas hydrophila JP101. Appl Environ Microbiol 67:5100–5106.
- Zhu YP, Pan JR, Qiu JZ and Guan X (2008) Isolation and characterization of a chitinase gene from entomopathogenic fungus Verticillium lecanii Braz J Microbiol 39:314–320.
Internet Resources
- Expasy, http://web.expasy.org/protparam/ (July 19, 2014).
» http://web.expasy.org/protparam/ - SignalP 4.0 server, http://www.cbs.dtu.dk/services/SignalP/ (July 19, 2014).
» http://www.cbs.dtu.dk/services/SignalP/ - BLASTP tool, http://www.ncbi.nlm.nih.gov/BLAST
» http://www.ncbi.nlm.nih.gov/BLAST - ClustalW, http://www.genome.jp/tools/clustalw/
» http://www.genome.jp/tools/clustalw/
Publication Dates
-
Publication in this collection
Sept 2015
History
-
Received
08 Jan 2015 -
Accepted
27 Apr 2015