Abstract
The phospholipase A2 superfamily encompasses 15 groups that are classified into: secreted PLA2 (sPLA2); cytosolic PLA2 (cPLA2); Ca2+-independent intracellular PLA2 (iPLA2); platelet-activating factor acetylhydrolase (PAF-AH); and lysosomal PLA2. Currently, approximately 700 PLA2 sequences are known, of which 200 are obtained from the venom gland of Crotalinae snakes. However, thus far, little information is available on cloning, purification and structural characterization of PLA2 from Crotalus durisssus cascavela venom gland. In the present work, we report the molecular cloning of a novel svPLA2 from C. d. cascavella (Cdc), a predominant rattlesnake subspecies in northeastern Brazil. The Cdc svPLA2 cDNA precursor is 689 nucleotides long and encodes a protein of 138 amino acid residues, with a calculated molecular mass of approximately 13,847 Da and an estimated isoelectric point of 5.14. Phylogenetic analysis of Crotalinae PLA2 reveals that Cdc PLA2 clustered with other acidic type IIA PLA2 homologues is also present in the venom of North American rattlesnakes. Hitherto, this study presents a novel PLA2 cDNA precursor from C. d. cascavella and data reported herein will be useful for further steps in svPLA2 purification and analysis.
molecular toxinology; Crotalus durissus cascavella; snake venom gland; cDNA library; acidic PLA2
Cloning of a novel acidic phospholipase A2 from the venom gland of Crotalus durissus cascavella (Brazilian northeastern rattlesnake)
Guarnieri MCI; Melo ESLI; Melo KMSII; Albuquerque-Modesto JCIII; Prieto-da-Silva ARBIV; Rádis-Baptista GII,V
IDepartment of Zoology, Federal University of Pernambuco, UFPE, Recife, Pernambuco State, Brazil
IIDepartment of Biochemistry, Federal University of Pernambuco, UFPE, Recife, Pernambuco State, Brazil
IIIDepartment of Biology, Federal University of Pernambuco, UFPE, Vitória de Santo Antão, Pernambuco State, Brazil
IVLaboratory of Genetics, Butantan Institute, São Paulo, São Paulo State, Brazil
VInstitute of Marine Sciences, Federal University of Ceará, UFC, Fortaleza, Ceará State, Brazil
Correspondence to Correspondence to: Ghandi Rádis-Baptista Instituto de Ciências do Mar, Labomar Universidade Federal do Ceará Av. da Abolição, 3207, Fortaleza, CE, 60165-081, Brasil. Phone: +55 85 3366 7000. Fax: +55 85 3242 8355. Email: gandhi.radis@ufc.br.
ABSTRACT
The phospholipase A2 superfamily encompasses 15 groups that are classified into: secreted PLA2 (sPLA2); cytosolic PLA2 (cPLA2); Ca2+-independent intracellular PLA2 (iPLA2); platelet-activating factor acetylhydrolase (PAF-AH); and lysosomal PLA2. Currently, approximately 700 PLA2 sequences are known, of which 200 are obtained from the venom gland of Crotalinae snakes. However, thus far, little information is available on cloning, purification and structural characterization of PLA2 from Crotalus durisssus cascavela venom gland. In the present work, we report the molecular cloning of a novel svPLA2 from C. d. cascavella (Cdc), a predominant rattlesnake subspecies in northeastern Brazil. The Cdc svPLA2 cDNA precursor is 689 nucleotides long and encodes a protein of 138 amino acid residues, with a calculated molecular mass of approximately 13,847 Da and an estimated isoelectric point of 5.14. Phylogenetic analysis of Crotalinae PLA2 reveals that Cdc PLA2 clustered with other acidic type IIA PLA2 homologues is also present in the venom of North American rattlesnakes. Hitherto, this study presents a novel PLA2 cDNA precursor from C. d. cascavella and data reported herein will be useful for further steps in svPLA2 purification and analysis.
Keywords: molecular toxinology, Crotalus durissus cascavella, snake venom gland, cDNA library, acidic PLA2.
INTRODUCTION
The superfamily of phospholipase A2 enzymes is currently subdivided into 15 groups based on their structures, source and localization. Distributed among these groups are the multiple forms of secreted PLA2s (sPLA2s groups I, II, III, V, IX, X, XI, XII, XIII and XIV), cytosolic PLA2s (cPLA2 group IV), Ca2+-independent intracellular PLA2s (iPLA2 group VI), platelet-activating factor acetylhydrolases (PAF-AH groups VII and VIII) and the lysosomal PLA2s (group XV) (1).
Secreted PLA2s are found in fungi, bacteria, plants, marine sponges, cnidarians, mollusks, starfishes, insects, reptiles and mammals (1-4). Essentially, these enzymes catalyze the hydrolysis of different membrane phospholipids at the sn-2 position, releasing free fatty acids such as arachidonic acid (AA) a precursor of bioactive eicosanoids and lysophospholipids (lyso-PL). Both products represent the first step in generating second messengers that play important physiological and pathological roles. Lyso-PL can be converted into lysophosphatidic acid (LPA), involved in cell proliferation, survival and migration, or into platelet activating factor (PAF), implicated specifically in inflammatory processes (5, 6). Eicosanoids affect body mechanisms including sleep regulation, immune response, inflammation and pain (7).
PLA2s from Viperidae venoms (vPLA2) belong to the IIsubgroup together with mammalian enzymes isolated from the spleen, mast cells, macrophages, arthritic synovial fluid and serum of patients with inflammatory diseases (8-10). This subgroup is characterized by low-molecular-mass enzymes (~14 kDa), with a rigid three-dimensional structure composed of seven disulfide bridges, whose catalytic mechanism utilizes a His-Asp dyad. These enzymes require a millimolar concentration of Ca2+ to exert their enzymatic action and, in contrast to cPLA2s, they have low specifity for arachidonic acid at the sn-2 position (11).
Approximately 700 sequences from type II PLA2s are known and compiled in databases. The diversity of snake venom PLA2 functions includes: neurotoxicity, cardiotoxicity, myotoxicity, edema, hypotension, hyperalgesia as well as activation and inhibition of platelet aggregation (12-19). The diversity of biological and pharmacological functions of PLA2 denotes that accelerated or positive Darwinian evolution has occurred and appears to confer a better fitness on the snake venom (10, 20). In fact, the venom PLA2 subgroup II is further subdivided into two other smaller subgroups, vPLA2s exhibiting enzymatic activity and a predominance of two types of amino acid residues at the catalytic site (position 49) Asp (D49) and Ser (S49) and non-enzymatic vPLA2s (that is, vPLA2 with extremely low enzymatic activity), whose residues, D49 or S49, were replaced not only with Lys (K49), but also Gln 49 (Q49), Ala (A49) and Asn (N-49) (21-26). Furthermore, D49 PLA2 also includes acidic and basic toxic components that are found in venoms as monomers or homo- and heterodimers (27).
In this work, we report a novel PLA2 cDNA precursor of Crotalus durissus cascavella venom, in which the predicted protein was clustered with acidic members of the type II subfamily of venom PLA2.
MATERIALS AND METHODS
Specimens of Snake Venom Gland
For the construction of the venom gland cDNA library, a pair of glands was excised from a male adult specimen of Crotalus durissus cascavella (2 kg weight and 125 cm length measured from rostrum to cloaca) captured in Cabaceira, Paraíba state, Brazil, and maintained from 1999 to 2006 in the Laboratory of Venomous Animals and Toxins (LAPTOX), Federal University of Pernambuco, Recife state, Brazil. The snake venom was extracted by standard procedures three days before the surgery for gland excision, with the aim of reaching the maximal level of RNA synthesis. Once surgically removed, the venom glands were kept at 80°C until the procedures for RNA purification and analysis.
Construction of C. d. cascavella Snake Venom cDNA Library
A Crotalus durissus cascavella venom cDNA library was constructed from 1 µg of total RNA, as follows: frozen venom glands were finely crushed in a mortar with a pestle under liquid nitrogen; then, total RNA was purified using Trizol® reagent (Invitrogen, USA), according to the manufacturer's instructions. The quality and yield of total RNA were verified by the integrity of 28S and 18S rRNA, through denaturing agarose gel electrophoresis using the spectrophotometric ratio 260/280 nm. Poly(A+)-RNA was purified from total RNA by a complex of oligo(dT)-biotin and streptavidin MagneSphere® paramagnetic particles (PolyATract® system, Promega, USA). Next, mRNA was quantified and employed for cDNA synthesis using the switching mechanism at the 5' end of RNA transcriptiom (SMART) protocol (Creator SMART cDNA Library Construction kit®, BD Biosciences, USA), which preferentially enriches the final library with full-length cDNA.
Cloning of Cdc PLA2 cDNA and Nucleotide Sequencing
The C. durissuscascavella venom gland cDNA library was then titered and pools of approximately 106 colony forming units (CFU) were used as a template in ten separate homology screening polymerase chain reactions (HR-PCR). Each reaction, in a final volume of 50 mL, in high fidelity PCR buffer (60 mM Tris-SO4, pH 8.4, 18 mM (NH4)2SO4, 2.5 mM MgSO4), consisted of 2.5 U of Platinum® Taq DNA polymerase (Invitrogen Life Technologies, USA), 2 mM MgCl2, 1 mM dNTPs, and 0.2 mM of each forward and reverse primer.
Of the two primers utilized to isolate the Cdc PLA2 cDNA, one (called Cdc_PLA2 sense primer, 5'-TGCACGACTGYTGYTAYGGA-3') anneals to the specific gene sequence, corresponding to the amino acids -FVHDCCYG-, which are conserved in most snake venom PLA2s, and the other oligonucleotide primer to the plasmid vector (M13 reverse, 5'-AACAGCTATGACCATGTTCA- 3'), which corresponds to the flanking region of insertion in the pDNR-LIB vector.
The cloned full length Cdc PLA2 was automatically sequenced by the dideoxy chain termination method, using the dye-terminator chemistry (DYEnamic ET Dye Terminator® kit, GE Healthcare, USA) and the MegaBACE 750 DNA Analysis System® (GE Healthcare, USA). The PLA2 gene was in silico translated, and both nucleotide and amino acid sequences were compared against a database of genes and proteins, maintained by the NCBI (http://www.ncbi.nlm.nih.gov).
Crotalus durissus cascavella (Cdc) PLA2 Aligment and Phylogenetic Analysis
The deduced amino acid sequence of C. d. cascavella PLA2 cDNA precursor (present study) was compared with the GenBank (http://www.ncbi.nlm.nih.gov) by using the BLAST program (28). This search retrieved 182 protein sequences corresponding to all Crotalinae PLA2s available in the database. The incomplete and redundant sequences were manually removed from the data set whereas the file with complete sequences was processed for alignment through the multialignment bioinformatic tool ClustalW2, available at the European Bioinformatic Institute website (http://www.ebi.ac.uk). The structural characteristics of the predicted PLA2 precursor were manually annotated based on data from the literature. Precursors of sequences from Crotalinae PLA2 toxins which presented higher PLA2 member scores in comparison with Cdc PLA2 were aligned with MUSCLE 3.6 using groups of amino acids GA, ST, MVLI, KR, EQDN, FWYH, C and P to determine the grade of similarity (29).
The evolutionary history was inferred using the neighbor-joining method by analyzing all sequences together, including not only higher score sequences, but also the most dissimilar PLA2s (30). Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are defined as collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches (31). The evolutionary distances were computed using the Dayhoff matrix-based method expressed as the number of amino acid substitutions per site (32). All positions containing gaps or missing data were eliminated from the dataset (complete deletion option). There was a total of 85 positions in the final dataset. Phylogenetic analyses were conducted with MEGA4 (33).
RESULTS
A Novel PLA 2 cDNA Precursor of Crotalus durissus cascavella Venom Gland
By a homology cloning method, a novel PLA2 precursor, called Cdc-PLA2, was retrieved from the venom gland cDNA library of C. d. cascavella. As indicated in Figure 1, the Cdc-PLA2 cDNA precursor is 689 nucleotides long, with an open reading frame (ORF) of 453 nucleotides. The ORF encodes a complete precursor of 138 amino acid residues, including a signal peptide of 16 residues (MRTLWIVAVLLLGVEG). The novel Cdc-PLA2 cDNA sequence was submitted to GenBank and received the accession number GQ466583.
Comparative Sequence Analysis of the Novel Cdc-PLA 2
The complete amino acid sequence of Cdc-PLA2 precursor was predicted from a cDNA sequence (Figure 1). Based on this deduced sequence, an isoeletric point of 5.14 and molecular mass of 13,846.81 Da was calculated.
The Cdc-PLA2 conserved residues involved in Ca2+ binding (Tyr28, Gly30, Gly32 and Asp49) and in the catalytic network (His48), characterizing the D-49 group, and maintained conserved sequence domains common to the group IIA PLA2, including the 14 cysteines responsible for disulfide bond formation.
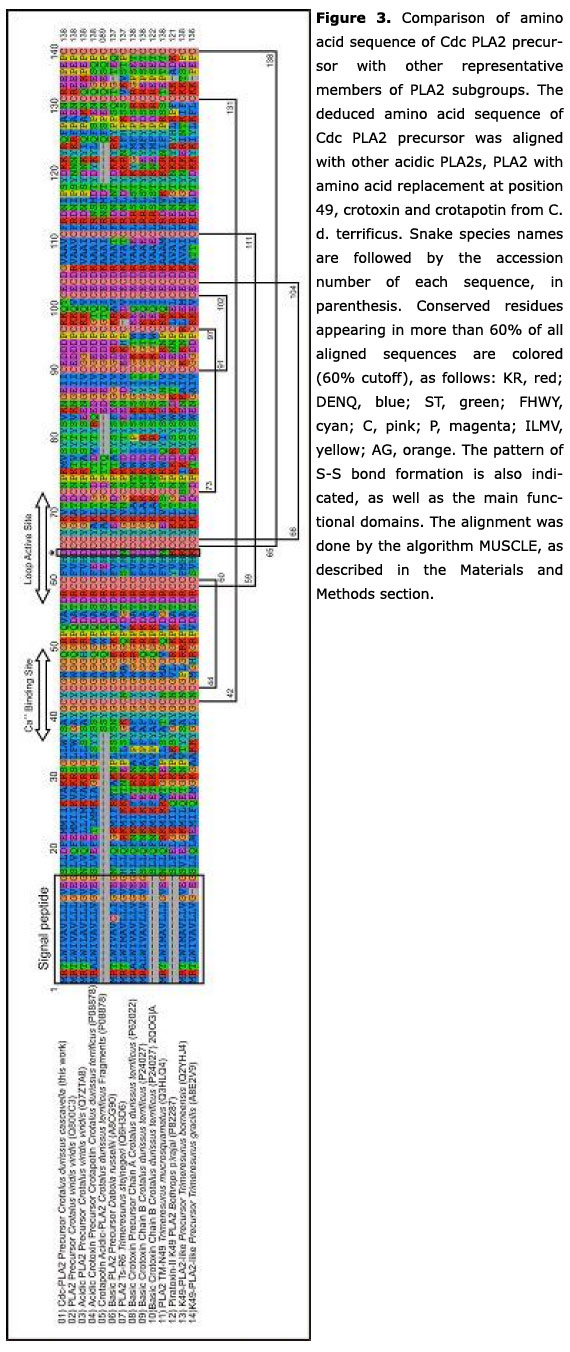
Phylogenetic analysis of Cdc-PLA2 and 54 precursors of Crotalinae PLA2 showed that the maximum grade of parental relationship of Cdc-PLA2 occurs with acidic PLA2 from North American snakes (Figure 2). In this case, best similarity values (identities in the range of 60 to 86%) are observed in North American rattlesnakes, for example Crotalus v. viridis (86%).
The sequence was aligned with precursors of other Crotalinae PLA2s, obtained by BLASTp search, which included PLA2 with amino acid replacement at the position 49, crotapotin from C. d. terrificus and the acidic subunit of crotoxin (CA) sequence from C. d. cascavella (34). Several residues were highly conserved in the monomeric/homodimeric acidic PLA2 analyzed in the present work as the N-terminal region (L2XXFE6), Ca2+ binding site (Y25GCYCGXGG33), active site (D42RCCFVHDCCYGK54) and C-terminal region (A101AXCFFDN108, Y112, Y117) (Figure 3). On the other hand, the residues A53, D79, S108 and G129 present in heterodimeric toxins (crotoxin A from C. d. terrificus,C. d. cascavella and C. s. scutulatus) are replaced in Cdc-PLA2 and in the majority of the mono/homodimeric acid PLA2s analyzed. Comparative analysis revealed that the hot spot of Cdc-PLA2 mutations were found at the residues D4, I10, A34, V39, V77, K78, E85, D86, T94, G99, R118 and R128.
DISCUSSION
Although studies involving snake venom acidic PLA2s have increased considerably in the recent years, only a few acidic PLA2s from Brazilian snake venoms were purified and cloned (18, 21, 35-41). Up to date, nothing was known about the expression of acidic (subgroup II) PLA2 in the venom gland of Crotalus durissus cascavella.
Cdc-PLA2 possesses high similarity with a subgroup of acidic D49-PLA2s which is expressed in the venom as monomers and/or as homodimers (42-45). In fact, the ability of an acidic glycosylated and phosphorylated PLA2s to co-exist in snake venom as monomer and homodimer was recently described by Sun et al. (27).
Experimental investigations with native toxins have shown that such group of PLA2 presents enzymatic activity and capacity of binding calcium ions for maximal catalysis, as seen by the conserved residues His48 and Asp49 in the primary sequences (39, 46-47). These acidic PLA2s can also induce myotoxicity, platelet aggregation inhibition, hypotension, prostaglandin I2 induction or paw edema (16, 18, 21, 23, 35, 37, 39, 42, 45, 48-50). Some residues associated with antiplatelet (W21, Y113, D114) and edema-forming activities (K78 and D85) are conserved in Cdc-PLA2 and in some very similar acidic PLA2 isoforms from C. v. viridis venom (43, 50-51). All analyzed acidic PLA2s presented Glu residue in the position 6, which seems an ancient condition of basic G6 and N6 PLA2 (21, 52).
Cdc-PLA2 possesses lower similarity with the other subgroup of acidic D49-PLA2s (particularly, A chain crotoxin-CA) which can make high stable complexes with basic F24N6 PLA2 (B chain crotoxin-CB) and increase the toxicity of CB in several folds (52, 53). Except for E124 residue, all amino acids that could be involved in the recognition and binding of a CA with CB (W36, E47, A53, D79, E124 and G129) are replaced in Cdc-PLA2, what consequently suggests, at a first glance, the impossibility of this toxin to be an A chain crotoxin precursor for heterodimer formation (52). However, this point deserves more attention and further functional and structural analysis.
Acidic IIA phospholipases A2 present multiple isoforms, generally associated with intra-specific geographic variation, as well as adaptation to prey diversity (43, 44, 54, 55). On the other hand, some PLA2 clones apparently not translated into venom proteins has been reported, since not-expressing toxin mRNA may be a repository for snake survival under an ever-changing environment (54, 55).
In this work, we report the molecular cloning of an acidic PLA2 type II from the venom gland of C. d. cascavella. Phylogenetic and structural analyses allowed us to make evident that the precursor, retrieved from the C. d. cascavella venom gland cDNA library, is a novel member of acidic PLA2 subgroup. Moreover, a global analysis has shown that the most ancestral member of all PLA2 precursors in the venom of Crotalinae snakes seems to be related to the crotoxin.
Altogether, the present data will be useful, for example, to drive steps of purification and structural analysis of such flexible and fast evolving snake venom molecule.
ACKNOWLEDGEMENTS
The authors are grateful to the National Council for Scientific and Technological Development (CNPq) from the Ministry of Science and Technology for the financial support. Part of experimental data reported herein were obtained during the course "Molecular techniques for the scientific investigation of biological and chemical diversity in terrestrial and marine organisms with potential biotechnological application", held in Recife, in 2008, in the context of the program of cooperation in biotechnology from the Brazilian and Argentinean governments (CBAB/MCT).
Received: November 22, 2008
Accepted: September 15, 2009
Abstract published online: October 9, 2009
Full paper published online: November 30, 2009
Conflicts of interest: There is no conflict.
- 1. Schaloske RH, Dennis EA. The phospholipase A2 superfamily and its group numbering system. Biochim Biophys Acta. 2006;1761(11):1246-59.
- 2. Nevalainen TJ, Quinn RJ, Hooper JN. Phospholipase A2 in porifera. Comp Biochem Physiol B Biochem Mol Biol. 2004;137(3):413-20.
- 3. Nevalainen TJ, Peuravuori HJ, Quinn RJ, Llewellyn LE, Benzie JAH, Fenner PJ, et al. Phospholipase A2 in cnidaria. Comp Biochem Physiol B Biochem Mol Biol. 2004;139(4):731-5.
- 4. Ota E, Nagai H, Nagashima Y, Shiomi K. Molecular cloning of two toxic phospholipases A2 from the crown-of-thorns starfish Acanthaster planci venom. Comp Biochem Physiol B Biochem Mol Biol. 2006;143(1):54-60.
- 5. Moolenaar WH, van Meeteren LA, Giepmans BN.The ins and outs of lysophosphatidic acid signaling. Bioessays. 2004;26(8):870-81.
- 6. Prescott SM, Zimmerman GA, Stafforini DM, McIntyre TM. Platelet-activating factor and related lipid mediators. Annu Rev Biochem. 2000;69:419-45.
- 7. Tsuboi K, Sugimoto Y, Ichikawa A. Prostanoid receptor subtypes. Prostaglandins Other Lipid Mediat. 2002;68-69:535-56.
- 8. Kudo I, Murakami M, Hara S, Inoue K. Mammalian non-pancreatic phospholipases A2 Biochim Biophys Acta. 1993;1170(3):217-31.
- 9. Nevalainen TJ. Serum phospholipases A2 in inflammatory diseases. Clin Chem. 1993;39(12):2453-9.
- 10. Jan VM, Guillemin I, Robbe-Vincent A, Choumet V. Phospholipase A2 diversity and polymorphism in European viper venoms: paradoxical molecular evolution in Viperinae. Toxicon. 2007;50(8):1140-61.
- 11. Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488(1-2):1-19.
- 12. Prijatelj P, Charnay M, Ivanovski G, Jenko Z, Pungercar J, Krizaj I, et al. The C-terminal and beta-wing regions of ammodytoxin A, a neurotoxic phospholipase A2 from Vipera ammodytes ammodytes, are critical for binding to factor Xa and for anticoagulant effect. Biochimie. 2006;88(1):69-76.
- 13. Valentin E, Lambeau G. What can venom phospholipase A2 tell us about the functional diversity of mammalian secreted phospholipase A2 Biochemie. 2000;82(9-10):815-31.
- 14. Angulo Y, Olamendi-Portugal T, Possani LD, Lomonte B. Isolation and characterization of myotoxin II from Atropoides (Bothrops) nummifer snake venom, a new Lys49 phospholipase A2 homologue. Int J Biochem Cell Biol. 2000;32(1):63-71.
- 15. Câmara PR, Esquisatto LC, Camargo EA, Ribela MT, Toyama MH, Marangoni S, et al. Inflammatory oedema induced by phospholipases A2 isolated from Crotalus durissus sp. in the rat dorsal skin: a role for mast cells and sensory C-fibers. Toxicon. 2003;41(7):823-9.
- 16. Adriăo-Escarso SH, Soares AM, Rodrigues VM, Angulo Y, Díaz C, Lomonte B, et al. Myotoxic phospholipases A2 in Bothrops snake venoms: effect of chemical modifications on the enzymatic and pharmacological properties of bothropstoxins from Bothrops jararacussu Biochimie. 2000;82(8):755-63.
- 17. Chacur M, Milligan ED, Sloan EM, Wieseler-Frank J, Barrientos RM, Martin D, et al. Snake venom phospholipase A2s (Asp49 and Lys49) induce mechanical allodynia upon peri-sciatic administration: involvement of spinal cord glia, proinflammatory cytokines and nitric oxide. Pain. 2004;108(1-2):180-91.
- 18. de Albuquerque-Modesto JC, Spencer PJ, Fritzen M, Valença RC, Oliva ML, da Silva MB, et al. BE-I-PLA2, a novel acidic phospholipase A2 from Bothrops erythromelas venom: isolation, cloning and characterization as potent anti-platelet and inductor of prostaglandin I2 release by endothelial cells. Biochem Pharmacol. 2006;72(3):377-84.
- 19. Higuchi DA, Barbosa CM, Bincoletto C, Chagas JR, Magalhaes A, Richardson M, et al. Purification and partial characterization of two phospholipases A2 from Bothrops leucurus (white-tailed-jararaca) snake venom. Biochimie. 2007;89(3):319-28.
- 20. Ogawa T, Nakashima K, Nobuhisa I, Deshimaru M, Shimohigashi Y, Fukumaki Y, et al. Accelerated evolution of snake venom phospholipase A2 isozymes for acquisition of diverse physiological functions. Toxicon. 1996;34(11-12):1229-36.
- 21. Cogo JC, Lilla S, Souza GH, Hyslop S, de Nucci G. Purification, sequencing and structural analysis of two acidic phospholipases A2 from the venom of Bothrops insularis (jararaca ilhoa). Biochimie. 2006;88(12):1947-59.
- 22. Polgár J, Magnenat EM, Peitsch MC, Wells TN, Clemetson KJ. Asp-49 is not an absolute prerequisite for the enzymic activity of low-M(r) phospholipases A2: purification, characterization and computer modeling of an enzymically active Ser-49 phospholipase A2, ecarpholin S, from the venom of Echis carinatus sochureki (saw-scaled viper). Biochem J. 1996;319(Pt 3):961-8.
- 23. Lomonte B, Angulo Y, Calderón L. An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon. 2003;42(8):885-901.
- 24. Bao Y, Bu P, Jin L, Hongxia W, Yang Q, An L. Purification, characterization and gene cloning of a novel phospholipase A2 from the venom of Agkistrodon blomhoffii ussurensis. Int J Biochem Cell Biol. 2005;37(3):558-65.
- 25. Liu CS, Kuo PY, Chen JM, Chen SW, Chang CH, Tseng CC, et al. Primary structure of an inactive mutant of phospholipase A2 in the venom of Bungarus fasciatus (banded krait). J Biochem. 1992;12(5):707-13.
- 26. Wei JF, Wei XL, Chen QY, Huang T, Qiao LY, Wang WY, et al. N49 phospholipase A2, a unique subgroup of snake venom group II phospholipase A2 Biochim Biophys Acta. 2006;1760(3):462-71.
- 27. Sun MZ, Liu S, Yang F, Greenaway FT, Xu Y. A novel phospholipase A2 from Agkistrodon blomhoffii ussurensis venom: purification, proteomic, functional and structural characterizations. Biochimie. 2009;91(4):558-67.
- 28. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389-402.
- 29. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113.
- 30. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-25.
- 31. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-91.
- 32. Schwarz RM, Dayhoff MO. Matrices for detecting distant relationships. In: Dayhoff M, editor. Atlas of protein sequences and structure. Washington: National Biomedical Research Foundation; 1979. p. 353-9.
- 33. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9.
- 34. de Oliveira DG, Toyama MH, Martins AM, Havt A, Nobre AC, Marangoni S, et al. Structural and biological characterization of a crotapotin isoform isolated from Crotalus durissus cascavella venom. Toxicon. 2003;42(1):53-62.
- 35. Santos-Filho NA, Silveira LB, Oliveira CZ, Bernardes CP, Menaldo DL, Fuly AL, et al. A new acidic myotoxic, anti-platelet and prostaglandin I2 inductor phospholipase A2 isolated from Bothrops moojeni snake venom. Toxicon. 2008;52(8):908-17.
- 36. Fuly AL, de Miranda AL, Zingali RB, Guimarăes JA. Purification and characterization of a phospholipase A2 isoenzyme isolated from Lachesis muta snake venom. Biochem Pharmacol. 2002;63(9):1589-97.
- 37. Serrano SM, Reichl AP, Mentele R, Auerswald EA, Santoro ML, Sampaio CA, et al. A novel phospholipase A2, BJ-PLA2, from the venom of the snake Bothrops jararaca: purification, primary structure analysis, and its characterization as a platelet-aggregation-inhibiting factor. Arch Biochem Biophys. 1999;367(1):26-32.
- 38. Andriăo-Escarso SH, Soares AM, Fontes MRM, Fuly AL, Correa FMA, Rosa JC, et al. Structural and functional characterization of an acidic platelet aggregation inhibitor and hypotensive phospholipase A2 from Bothrops jararacussu snake venom. Biochem Pharmacol. 2002;64(4):723-32.
- 39. Ketelhut DF, de Melo MH, Veronese EL, Esmeraldino LE, Murakami MT, Arni RK, et al. Isolation, characterization and biological activity of acidic phospholipase A2 isoforms from Bothrops jararacussu snake venom. Biochimie. 2003;85(10):983-91.
- 40. Roberto PG, Kashima S, Soares AM, Chioato L, Faca VM, Fuly AL, et al. Cloning and expression of an acidic platelet aggregation inhibitor phospholipase A2 cDNA from Bothrops jararacussu venom gland. Protein Expr Purif. 2004;37(1):102-8.
- 41. Rodrigues RS, Izidoro LF, Teixeira SS, Silveira LB, Hamaguchi A, Homsi-Brandeburgo MI, et al. Isolation and functional characterization of a new myotoxic acidic phospholipase A2 from Bothrops pauloensis snake venom Toxicon. 2007;50(1):153-65.
- 42. Chen YC, Maraganore JM, Reardon I, Heinrikson RL. Characterization of the structure and function of three phospholipases A2 from the venom of Agkistrodon halys pallas. Toxicon. 1987;25(4):401-9.
- 43. Tsai IH, Wang YM, Chen YH, Tu AT. Geographic variations, cloning, and functional analyses of the venom acidic phospholipases A2 of Crotalus viridis viridis Arch Biochem Biophys. 2003;411(2):289-96.
- 44. Tsai IH, Wang YM, Chen YH, Tsai TS, Tu MC. Venom phospholipases A2 of bamboo viper (Trimeresurus stejnegeri): molecular characterization, geographic variations and evidence of multiple ancestries. Biochem J. 2004;377(Pt 1):215-23.
- 45. Magro AJ, Murakami MT, Marcussi S, Soares AM, Arni RK, Fontes MR. Crystal structure of an acidic platelet aggregation inhibitor and hypotensive phospholipase A2 in the monomeric and dimeric states: insights into its oligomeric state. Biochem Biophys Res Commun. 2004;323(1):24-31.
- 46. Gu L, Zhang H, Song S, Zhou Y, Lin Z. Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus Acta Crystallogr D Biol Crystallogr. 2002;58(pt 1):104-10.
- 47. Jasti J, Paramasivam M, Srinivasan A, Singh TP. Structure of an acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) at 2.6 Ĺ resolution reveals a novel intermolecular interaction. Acta Crystallogr D Biol Crystallogr. 2004;60(1):66-72.
- 48. Fuly AL, Calil-Elias S, Martinez AM, Melo PA, Guimarăes JA. Myotoxicity induced by an acidic Asp-49 phospholipase A2 isolated from Lachesis muta snake venom: comparison with lysophosphatidylcholine. Int J Biochem Cell Biol. 2003;35(10):1470-81.
- 49. Fuly AL, Machado OL, Alves EW, Carlini CR. Mechanism of inhibitory action on platelet activation of a phospholipase A2 isolated from Lachesis muta (Bushmaster) snake venom. Thromb Haemost. 1997;78(5):1372-80.
- 50. Liu X, Wu X, Zhou Y. Identification of key residues responsible for enzymatic and platelet-aggregation-inhibiting activities of acidic phospholipase A2 from Agkistrodon halys pallas. J Nat Toxins. 2001;10(1):43-55.
- 51. Wang XQ, Yang J, Gui LL, Lin ZJ, Chen YC, Zhou YC. Crystal structure of an acidic phospholipase A2 from the venom of Agkistrodon halys Pallas at 2.0 a resolution. J Mol Biol. 1996;255(5):669-76.
- 52. Chen YH, Wang YM, Hseu MJ, Tsai IH. Molecular evolution and structure-function relationships of crotoxin-like and asparagine-6-containing phospholipases A2 in pit viper venoms. Biochem J. 2004;381(Pt 1):25-34.
- 53. Choumet V, Lafaye P, Demangel C, Bon C, Mazié JC. Molecular mimicry between a monoclonal antibody and one subunit of crotoxin, a heterodimeric phospholipase A2 neurotoxin. Biol Chem. 1999;380(5):561-8.
- 54. Tsai IH, Wang YM, Au LC, Ko TP, Chen YH, Chu YF. Phospholipases A2 from Callosellasma rhodostoma venom gland: cloning and sequencing of 10 of the cDNAs, three-dimensional modeling and chemical modification of the major isozyme. Eur J Biochem. 2000;267(22):6684-91.
- 55. Gibbs HL, Rossiter W. Rapid evolution by positive selection and gene gain and loss: PLA(2) venom genes in closely related Sistrurus rattlesnakes with divergent diets. J Mol Evol. 2008;66(2):151-66.
Publication Dates
-
Publication in this collection
27 Nov 2009 -
Date of issue
2009
History
-
Accepted
30 Nov 2009 -
Reviewed
15 Sept 2009 -
Received
22 Nov 2008