Abstract
Thirty-eight strains of Shiga toxin-producing Escherichia coli (STEC) were characterised in terms of biochemical properties, enterohaemolysin production and plasmid carriage. A wide variation in the biochemical properties was observed among the STEC, with 14 distinct biotypes identified. Biotype 1 was the most common, found in 29% of the strains. Enterohaemolysin production was detected in 29% of the strains. Most of the bacterial strains (95%) carried one or more plasmids and considerable heterogeneity in size and combinations was observed. Seven distinct plasmid profiles were identified. The most common profile, characterised by the presence of a single plasmid of ~90 kb, was found in 50% of these strains. These data indicate extensive diversity among STEC strains. No correlation was found among biotype, serotype, enterohaemolysin production and plasmid profile.
Escherichia coli; STEC; plasmid profile; enterohaemolysin; biochemical properties
ARTICLES
Biochemical properties, enterohaemolysin production and plasmid carriage of Shiga toxin-producing Escherichia coli strains
Mario RSM SouzaI; Giseli KlassenII; Fabiana De ToniI; Liu U RigoIII; Caroline HenkesI; Caroline P PigattoIV; Cibelle de Borba DalagassaI; Cyntia MT Fadel-PichethI, + + Corresponding author: fpicheth@ufpr.br
IDepartamento de Patologia Médica, Universidade Federal do Paraná, Av. Lothário Meissner 632, 80210-170 Curitiba, PR, Brasil
IIDepartamento de Patologia Básica, Universidade Federal do Paraná, Av. Lothário Meissner 632, 80210-170 Curitiba, PR, Brasil
IIIDepartamento de Bioquímica e Biologia Molecular, Universidade Federal do Paraná, Av. Lothário Meissner 632, 80210-170 Curitiba, PR, Brasil
IVDepartamento Medicina Veterinária Preventiva, Universidade Federal do Tocantins, Araguaína, TO, Brasil
ABSTRACT
Thirty-eight strains of Shiga toxin-producing Escherichia coli (STEC) were characterised in terms of biochemical properties, enterohaemolysin production and plasmid carriage. A wide variation in the biochemical properties was observed among the STEC, with 14 distinct biotypes identified. Biotype 1 was the most common, found in 29% of the strains. Enterohaemolysin production was detected in 29% of the strains. Most of the bacterial strains (95%) carried one or more plasmids and considerable heterogeneity in size and combinations was observed. Seven distinct plasmid profiles were identified. The most common profile, characterised by the presence of a single plasmid of ~90 kb, was found in 50% of these strains. These data indicate extensive diversity among STEC strains. No correlation was found among biotype, serotype, enterohaemolysin production and plasmid profile.
Key words:Escherichia coli - STEC - plasmid profile - enterohaemolysin - biochemical properties
Shiga toxin-producing Escherichia coli (STEC) strains were recognised as human pathogens in 1982 when the O157:H7 serotype was implicated in two outbreaks of haemorrhagic colitis. Since then, these bacteria have been considered an important group of food-borne pathogens associated with diseases varying from diarrhoea and haemorrhagic colitis to haemolytic uraemic syndrome. They are considered a major public health problem (Bettelheim & Beutin 2003). While O157:H7 is the most common serotype in many parts of the world, STEC strains belonging to a diverse range of serotypes have been isolated with increasing frequency and predominate in many countries (Karch et al. 1999, Bettelheim & Beutin 2003). STEC are characterised by their ability to produce Shiga toxins, which exhibit cytotoxic activity on Vero cells as well as on a number of cell types in the human body (Karch et al. 1999). In addition to Shiga toxins, which constitute their main virulence factor, other virulence properties may be exhibited by STEC. One accessory virulence factor is enterohaemolysin, encoded by ehxA, a plasmid-borne gene. Enterohaemolysin belongs to the RTX family of pore-forming cytolysins and its presence apparently increases the ability of STEC to cause extraintestinal complications (Schmidt et al. 1995). STEC can be found in the gut of several animal species and cattle are considered the major reservoir of these organisms and the most important source of human infections caused by these pathogens. In addition to foods of bovine origin, disease in humans has also been associated with ingestion of contaminated water, fruit and vegetables, as the dispersion of untreated manure in the environment can cause contamination of other edible substances, which can then act as secondary vehicles of human infections (Caprioli et al. 2005). A high prev-alence of STEC in stools of healthy cattle was found in the states of São Paulo (Irino et al. 2005), Rio de Janeiro (Cerqueira et al. 1999), Rio Grande do Sul (Moreira et al. 2003, Timm et al. 2007) and Paraná (PR) (Farah et al. 2007, Pigatto et al. 2008) and a prevalence of 1-2% of STEC in cases of diarrhoea in humans was reported (Vaz et al. 2004, De Toni et al. 2009). These studies have also shown that STEC strains belonging to non-O157 serotypes predominate in Brazil. In the light of the increasing importance of STEC as food-borne pathogens, the aim of this paper was to analyse the diversity among STEC strains in their biochemical properties, enterohaemolysin production and plasmid carriage.
MATERIALS AND METHODS
Bacteria - Thirty-eight STEC isolated from stools of healthy cattle (37 strains) and children with diarrhoea (1 strain) in PR were analysed. These bacteria had been previously identified using screening tests for E. coli (Pigatto et al. 2008) and had then been tested for Shiga toxin production using a cytotoxicity assay in Vero cells and for stx genes using a PCR assay according to Lin et al. (1993) or De Toni et al. (2009). The strains were also serotyped and classified in 24 distinct serotypes (Pigatto et al. 2008, De Toni et al. 2009). The STEC strains were grown overnight in tryptic soy broth (TSB) at 36°C and maintained at -20°C in TSB with 20% glycerol.
Biochemical characteristics - The biochemical characteristics of the STEC strains were analysed using 28 distinct tests prepared according to the classical methods described in MacFaddin (2000): arginine decarboxylase; adonitol, arabinose, cellobiose, dulcitol, glucose, inositol, lactose, maltose, melibiose, raffinose, rhamnose, salicin, sorbitol, sucrose, threhalose and xylose (carbohydrate fermentation tests), citrate, gas production, hydrogen sulphide, indole, lysine decarboxylase, methyl red, motility, ornithine decarboxylase, phenylalanine deaminase, urease and Voges-Proskauer tests.
Enterohaemolytic activity - The enterohaemolytic activity of the strains was determined on blood agar plates containing 5% defibrinated, washed sheep erythrocytes supplemented with 10 mM CaCl2 as described elsewhere (Beutin et al. 1989).
Plasmid preparations - Plasmid DNA was purified by the alkaline lysis method (Sambrook et al. 1989). At least two plasmid preparations were performed for each strain. The approximate size of plasmids was determined based on migration on 0.7% agarose gel compared with plasmids of known sizes.
RESULTS
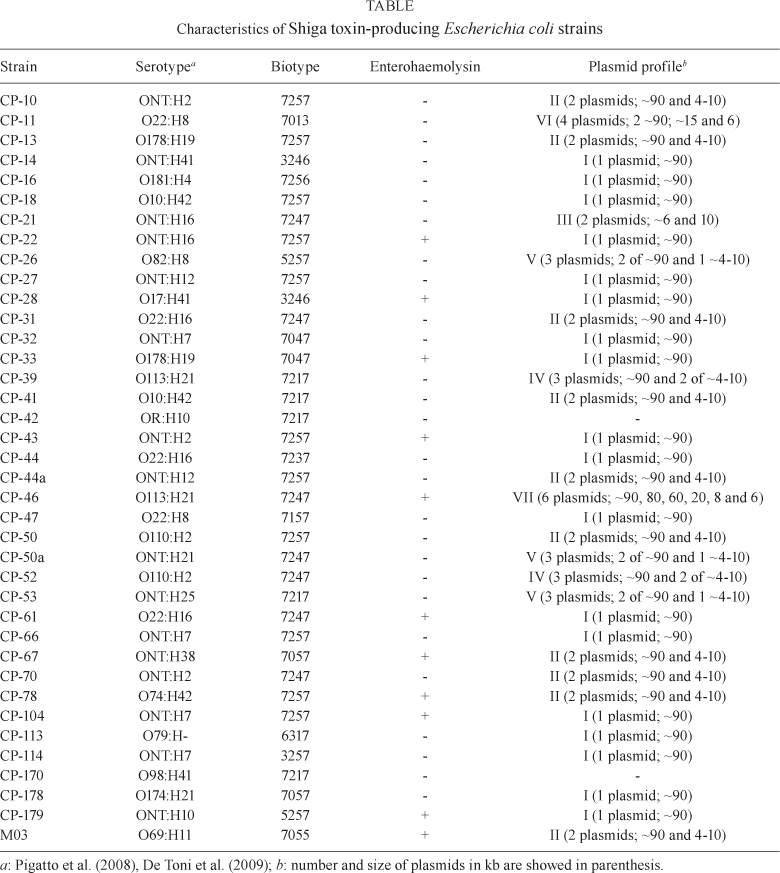
All 38 strains were positive for the following tests: lysine decarboxylase, arabinose, glucose, lactose, melibiose, threhalose, xylose, indole and methyl red. The 38 strains were negative for arginine decarboxylase, adonitol, citrate, hydrogen sulphide, phenylalanine deaminase, urease and Voges-Proskauer tests. Variable reactions were observed in all other tests, which were used to biotype the strains. The respective frequencies of positive tests among this latter group were as follows: maltose (98%), gas (95%), motility (90%), ornithine decarboxylase (88%), raffinose (88%), sorbitol (81%), dulcitol (76%), sucrose (74%), cellobiose (5%), inositol (3%), rhamnose (3%) and salicin (3%). All strains were confirmed as E. coli by their biochemical properties and classified in 14 different biotypes based on these characteristics (Table). The biotype was determined by a numeric code in which each test was represented by a number (shown in parentheses below) if the test was positive. For negative results the test value was zero. The tests were grouped in four classes in the following order: motility (1), gas (2) and ornithine decarboxylase (4), cellobiose (1), dulcitol (2) and inositol (4), sucrose (1), salicin (2) and sorbitol (4), raffinose (1), rhamnose (2) and maltose (4). The sum of the numbers of the tests of each class resulted in a biotype containing four digits (Table). The biotype most frequently found in the STEC strains analysed was 7257, which was exhibited by 11 strains (29%). Enterohaemolysin production was observed in only 11 (29%) of the strains and was not associated with specific biotypes or serotypes (Table). Most of the strains (95%) carried one or more plasmids. The number of plasmids carried in these strains varied from 1-6. Three classes of plasmids were found according to their approximate size: from 4-~10 kb, greater than 10-~22 kb and greater than 43-~90 kb. No plasmids were found with sizes in the range of 22-43 kb. According to the distribution of the plasmids by size and combination, seven major plasmid profiles were found (Fig. 1, Table). Plasmids larger than 43 kb were found in 35/36 STEC (97%). Eighteen strains (50%) contained single plasmids of approximately 90 kb (profile I). The other 18 strains (50%) contained different combinations of plasmids. Of these, 10 strains (28%) bore two plasmids: one of approximately 90 kb and another whose size varied among the strains between 4-10 kb (profile II). Another strain also presented two plasmids, with sizes of approximately 6 and 10 kb (profile III). Two strains belonged to profile IV, in which three plasmids were found: one of approximately 90 kb and two between 4-10 kb. Another three strains also bore three plasmids: two of them around 90 kb and another between 4-10 kb (profile V). Four plasmids were found in one strain, two of approximately 90 kb, another around 15 kb and one around 6 kb (profile VI). Profile VII was found in one strain, characterised by the presence of six plasmids of approximately 90, 80, 60, 20, 8 and 6 kb.
DISCUSSION
In most of the studies on STEC, only a few tests are used for E. coli identification (Irino et al. 2005, Farah et al. 2007, Pigatto et al. 2008). In the present study 28 biochemical tests were used to characterise the strains and the results indicate a wide variation in the metabolic traits of the STEC. Although only 38 strains were examined, 14 distinct biotypes were identified among them. Biotype 1 was the most common, found in 29% of the strains (Table). These results confirm that STEC strains present a high diversity in their biochemical behaviour and are phenotypically indistinguishable from other E. coli of the normal microbiota of the gut. Additionally, most of the strains were positive for sorbitol fermentation, indicating that MacConkey-Sorbitol, which was developed for detection of the sorbitol-negative STEC O157:H7 (Karch et al. 1999), is not useful for STEC screening in regions in which non-O157 serotypes predominate. Therefore, STEC identification should be done by detecting Shiga toxin production using immunological methods or the cytotoxicity assay in Vero cells or using molecular methods like PCR for stx gene detection. Enterohaemolysin production was observed only in 29% of the strains. This is in contrast with data reported by Irino et al. (2005) and Farah et al. (2007), who found 88% and 61%, respectively, of strains produced enterohaemolysin. These data suggest that the strains analysed in this study may have a lesser degree of virulence. The enterohaemolysin-producing strains presented plasmid profiles I, II and VII, all containing a large plasmid of around 90 kb (Table). These data suggest the presence of the ehxA gene in those plasmids and also that the large plasmids found in the STEC strains analysed are genetically heterogeneous, as only a portion of them were associated with enterohaemolysin production. Our data are in agreement with other studies that showed that the presence of large plasmids is common in STEC strains and that these elements are also heterogeneous in gene composition (Zhang et al. 2000, Karama et al. 2008). A considerable degree of heterogeneity was found in the plasmid carriage in the strains analysed in this study, which might reflect the degree of horizontal transfer events in these strains. This may also affect the virulence properties of these strains, as it is known that several of these characteristics are codified in plasmids. The use of the plasmid profile to analyse non-O157 STEC has revealed a high degree of diversity in these strains and shown strong discriminatory potential as a subtyping method. Extensive heterogeneity has been found in the plasmid profiles of STEC strains belonging to the O26 and O145 serotypes (Zhang et al. 2000, Sonntag et al. 2004). High diversity in the plasmid profile has also been found in STEC strains belonging to the O103:H2 serotype, for which a correlation between their human or animal source and geographic origin was found (Karama et al. 2008). No correlation was found among biotype, serotype, enterohaemolysin production and plasmid profile in this study.
In conclusion, considerable diversity was found in the biochemical characteristics and the presence of plasmids in STEC strains isolated in PR, Brazil. A broad distribution of plasmids was observed, with the larger plasmids being found most often.
Received 9 October 2009
Accepted 26 March 2010
Financial support: Fundação Araucária
- Bettelheim KA, Beutin L 2003. Rapid laboratory identification and characterization of verocytotoxigenic (Shiga toxin producing) Escherichia coli (VTEC/STEC). J Appl Microbiol 95: 205-217.
- Beutin L, Montenegro MA, Orskov F, Prada J, Zimmermann S, Stephan R 1989. Close association of Verotoxin (Shiga-like toxin) production with enterohaemolysin production in strains of Escherichia coli. J Clin Microbiol 27: 2559-2564.
- Caprioli A, Morabito S, Brugère H, Oswald E 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res 36: 289-311.
- Cerqueira AMF, Guth BEC, Joaquim RM, Andrade JRC 1999. High occurrence of Shiga toxin-producing Escherichia coli (STEC) in healthy cattle in Rio de Janeiro state, Brazil. Vet Microbiol 70: 111-121.
- De Toni F, Souza EM, Pedrosa FO, Klassen K, Irino K, Rigo LU, Steffens MBR, Fialho OB, Farah SMSS, Fadel-Picheth CMT 2009. A prospective study on Shiga toxin-producing Escherichia coli in children with diarrhoea in Paraná state, Brazil. Lett Appl Microbiol 48: 645-647.
- Farah SMSS, Souza EM, Pedrosa FO, Irino K, Silva LR, Rigo LU, Steffens MBR, Pigatto CP, Fadel-Picheth, CMT 2007. Phenotypic and genotypic traits of Shiga toxin-producing Escherichia coli strains isolated from beef cattle from Paraná state, Southern Brazil. Lett Appl Microbiol 44: 607-612.
- Irino K, Kato MAMF, Vaz TMI, Ramos II, Souza MAC, Cruz AS, Gomes TAT, Vieira MAM, Guth BEC 2005. Serotypes and virulence markers of Shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in São Paulo state, Brazil. Vet Microbiol 105: 29-36.
- Karama M, Johnson RP, Holtslander R, Gyles CL 2008. Phenotypic and genotypic characterization of verotoxin-producing Escherichia coli O103:H2 isolates from cattle and humans. J Clin Microbiol 46: 3569-3575.
- Karch H, Bielaszewska M, Bitzan M, Schmidt H 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn Microbiol Infect Dis 34: 229-243.
- Lin Z, Kurazono H, Yamasaki S, Takeda Y 1993. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol Immunol 37: 543-548.
- MacFaddin JF 2000. Biochemical tests for identification of medical bacteria, 2nd ed., Lippincott Williams & Wilkins, Philadelphia, 912 pp.
- Moreira CN, Pereira MA, Brod CS, Rodrigues DP, Carvalhal JB, Aleixo JAG 2003. Shiga toxin-producing Escherichia coli (STEC) isolated from healthy dairy cattle in Southern Brazil. Vet Microbiol 93: 179-183.
- Pigatto CP, Schocken-Iturrino RP, Souza EM, Pedrosa FO, Comarella L, Irino K, Kato MAMF, Farah SMSS, Warth JF, Fadel-Picheth CMT 2008. Virulence properties and antimicrobial susceptibility of Shiga toxin-producing Escherichia coli strains isolated from healthy cattle from Paraná state, Brazil. Can J Microbiol 54: 588-593.
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, New York, 1200 pp.
- Schmidt H, Beutin L, Karch H 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun 63: 1055-1061.
- Sonntag AK, Prager R, Bielaszewska M, Zhang W, Fruth A, Tschäpe H, Karch H 2004. Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J Clin Microbiol 42: 954-962.
- Timm CD, Irino K, Gomes TAT, Vieira MM, Guth BEC, Vaz TMI, Moreira CN, Aleixo JAG 2007. Virulence markers and serotypes of Shiga toxin-producing Escherichia coli, isolated from cattle in Rio Grande do Sul, Brazil. Lett Appl Microbiol 44: 419-425.
- Vaz TMI, Irino K, Kato MAMF, Dias AMG, Gomes TAT, Medeiros MIC, Rocha MMM, Guth BEC 2004. Virulence properties and characteristics of Shiga toxin-producing Escherichia coli in São Paulo, Brazil, from 1976 through 1999. J Clin Microbiol 42: 903-905.
- Zhang WL, Bielaszewska M, Liesegang A, Tschäpe H, Schmidt H, Bitzan M, Karch H 2000. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J Clin Microbiol 38: 2134-2140.
Publication Dates
-
Publication in this collection
21 May 2010 -
Date of issue
May 2010
History
-
Accepted
26 Mar 2010 -
Received
09 Oct 2009