Abstracts
The recently described fish species, Astyanax altiparanae (tetra) is common in the upper rio Paraná basin, and has been reported in the Iguaçu basin. However, its natural origin in the rio Iguaçu is questionable. In the present work, karyotypical and morphological features of two populations of Astyanax altiparanae from the upper rio Tibagi and upper rio Iguaçu were compared. Both populations showed 2n=50 chromosomes and differences in their karyotype formula, NOR-bearing chromosomes and location of heterochromatin. Morphometric data from both populations were analyzed through free-size canonical variables. Cytogenetic and morphological results were mostly coincident showing exclusive markers that reflect their degree of populational isolation. In addition to other geographic, morphological and molecular data for A. altiparanae populations from the lower rio Iguaçu and rio Paraná tributaries upstream from the Itaipu Dam (South Brazil), the present results indicate that the two populations analyzed in this study belong to different stocks. The presence of this species along the rio Iguaçu basin would be a consequence of a complex and poorly understood evolutionary history.
Tetra; Karyotype evolution; Distribution; Morphometry
Astyanax altiparanae, recentemente descrita, é comum na bacia do alto rio Paraná, tendo sido registrada recentemente na bacia do rio Iguaçu. Entretanto, sua origem natural no rio Iguaçu é questionável. No presente trabalho algumas características cariotípicas e morfológicas de duas populações de Astyanax altiparanae do rio alto rio Tibagi e alto rio Iguaçu foram comparadas. Ambas as populações mostraram 2n=50 cromossomos e diferenças nas suas fórmulas de cariotípicas, cromossomos portadores das regiões organizadoras de nucléolos e localização da heterocromatina. Dados morfométricos das duas populações foram analisados para variáveis canônicas livres de tamanho. Os dados citogenéticos e morfológicos foram coincidentes e mostraram marcas exclusivas que refletem o grau de isolamento populacional. Em adição a outros dados geográficos, morfológicos e moleculares em populações de A. altiparanae do baixo rio Iguaçu e tributários do rio Paraná acima da represa de Itaipu (Sul do Brasil), os presentes resultados indicam que as duas populações analisadas neste estudo pertençam a diferentes estoques. A presença desta espécie ao longo da bacia do rio Iguaçu pode ser uma conseqüência de uma história evolutiva complexa e pouco conhecida.
Cytogenetic and comparative morphology of two allopatric populations of Astyanax altiparanae Garutti & Britski, 2000 (Teleostei: Characidae) from upper rio Paraná basin
Manoela de S. DominguesI; Marcelo R. VicariII; Vinícius AbilhoaIII; João P. WamserIV; Marta M. CestariI; Luiz A. C. BertolloII; Mara C. de AlmeidaIV; Roberto F. ArtoniIV
IUniversidade Federal do Paraná. Programa de Pós-Graduação em Genética. Centro Politécnico, Jardim das Américas. 81531-990 Curitiba, PR, Brazil
IIUniversidade Federal de São Carlos. Programa de Pós-Graduação em Genética e Evolução. Via Washington Luís, km 235. 13565-905 São Carlos, SP, Brazil
IIIMuseu de História Natural Capão da Imbuia. Prefeitura de Curitiba. Rua Prof. Benedito Conceição, 407. 82810-080 Curitiba, PR, Brazil
IVUniversidade Estadual de Ponta Grossa. Departamento de Biologia Estrutural, Molecular e Genética. Av. Carlos Cavalcanti, 4748, 84030-900 Ponta Grossa, PR, Brazil. rfartoni@pesquisador.cnpq.br
ABSTRACT
The recently described fish species, Astyanax altiparanae (tetra) is common in the upper rio Paraná basin, and has been reported in the Iguaçu basin. However, its natural origin in the rio Iguaçu is questionable. In the present work, karyotypical and morphological features of two populations of Astyanax altiparanae from the upper rio Tibagi and upper rio Iguaçu were compared. Both populations showed 2n=50 chromosomes and differences in their karyotype formula, NOR-bearing chromosomes and location of heterochromatin. Morphometric data from both populations were analyzed through free-size canonical variables. Cytogenetic and morphological results were mostly coincident showing exclusive markers that reflect their degree of populational isolation. In addition to other geographic, morphological and molecular data for A. altiparanae populations from the lower rio Iguaçu and rio Paraná tributaries upstream from the Itaipu Dam (South Brazil), the present results indicate that the two populations analyzed in this study belong to different stocks. The presence of this species along the rio Iguaçu basin would be a consequence of a complex and poorly understood evolutionary history.
Key words: Tetra, Karyotype evolution, Distribution, Morphometry.
RESUMO
Astyanax altiparanae, recentemente descrita, é comum na bacia do alto rio Paraná, tendo sido registrada recentemente na bacia do rio Iguaçu. Entretanto, sua origem natural no rio Iguaçu é questionável. No presente trabalho algumas características cariotípicas e morfológicas de duas populações de Astyanax altiparanae do rio alto rio Tibagi e alto rio Iguaçu foram comparadas. Ambas as populações mostraram 2n=50 cromossomos e diferenças nas suas fórmulas de cariotípicas, cromossomos portadores das regiões organizadoras de nucléolos e localização da heterocromatina. Dados morfométricos das duas populações foram analisados para variáveis canônicas livres de tamanho. Os dados citogenéticos e morfológicos foram coincidentes e mostraram marcas exclusivas que refletem o grau de isolamento populacional. Em adição a outros dados geográficos, morfológicos e moleculares em populações de A. altiparanae do baixo rio Iguaçu e tributários do rio Paraná acima da represa de Itaipu (Sul do Brasil), os presentes resultados indicam que as duas populações analisadas neste estudo pertençam a diferentes estoques. A presença desta espécie ao longo da bacia do rio Iguaçu pode ser uma conseqüência de uma história evolutiva complexa e pouco conhecida.
Introduction
Excepting putative anthropic introductions, the rio Tibagi basin in southern Brazil probably maintains most of its original ichthyofauna since it was isolated from the rio Paraná after the construction of dams for hydroelectric power (Shibatta et al., 2002). On the other hand, the rio Iguaçu, although geologically old, has been renewed by several uplifting events giving rise to a sort of falls, alternating with wide curvatures and extended flooded areas. Due to the rise of Iguaçu waterfalls, nearly 22 million years ago, fish populations of the rio Iguaçu basin were isolated from the upper rio Paraná basin (Severi & Cordeiro, 1994). Such fact might have favored a speciation process, which would be responsible for the remarkable level of endemism of its ichthyofauna (Ingenito et al., 2004).
The genus Astyanax comprises about a hundred species, widespread throughout Brazilian hydrographic basins, and is considered an incertae sedis genus within Characidae (Reis et al., 2003). Garutti & Britski (2000) revised some Astyanax species and described A. altiparanae, a new species formerly reported as A. bimaculatus, to the upper rio Paraná basin. The species A. altiparanae is found in tributaries of the rio Tibagi basin and shows high feeding flexibility and opportunistic behavior (Shibatta et al., 2002).
Although the headwaters of the Iguaçu and Tibagi rivers are close to each other, the occurrence of A. altiparanae in the rio Iguaçu basin has just recently been reported (Ingenito et al., 2004), leading to the following question: Was this species never collected before, or introduced by humans, or did it reach the main river from a tributary where it naturally occurs? Prioli et al. (2002) studied a population of A. altiparanae from the ower rio Iguaçu and did not find evidence of,endemism. There was a strong genetic similarity and high gene flow among A. altiparanae populations from the upper rio Paraná (and its tributaries) and lower rio Iguaçu, as demonstrated by RAPD and mitochondrial DNA markers (Prioli et al. op. cit.). Morphological studies have reinforced this hypothesis, although they have not been conclusive about the origin of this species in the rio Iguaçu (Graça & Pavanelli, 2002).
Astyanax representatives have shown a great karyotypic diversity, with a chromosome number ranging from 2n=36 in Astyanax schubarti (Morelli et al., 1983) to 2n=50 in Astyanax scabripinis, Astyanax bimaculatus, Astyanax altiparanae (Oliveira et al., 1988) and others. Diploid (2n) and fundamental numbers (FN), associated with other chromosomal markers, have allowed the establishment of cytotaxonomic and evolutionary inferences for these fishes. Sympatric and allopatric karyotypic conditions are verified in Astyanax scabripinnis (Souza & Moreira-Filho, 1995a; b). For instance, species-specific cytotypes were identified in A. parahybae, A. intermedius and A. giton from the rio Paraíba do Sul basin (Kavalco & Moreira-Filho, 2003). The presence of B-chromosomes is also a remarkable feature amongst Astyanax species (Moreira-Filho et al., 2004).
The integration of cytogenetic data and morphological analyses is uncommon in studies of the evolutionary biology of Neotropical fishes. Moreira-Filho & Bertollo (1991), in studying six Astyanax scabripinnis populations, demonstrated that some of them could be identified through morphological and/or karyotypic analyses, while others overlapped, independent of their geographical distribution along the hydrographic basins selected.
The present study describes trends in karyotype evolution and cytotaxonomy based on chromosomal and morphological data of two populations of A. altiparanae from the upper rio Tibagi and upper rio Iguaçu.
Material and Methods
Characterization of study area and sampling
The two populations of Astyanax altiparanae from the upper rio Tibagi and upper rio Iguaçu (Fig. 1) were studied cytogenetically and morphologically. For the karyotypic analyses 65 specimens of the upper rio Tibagi and 66 specimens of the upper rio Iguaçu were sampled. For the morphologic analyses 36 specimens of the upper rio Tibagi and 37 specimens of the upper rio Iguaçu were sampled. The specimens examined were deposited in the Museu de História Natural Capão da Imbuia, Curitiba, Brazil: MHNCI 11458, 21, and MHNCI 11459, 17, Brazil, Paraná, Ponta Grossa, Vila Velha State Park, lagoa Tarumã, Guabiroba river basin, upper rio Tibagi. MHNCI 11460, 30, and MHNCI 11461, 7, Brazil, Paraná, Piraquara, Canguiri Farm - UFPR, rio Irai tributary, upper rio Iguaçu.
Chromosomal markers
Chromosome preparations were obtained according to Bertollo et al. (1978), using a cell suspension from the anterior kidney after in vivo treatment with colchicine (1ml/100g body weight). The nucleolar organizing regions (NORs) and heterochromatin location were detected according to Howell & Black (1980) and Sumner (1972), respectively. Chromomycin A3 staining was performed according to Schmid (1980). The location of the ribosomal genes on the chromosomes was obtained by FISH, with an 18S rDNA probe obtained by PCR from the nuclear DNA of Prochilodus argenteus (Hatanaka & Galetti Jr., 2004) using the primers NS1 5'-GTAGTCATATGCTTGTCTC 3' and NS8 5'-TCCGCAGGTTCACCTACGGA3' (White et al., 1990) and 5S rDNA probe amplified using A (5'-TACGCCCGATCTCGTCCGATC-3') and B (5'-GCTGGTATGGCCGTAGC-3') primers of Leporinus elongatus (Martins & Galetti Jr., 1999). The probes were labelled with 14-dATP biotin by nick translation, according to the manufacturer's instructions (Bionick Labelling System Invitrogen). The chromosomes were treated with RNAse (40µg/ml in 2xSSC) at 37ºC for one hour and with pepsin (0.005% in 10mM HCl) at 37ºC for 10 min, and denatured in 70% formamide/2xSSC for 5 min. The hybridization solution consisted of 50% formamide, 2xSSC, dextran sulfate (10%) and also the denatured probe. After an overnight hybridization at 37ºC, the slide was washed in 50% formamide at 42ºC for 20 min and 0.1xSSC at 60ºC for 15 min. The hybridization signal was detected using conjugated avidin-fluorescein (FITC) and biotinylated anti-avidin antibody. The chromosomes were counterstained with propidium iodide (50µg/ml) and analyzed with an Olympus BX50 epifluorescence microscope. The metaphases were captured by the CoolSNAP-Pro software (Media Cybernetic). For karyotyping, the chromosomes were ordered in decreasing size and classified as metacentric (m), submetacentric (sm) and subtelocentric (st), according to their arms ratio following the method of Levan et al. (1964).
Morphometric analysis
Thirty-six specimens from the rio Tibagi and 37 from the rio Iguaçu were measured. Measurements were taken following Fink & Weitzmann (1974). All measurements were taken as straight-line distances using a manual caliper of 0.05mm precision. For morphometric analysis, point-to-point measurements were peformed for standard length (SL), pre-dorsal length (PDL), pre-ventral length (PVL), pre-pectoral length (PPL), pre-anal length (PAL), body height (BH), caudal peduncle height (CPH), caudal peduncle length (CPL), dorsal fin length (DFL), ventral fin length (VFL), pectoral fin length (PFL), anal fin length (AFL), head length (HL), snout length (SL), upper jaw length (JL), interorbital distance (ID) and orbit diameter (OD) were performed.
Morphometric data were explored using multivariate analysis. Size-free canonical discriminant analysis was used on the covariance matrix of log-transformed measurements to assess morphometric variation between samples. This technique consists of removing the effect of within-group ontogenetic variation, regressing each character separately on the within-group first principal component, which is a multivariate estimate of size (Reis et al., 1990). The analyses were carried out using the software PAST v. 1.3 (Hammer & Harper, 2004) and BAC v.2 (Dujardin, 2002).
Results
Chromosomal variability
The two samples analyzed, called the Tibagi and Iguaçu populations in the present report, showed the same diploid number (2n=50) and different karyotypic macrostructure, with FN=92, showing 6 metacentric (m), 28 submetacentric (sm), 8 subtelocentric (st) and 8 acrocentric (a) chromosomes in the Tibagi population, and FN=94, showing 6 metacentric (m), 30 submetacentric (sm), 8 subtelocentric (st) and 6 acrocentric (a) chromosomes in the Iguaçu population. The karyotypes were identical between sexes, without any evidence of heteromorphic sex chromosomes (Fig. 2 A; D).
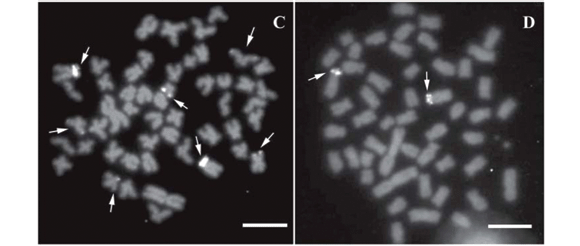
Most chromosomes showed a small amount of pericentromeric heterochromatin. In some cases, equilocal centromeric and nearly interstitial C-bands on long arms could be observed in both populations. Several chromosomes bearing entirely heterochromatic short arms were observed only in the Tibagi population (Fig. 2 C; F). The most frequent chromosome pair stained by silver nitrate, comparable in the two populations, showed nucleolar organizer regions (NORs) coincident with C-bands (Fig. 2 B; E).
All nucleolar organizer regions demonstrated by silver nitrate (Ag-NORs) were coincident with positive GC-rich staining by chromomycin A3/DAPI staining, revealing a single pattern in the Iguaçu population and a multiple pattern in the Tibagi population. The former population showed NORs located in the telomeric region of a submetacentric chromosome pair, and the latter displayed two NOR-bearing chromosomes in the karyotype, one metacentric and the other submetacentric (Figs. 2 B; E and 3 A ; B ). Two other GC-rich regions were detected in A. altiparanae from the rio Tibagi, at the telomeric position on a metacentric chromosome and at the terminal position on the long arms of a subtelocentric chromosome (Fig. 3 A ).
Fluorescent in situ hybridization with major 18S rDNA probes followed invariably the pattern observed with chromomycin A3, being coincident with GC-rich heterochromatin sites in all metaphases analyzed for the Tibagi populations. Up to seven loci of major ribosomal DNA were demonstrated in the Tibagi population, while the Iguaçu population showed up to two chromosomal regions bearing 18S rDNA to the detriment of a larger number of the heterochromatin GC-rich (Fig. 3 C; D). Fluorescent in situ hybridization with 5S rDNA probes revealed positive signals in the pericentromeric region of a submetacentric pair in both populations analyzed (Fig. 3 E; F).
Morphological variability
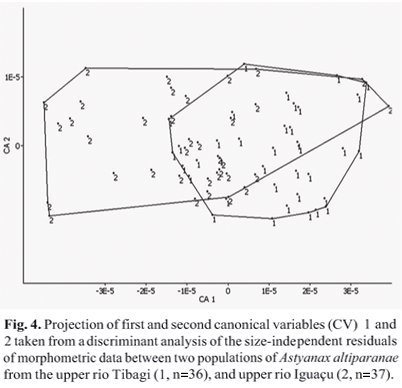
Size-free canonical discriminant analysis (SF-CDA) in Astyanax altiparanae populations from the upper parts of the rio Tibagi and upper rio Iguaçu showed that the cumulative variance of the first two variables represented acceptably the total variation (57.46%.). The first canonical variable (CV I) explained 45.79% of the morphometric variation, while the second canonical variable (CV II) explained 11.67% (Table 1). The morphometric measurements responsible for the variability in the canonical analysis were caudal peduncle height, caudal peduncle length, snout length and upper jaw length.
No clear morphological discrimination based on these measurements between the Iguaçu and Tibagi populations was evident. In SF-CDA analysis, the projection of the individual scores in the space of the first and second canonical axis showed no clear distinction between the groups (Fig. 4), revealing a non-significant divergence in shape. Conversely, univariate statistical comparisons between caudal peduncle height (t=4.47; p<0.05) and upper jaw length (t=2.19; p<0.05) revealed significant differences, indicating lower mean caudal peduncle height and upper jaw length values in the Tibagi sample. Although these significant differences were evident, within-group clusters overlapped considerably. Due to the high overlapping, the Iguaçu and Tibagi rivers populations analyzed were undistinguishable from each other using morphological multivariate analysis.
Discussion
Karyotypic comparisons in Astyanax altiparanae
The karyotypic data obtained from both populations indicate a conservativeness in diploid number. Similar to previous cytogenetic studies in Astyanax altiparanae (Fernandes & Martins-Santos, 2004), the present report also verified a diploid number of 50 chromosomes for these two populations. Thus, this feature should be considered primitive for this species and very relevant for the polarization of karyotypic data in phylogenetic comparisons within the genus Astyanax.
No chromosomal heteromorphism concerning sex differentiation has been foundiin any population analyzed so far. On the other hand, the karyotypic formula and the fundamental number of both populations showed a disruption of the main evolutionary conservativeness. Inter-populational karyotypic differences related to chromosomal morphology, especially concerning submetacentric and acrocentric chromosomes, could be observed in the populations from the upper rio Tibagi and upper rio Iguaçu, with a fundamental number equal to 92 and 94, respectively. Therefore, the role of non-robertsonian events in karyotypic diversification, such as inversions (especially pericentric ones) could be discussed in the karyotypic evolution of this species. Other studies have demonstrated inter-populational variations in the karyotypic macrostructure of Astyanax altiparanae (also Astyanax bimaculatus) (Morelli et al., 1983; Paganelli, 1990; Pacheco et al., 2001; Fernandes & Martins-Santos, 2004). Some biological features of Astyanax altiparanae can partially explain such level of karyotypic variability. This species shows wide adaptive abilities, using particular strategies of population structuring (Orsi et al., 2002). The high feeding versatility of Astyanax altiparanae, as well as its ability to reproduce in several environments, favors its successful colonization of new habitats. However, the species shows a preference for calm waters, since this fish occurs in several different microenvironments and since the populations are not morphologically homogeneous (Garutti & Britski, 2000). These adaptations could facilitate the maintenance of regional variations in karyotype formula and fundamental number in different populations of Astyanax altiparanae from the Tibagi and Iguaçu river basins in the present report.
The hypothesis of gene flow or reproductive isolation proposed by Prioli et al. (2002) for A. altiparanae populations from the lower and middle rio Iguaçu and another population from the rio Paraná does not refute the possibility that anthropic introduction, headwater capture or gene introgression might have occurred in the rio Iguaçu basin and influenced the expected diversity, either by vicariance or dispersal. Apart from possible vicariance effects over karyotypic evolution of species from distinct large hydrographic basins such as those of the Tibagi and Iguaçu rivers, the distribution pattern of Astyanax altiparanae along these basins indicates a high adaptive radiation by dispersal. Different cytotypes can be identified throughout these basins, especially involving karyotype formulae and fundamental numbers (FN). The vicariance and dispersal hypothesis may eventually be tested by cladistic or similar methods using molecular markers in addition to those used in this study.
Despite the inter-populational differences found in the karyotype macrostructure, diploid number stability is maintained in A. altiparanae, with 2n = 50 chromosomes. The plesiomorphic character may be a consequence of species migratory behavior. Even within short distances (Orsi et al., 2002), migration may lead to homogenization of diploid number in this fish, widely distributed over the upper rio Paraná basin (Garutti & Britski, 2000).
Microevolutionary processes in karyotype evolution, such as patterns of location and distribution of highly repetitive (e.g. heterochromatin) and moderately repetitive (e.g. nucleolus organizer regions) DNA sequences, do not rely only on external populational factors or behavior. The heterochromatin distribution, demonstrated in both A. altiparanae populations by C-banding and chromomycin A3, indicate some distinct features in relation to other Astyanax species, such as A. scabripinnis and A. fasciatus, which usually show conspicuous blocks, especially on acrocentric chromosomes, comprising different heterochromatin classes of variable origin. In the present study, the heterochromatin was mainly distributed in the pericentromeric regions on two-armed chromosomes, and sometimes, at the interstitial position on long arms of these chromosomes. Such pericentromeric and interstitial pattern of heterochromatin localization seems to be conserved among the few Astyanax altiparanae populations previously characterized by C-banding (Daniel-Silva & Almeida-Toledo, 2001, Fernandes & Martins-Santos, 2004). Comparatively, heterochromatic staining on short arms of some chromosomes in A. altiparanae from upper rio Tibagi would represent a cytotaxonomic marker for this population in relation to that from the upper rio Iguaçu.
The nature of heterochromatin was not differentiated by fluorochrome staining, excepting the GC-rich heterochromatic region associated with NORs. A specific chromosome pair stained with CMA3 in the rio Iguaçu population was neither coincident with Ag-NOR staining nor rDNA probes. NORs in the population from the upper rio Tibagi were located at the terminal position on seven bi-armed chromosomes (multiple NORs), while the population from the upper rio Iguaçu had single NORs on a subtelocentric chromosome pair, also in the telomeric region. The occurrence of a single nucleolar pair, a clear primitive condition, is demonstrated for the first time in Astyanax altiparanae through the localization of major rDNA. The presence of multiple NORs involving several chromosomes seems to be more common, as revealed in the A. altiparanae population from the upper rio Tibagi and also at other sites within the same hydrographic basin (Pacheco et al., 2001) or not (Daniel-Silva & Almeida-Toledo 2001). Fernandes and Martins-Santos (2004) revealed some GC positive signals after chromomycin staining in a population of A. altiparanae from the rio Paraná, indicating that more than a single NOR-bearing pair, although inactive, could be present. However, this suggestion still needs to be tested through in situ hybridization with major rDNA probes.
The correlation between NORs and GC-rich sites have been commonly reported in fishes, although GC-specific fluorochrome staining can not be considered a direct method of determining ribosomal genes, but only GC-rich heterochromatin (Artoni et al., 1999). Despite the inter-populational variation regarding the location of major ribosomal DNA, both populations showed only a single and comparable submetacentric chromosome pair bearing 5S rDNA in the pericentromeric region. On the other hand, the location of 18S ribosomal genes indicates that processes of karyotypic diversification, such as transposition and translocation, might have differently influenced the populations studied, reinforcing the hypothesis of recent allopatry. Conversely, the distribution of the 5S ribosomal DNA family indicates a clear conservativeness of these genes, probably due to its pericentromeric location on long arms of chromosomes carrying these cistrons (Mantovani et al., 2000).
Morphometric comparisons and integration with karyotypic data
Size-free canonical discriminant analysis (SF-CVA) in Astyanax altiparanae populations from the upper rio Tibagi and upper rio Iguaçu revealed an overlapping of individual scores based on morphometric measurements in the space defined by the first two canonical axis. These data suggest that both populations showed no relevant morphological differences. Multivariate analysis indicated that both populations belong to the same species, and the differences could be due to inter-populational variations. Smaller variations (caudal peduncle height t=4.47; p<0.05 and upper jaw length t=2.19; p<0.05) may be attributed to environmental oscillations, as well as to a populational polymorphism, as observed in other populations of the same species from different locations. In contrast, Shibatta & Artoni (2005) demonstrated the presence of a species complex through the analysis of morphological canonical variables in different Astyanax aff. fasciatus populations from the upper rio Tibagi which was corroborated by the karyotypic analysis of these populations (Artoni et al., 2006).
The characterization of both Astyanax altiparanae populations in the present work, combining cytogenetic and morphological data, indicates the occurrence of distinct population stocks from the same species. Priolli et al. (2002), through mitochondrial DNA sequences and RAPD markers, concluded that a sample of Astyanax altiparanae from the third plateau of the rio Iguaçu in the state of Paraná, Brazil, did not differ from samples in tributaries from the upper rio Paraná. Similarly, morphological analyses of these populations confirmed that they represent a single species and have also increased the distribution range of A. altiparanae (Graça & Pavanelli, 2002). However, there is a divergence between these authors regarding the occurrence of A. altiparanae in the rio Iguaçu basin. The former authors suggest that its presence is due to recent introduction, while the latter authors revealed neither anthropic introduction nor common origin. Based on the present report, we present another hypothesis; the occurrence of different stocks of A. altiparanae along the rio Iguaçu basin could be a consequence of the formation history of this basin, its ancient lakes and connections with adjacent basins such as the upper Tibagi, Ribeira de Iguape and Paraná rivers. This hypothesis seems to be in agreement with the data found by Paiva et al. (2006) for Astyanax bimaculatus from the rio Doce basin. In this case, genetic and morphological variations do not reflect exclusively events of allopatric speciation but indicate divergence at the population level. The findings of Ingenito et al. (2004), especially involving reports on A. altiparanae in the upper rio Iguaçu, support that samplings in the rio Iguaçu and adjacent basins need to be increased in order to achieve a more accurate interpretation of both regional biogeography and distribution of these fishes. Within the rio Tibagi basin, cytogenetic studies have pointed to populational differences in Astyanax altiparanae. The utilization of other tools in evolutionary biology, as well as morphologic and molecular markers, may afford a better characterization and interpretation of this report for the rio Tibagi and rio Iguaçu basins.
Acknowledgments
The authors are grateful to IAP (Instituto Ambiental do Paraná) and IBAMA (Instituto Brasileiro do Meio Ambiente proc. IBAMA/MMA nº 02017.000686/00-21), for license to capture fish in the Vila Velha State Park. This study was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná). We also thank Miguel A Carvalho for field and laboratory assistance.
Literature Cited
Received November 2006
Accepted February 2007
- Artoni, R. F., O. A. Shibatta, M. C. Gross, C. H. Schneider, M. C. Almeida, M. R. Vicari & L. A. C. Bertollo, L. A. C. 2006. Astyanax aff. fasciatus Cuvier, 1819 (Teleostei; Characidae): evidences of a species complex in the upper rio Tibagi basin (Paraná, Brazil). Neotropical Ichthyology, 4(2): 197-202.
- Artoni, R. F., W. F. Molina, L. A. C. Bertollo & P. M. Galetti Jr. 1999. Heterochromatin analysis in the fish species Liposarcus anisitsi (Siluriformes) and Leporinus elongatus (Characiformes). Genetics and Molecular Biology, 22: 1-6.
- Bertollo, L. A. C., C. S. Takahashi & O. Moreira-Filho. 1978. Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics, 1: 103-120.
- Daniel-Silva, M. de F. Z. & L. F. Almeida-Toledo. 2001. Chromosome R-banding pattern and conservation of a marker chromosome in four species, genus Astyanax (Characiformes, Tetragonoopterinae). Caryologia, 54: 209-215.
- Dujardin, J. P. 2002. BAC software. Institut de Recherches pour le Développement (IRD, France) (http://www.fsf.org/copyleft/gpl.html).
- Fernandes, C. A. & I. C. Martins-Santos. 2004. Cytogenetic studies in two populations of Astyanax altiparanae (Pisces, Characiformes). Hereditas, 141: 1-5.
- Fink, W. L. & S. H. Weitzmann. 1974. The so-called cheirodontin fishes of Central America with description of two new species (Pisces, Characidae). Smithsonian Contributions to Zoology, 172: 1-46.
- Garutti, V. & H. A. Britski. 2000. Descrição de uma espécie nova de Astyanax (Teleostei: Characidae) da bacia do alto rio Paraná e considerações sobre as demais espécies do gênero na bacia. Comunicações do Museu de Ciência e Tecnologia da PUCRS Série Zoologia, 13: 65-88.
- Graça, W. J. & C. S. Pavanelli. 2002. Astyanax altiparanae Garutti & Britski, 2000 (Osteichthyes, Characidae) in the Iguaçu River basin. Acta Scientarium, 24: 451-453.
- Hammer, O. & D. A. T. Harper. 2004: Past version 1.3. (http://folk.uio.no/ohammer/past).
- Hatanaka, T. & P. M. Galetti Jr. 2004. Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica, 122: 239-244.
- Howell, W. M. & D. A. Black. 1980. Controlled silver-stining of nucleolus organizer regions with a protective colloidal developer: a 1- step method. Experientia, 36: 1014-1015.
- Ingenito, L. F. S., L. F. Duboc & V. Abilhoa. 2004. Contribuição ao conhecimento da ictiofauna da bacia do alto rio Iguaçu, Paraná, Brasil. Arquivos de Ciência Veterinária e Zoologia da UNIPAR, 7: 23-26.
- Kavalco, K. F. & O. Moreira-Filho. 2003. Cytogenetical analyses in four species of the genus Astyanax (Pisces, Characidae) from Paraíba do Sul River Basin. Caryologia, 56: 453-461.
- Levan, A., K. Fredga & A. A. Sandberg. 1964. Nomenclature for centromeric positions on chromosomes. Hereditas, 52: 201-220.
- Mantovani, M., L. D. S. Abel, C. A. Mestriner & O. Moreira-Filho. 2000. Accentuated polymorphism of heterochromatin and nucleolar organizer regions in Astyanax scabripinnis (Pisces, Characidae): tools for understanding karyotypic evolution. Genetica, 109: 161-168.
- Martins, C. & P. M. Galetti Jr. 1999. Chromosomal localization of 5S rDNA genes in Leporinus Fish (Anostomidae, Characiformes). Chromosome Research, 7: 363-367.
- Moreira-Filho, O. & L. A. C. Bertollo. 1991. Astyanax scabripinnis (Pisces, Characidae): a species complex. Brazilian Journal of Genetics, 14: 331-357.
- Moreira-Filho, O., P. M. Galetti Jr. & L. A. C. Bertollo. 2004. B chromosomes in the fish Astyanax scabripinnis (Characidae, Tetragonoopterinae): An overview in natural populations. Cytogenetics and Genome Research, 106: 230-234.
- Morelli, S., L. A. C. Bertollo, F. Foresti, O. Moreira-Filho & S. A. Toledo-Filho. 1983. Cytogenetic considerations on the genus Astyanax (Pisces, Characidae). I. Kariotypic Variability. Caryologia, 36: 235-244.
- Oliveira, C., L. F. Almeida-Toledo, F. Foresti, H. A. Britski & S. A. Toledo-Filho. 1988. Chromosome formulae of Neotropical freshwater fishes. Brazilian Journal of Genetics, 11: 577-624.
- Orsi, M. L., O. A. Shibatta & A. T. Silva-Souza. 2002. Caracterização biológica de populações de peixes do rio Tibagi, localidade de Sertanópolis. Pp. 425-432. In: Medri, M. E., E. Bianchini, O. A. Shibatta & J. A. Pimenta (Eds.). A bacia do rio Tibagi. Londrina, 595p.
- Pacheco, R. B., L. Giuliano-Caetano & A. L. Dias. 2001. Cytotypes and Multiple NORs in an Astyanax altiparanae population (Pisces, Tetragonopterinae). Chromosome Science, 5: 109-114.
- Paiva, S. R., Dergam, J. A. & Machado, F. 2006. Determining management units in southeastern Brazil: the case of Astyanax bimaculatus (Linnaeus, 1758) (Teleostei: Ostariophysi: Characidae). Hydrobiologia 560: 393-404.
- Prioli, S. M. A. P., A. J. Prioli, H. F. Júlio Jr., C. S. Pavanelli, A. V. Oliveira, H. Carrer, D. M. Carraro & L. M. Prioli. 2002. Identification of Astyanax altiparanae (Teleostei, Characidae) in the Iguaçu river, Brazil, based on mitochondrial DNA and RAPD markers. Genetics and Molecular Biology, 25: 421-430.
- Reis, S.F., L. M. Pessôa & R. E. Strauss. 1990. Application of size-free canonical discriminant analysis to studies of geographic differentiation. Brazilian Journal of Genetics, 13: 509-520.
- Reis, R. E., S. O. Kullander & C. Ferraris. 2003. Check List of Freshwater Fishes of South and Central America. Porto Alegre, Edipucrs, 729 p.
- Severi, W. & A. A. M. Cordeiro. 1994. Catálogo de peixes da bacia do Rio Iguaçu. Curitiba, 118 p.
- Schmid, M. 1980. Chromosome banding in Amphibia. IV. Differentiation of GC- and AT-rich chromosome region in Anura Chromosoma, 77: 83-103.
- Shibatta, O. A. & R. F. Artoni. 2005. Sobre a identidade das populações alopátricas de Astyanax (Characiformes, Characidae) das formações Furna 1 e Furna 2 do Parque Estadual de Vila Velha, Ponta Grossa, Paraná, Brasil. Publicatio UEPG Biological and Health Science, 11: 7-12.
- Shibatta, O. A., M. L. Orsi, S. T. Bennemann & A. T. Silva-Souza. 2002. Diversidade e distribuição de peixes na bacia do rio Tibagi. Pp. 403-423. In: Medri, M. E., E. Bianchini, O. A. Shibatta & J. A. Pimenta (Eds.). A bacia do rio Tibagi. Londrina, 595p.
- Souza, I. L. & O. Moreira-Filho. 1995a. Cytogenetic diversity in the Astyanax scabripinnis species complex (Pisces, Characidae). I. Allopatric distribution in a small stream. Cytologia, 60: 1-11.
- Souza, I. L. & O. Moreira-Filho. 1995b. Cytogenetic diversity in the Astyanax scabripinnis species (Pisces, Characidae) complex. II. Different cytotypes living in simpatry. Cytologia, 60: 273-281.
- Sumner, A. T. 1972. A simple technique for demosntrating centromeric heterochromatin. Experimental Cell Research, 74: 304-306.
- White, T. J., T. Bruns, S. Lee & L. Taylor. 1990. Amplification and direct sequencing of fungal ribossomal RNA genes for phylogenetics. PCR Protocools: A guide to methods and applications. Academic Press, New York/London. 482p.
Publication Dates
-
Publication in this collection
10 July 2007 -
Date of issue
Mar 2007
History
-
Accepted
Feb 2007 -
Received
Nov 2006