Abstracts
In this study, phylogenetic and phylogeographic analyses of populations identified as Hypostomus strigaticeps from the upper Paraná River basin were conducted in order to test whether these different populations comprises cryptic species or structured populations and to assess their genetic variability. The sequences of the mitochondrial DNA ATP sintetase (subunits 6/8) of 27 specimens from 10 populations (one from Mogi-Guaçu River, five from Paranapanema River, three from Tietê River and one from Peixe River) were analyzed. The phylogeographic analysis showed the existence of eight haplotypes (A-H), and despite the ancestral haplotype includes only individuals from the Tietê River basin, the distribution of H. strigaticeps was not restricted to this basin. Haplotypes A, B and F were the most frequent. Haplotypes D, E, F, G, and H were present in the sub-basin of Paranapanema, two (A and B) were present in the sub-basin of the Tietê River, one (C) was exclusively distributed in the sub-basin of the Peixe River, and one (B) was also present in the sub-basin of the Grande River. The phylogenetic analysis showed that the populations of H. strigaticeps indeed form a monophyletic unit comprising two lineages: TG, with representatives from the Tietê, Mogi-Guaçu and Peixe Rivers; and PP, with specimens from the Paranapanema River. The observed degree of genetic divergence within the TG and PP lineages was 0.1% and 0.2%, respectively, whereas the genetic divergence between the two lineages themselves was approximately 1%. The results of the phylogenetic analysis do not support the hypothesis of existence of crypt species and the phylogeographic analysis confirm the presence of H. strigaticeps in other sub-basins of the upper Paraná River: Grande, Peixe, and Paranapanema sub-basins.
ATPase; Bayesian Inference; Haplotype; Nested clade; Phylogeny
Neste estudo, foram conduzidas análises filogenéticas e filogeográficas de populações identificadas como Hypostomus strigaticeps na bacia do alto rio Paraná a fim de testar se essas populações compreendem espécies crípticas ou populações estruturadas e avaliar a variabilidade genética das mesmas. Foram analisadas sequências do DNA mitocondrial ATP sintetase (subunidades 6/8) de 27 espécimes de 10 populações (uma do rio Mogi-Guaçu, cinco do rio Paranapanema, três do rio Tietê e uma do rio do Peixe). A análise filogeográfica mostrou a existência de oito haplótipos (A-H), e apesar do haplótipo ancestral incluir apenas indivíduos da bacia do rio Tietê, a distribuição de H. strigaticeps não se restringe a esta bacia. Os haplótipos A, B e F foram os mais frequentes. D, E, F, G e H estão presentes na sub-bacia do rio Paranapanema, dois (A e B) estão presentes na sub-bacia do rio Tietê, um (C) está exclusivamente distribuído na sub-bacia do rio do Peixe, e um (B) também está presente na sub-bacia do rio Grande. A análise filogenética mostrou que as populações de H. strigaticeps realmente formam uma unidade monofilética que compreende duas linhagens: TG, com representantes do rio Tietê, rio Mogi-Guaçu e rio do Peixe, e PP, com espécimes do rio Paranapanema. O grau de divergência genética observada nas linhagens de TG e PP foram de 0,1% e 0,2%, respectivamente, enquanto que a divergência genética entre as duas linhagens foi de aproximadamente 1%. Os resultados da análise filogenética não suportam a hipótese da existência de espécies crípticas e a análise filogeográfica confirma a presença de H. strigaticeps em outras sub-bacias do alto rio Paraná: sub-bacias do rio Grande, rio do Peixe e rio Paranapanema.
Introduction
The Neotropical freshwater fish fauna is extremely diverse, comprising approximately 71 families and 4,475 valid species of fish characterized by high endemicity (Reis et al., 2003). The order Siluriformes represents a large and diverse group of bony fishes with 37 families and more than 2500 species identified (Burgess, 1989; Ferraris, 1995; Britto, 2003). One of the most species-rich families of siluriforms is the family Loricariidae whose members are known as suckermouth armored catfishes. The loricariids are characterized by a body completely covered by dermal plates, small bone-shaped structures (thorn odontodes) in the head and by a sucker-like mouth with fleshy papillae (Reis et al., 2003).
The genus Hypostomus is the most diverse group within the family (Weber, 2003; Hollanda Carvalho et al., 2010), with 126 species distributed from Central America to southern South America (Zawadzki et al., 2010). The relative morphological polymorphism of the Hypostomus species make difficult to identify species-specific characters for the genus (Armbruster, 2004). This extensive morphological variation combined with the cytogenetic diversity observed, with diploid numbers ranging from 2n = 52 in Hypostomus emarginatus (Artoni & Bertollo, 2001) to 2n = 84 in Hypostomus sp. (Cereali et al., 2008) evidence the great taxonomic diversity of group.
Studies related to phylogeography and genetic variability in the genus Hypostomus, are very scarce. A molecular study on the genus was conducted by Montoya-Burgos (2003) inferring about the biogeographic patterns of genus Hypostomus showed that in the upper Paraná River basin the genus represent a polyphyletic group. More recent studies involving the genetic characterization of Hypostomus populations used RAPD by Sofia et al. (2008) and Allozymes by Zawadzki et al. (2008) were conducted and these studies show a great genetic variability in some species, however not mentioned any phylogenetic relationships.
Hypostomus strigaticeps (Fig. 1) was described by Regan (1908) from specimens of the Piracicaba River a sub-basin of the Tietê River. According to Weber (2003) and Reis et al. (2003) it is restricted to the sub-basin of the Tietê River, in the upper Paraná River basin. This species was not mentioned in any other sub-basin of the upper Paraná River basin. Observed morphological variation in the Hypostomus populations along the sub-basins of the Paraná River (Paranapanema and Rio Grande sub-basins), raises the possibility that these morphotypes represent either polymorphic populations of H. strigaticeps or cryptic species morphologically similar to H. strigaticeps.
Dorsal, lateral and ventral views of Hypostomus strigaticeps. MZUSP 79646, Tietê River, São Paulo State, Brazil. 127.6 mm SL.
In order to test whether these different populations comprise cryptic species or structured populations, in this study we sequenced the mitochondrial ATPase 6/8 gene in different populations of Hypostomus strigaticeps from the Tietê river to assess their genetic variability. We also analyzed the genetic variability of Hypostomus specimens from three adjacent sub-basins of the Paraná River: the Paranapanema, Peixe and Grande rivers (Mogi-Guaçu River) sub-basins, to determine the current range of H. strigaticeps in the Paraná River, and to identify the historical process and dispersal patterns of this species.
Material and Methods
A total of twenty-seven Hypostomus specimens from four sub-basins of the Paraná River (Fig. 2) were examined. Specimens from 10 populations from four sub-basins were examined: three from the Tietê, five from Paranapanema, one from Grande (Mogi-Guaçu) and one from Peixe (Table 1). Hypostomus ancistroides, H. paulinus, and H. iheringii from the upper Paraná River basin were used as close outgroups, whereas Rhamdia quelen (Heptapteridae) was used as distant outgroup. The specimens were deposited in the collections of the Laboratório de Citogenética of Universidade Estadual Paulista (UNESP, Rio Claro, Brazil), and in the ichthyology collection of Nupelia (NUP) of Universidade Estadual de Maringá (UEM, Maringá, Brazil).
Map of the river system in Brazil, highlighting the upper rio Paraná basin and the collection points.
Identification of Hypostomus strigaticeps specimens collected, respective collection points at the upper Paraná River basin and their haplotypes and mtDNA lineage
DNA was extracted with the phenol-chloroform-isoamyl alcohol technique (Sambrook & Russell, 2001). PCR amplification of the ATPase 6/8 gene region was obtained from a volume of 13.5 mL of a solution containing 6.25 mL of PCR Mix (Quiagen), 5.25 mL of Milli Q water, 0.5 mL primer F (10 mM), 0.5 mL of primer R (10 mM) and 1.0 mL of template DNA (200 ng). PCR was performed in a thermocycler (Eppendorf Mastercycler) and consisted of the following cycling conditions: an initial cycle of denaturation at 94 °C for 40 s, followed by 35 cycles at 94 °C for 30 s, annealing at 55 °C for 40 s, chain extension at 68°C. Check temperature extension for 2 min, and a final extension at 72°C for 5 min. The following set of primers was used: L8331 (5'-AA GCR TYR GCC TTT TAA GC-3') and H9236 (5'-GTT AGT GGT CAK GGG CTT GGR TC-3') (Perdices et al., 2002), and resulted in an amplification of about 850 bp. The amplified DNA segments were run on 1% agarose gel stained with 2 mL SYBR Safe (10,000×) (Invitrogen(r)) in 1 mL of blue juice running buffer (10×) and visualized in ultraviolet light. The quantification of the amplified gene products was performed using the molecular weight marker Lambda (l) DNA 50 ng and 30 ng. For the sequence analysis, the amplified DNA was purified with the EXOSAP enzyme and subsequently sequenced (MacroGen, Korea).
Model Test 3.06 (Posada & Crandall, 1998) was used to find the best fitting evolution model for our data. The model selected by the Akaike criterion was GTR+I+G (empirical base frequencies A=0.3338, C=0.3053, G=0.1023, T=0.2586; substitution rates: A-C=1.77, A-G=36.7617, A-T=0.5967, C-G=2.1775, C-T=17.2043, G-T=1.0). These parameters were used to estimate Maximum Likelihood (ML) distances in the Neighbor-Joining (NJ) analyses in PAUP*, with 1,000 bootstrap replicates. The maximum parsimony (MP) analysis was performed using heuristic searches with tree-bisection-reconnection (TBR) branch swapping and 10 random-taxon-addition replicates. Only minimal trees were retained and zero length branches were collapsed. A total of 1,000 bootstrap replicates were used to assess the relative robustness of the branches of the MP trees in PAUP*. The saturation analysis of data was performed in DAMBE 4.5.8 (Xia & Xie, 2001).
Bayesian inference (BI) phylogenies were inferred using MrBayes v. 3.1.2 (Huelsenbenck & Ronquist, 2001). Four simultaneous Monte Carlo Markov Chains (MCMC) were run for 2 million generations under the General Time Reversible (GTR) model of evolution, with sample frequency of 100 generations and chain temperature set to 0.2. Log-likelihood stability was reached after approximately 80,000 generations (excluding the first 80,000 trees). The remaining trees were used to compute a 50% majority-rule consensus tree. Posterior probability values were calculated to determine the level of support to the Bayesian topology.
We identified the haplotypes network (nested clade) to evaluate the association between haplotypes and their geographical distribution. This analysis was constructed using the TCS software package (Clement et al., 2000), which employs the method of Templeton et al. (1992).
We used the mtDNA clock calibration of 1.30% sequence divergence per million years (calculate using uncorrected p-genetic distance) for marine fishes separated by the Pliocene closure of the Isthmus of Panama (Bermingham et al., 1997) and previously applied for the genus Rhamdia from Central America (Perdices et al., 2002).
Results
The alignment of the ATPase 6/8 gene sequences revealed 828 conserved and 14 variable base pairs, of which 11 were informative for the Maximum Parsimony analysis. Mean nucleotide base composition in the sequences was as follows: 28.7 (T), 29.5 (C), 30.1 (A) and 11.7 (G), the data obtained were not saturated.
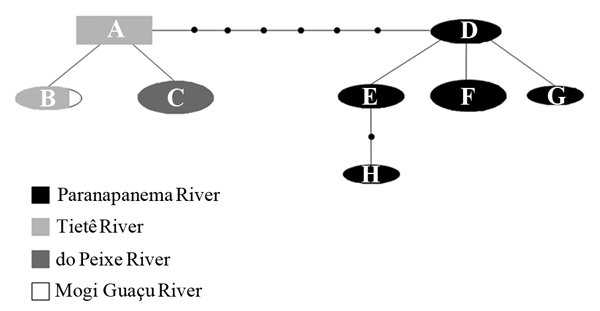
All phylogenies (NJ, MP and BI) were congruent differing only in their branch support. The phylogenetic analysis showed the analyzed Hypostomus specimens from the upper Paraná River as a monophyletic group organized into two major mtDNA lineages: Lineage TG, with representatives from the Tietê, Grande (Mogi-guaçu) and Peixe River populations; and Lineage PP, with specimens from the Paranapanema River basin populations (Fig. 3) (Table 1). These lineages were highly supported by bootstrap values (>86%) and posterior probability values ( >80%). The mean genetic distance between TG and PP lineages was 0.94% ± 0.20 whereas among individuals within the TG and PP lineages was 0.11% ± 0.08 and 0.27% ± 0.30, respectively. The genetic distance observed between H. strigaticeps and other species of the genus analyzed, H. iheringii, H. paulinus, and H. ancistroides was 3.80% ± 0.09, 4.06% ± 0.10 and 5.82% ± 0.10, respectively.
Phylogenetic tree based on the ATPase 6/8 mitochondrial gene using NJ, MP and BI methods. Values on branches represent bootstrap values for NJ, MP and posterior probabilities for BI.
The phylogeographic analysis revealed eight haplotypes (A-H). Haplotype A was considered the ancestral haplotype, and it was present in two of the three populations analyzed from the Tietê River sub-basin (Araquá and Conchas Rivers). The individuals of the Corumbataí River (Tietê River sub-basin) showed haplotype B shared with individuals inhabiting água Boa stream (Grande River) (Fig. 4). Haplotype C was exclusively found in the Peixe sub-basin whereas Haplotypes D, E, F, G, and H were exclusive of all rivers of the Paranapanema sub-basin (Table 1). Haplotypes A, C, and F had the highest frequency (18.51%).
Discussion
Due to the high degree of morphological similarity between the representatives of the genus Hypostomus, the use of molecular markers to identify and characterize its populations and species is extremely valuable. Our data supported the hypothesis that the different ingroup Hypostomus populations analyzed represented the same species, Hypostomus strigaticeps, as evidenced by the small genetic distances among the analyzed populations. The phylogenetic analysis showed that these populations form two major mtDNA lineages: TG, distributed in the sub-basins of the Tietê, Grande (Mogi-Guaçu) and Peixe Rivers; and PP, represented in the sub-basin of the Paranapanema River. Similarly, the phylogeographic analysis also showed two clades: Tietê-Peixe-Grande and Paranapanema, corresponding to lineages TG and PP, respectively, in the phylogenetic analysis. Although all samples analyzed were H. strigaticeps individuals, our phylogenetic and phylogeographic results consistently showed a clear separation between the sub-basins. The phylogeographic analysis showed that the ancestral haplotype of H. strigaticeps was distributed in the Tietê River sub-basin. The observed network of haplotypes related haplotypes found in the Tietê River sub-basin and Grande River (Mogi-Guaçu) and Peixe River, and clearly separated haplotypes found in the Paranapanema sub-basin. Although further analysis with more populations and sub-basins of the Paraná River basin should be performed to have more consistent results, we found the ancestral haplotypes of H. strigaticeps distributed in the Tietê River sub-basin related to the adjacent sub-basins Peixe and Grande what suggests a recent gene flow.
Considering a rate of mutation of the ATPase 6/8 gene in fish of 1.3%/Ma (Bermingham et al., 1997; Perdices et al., 2002) and the observed genetic divergence of 0.94% between lineages TG and PP, we calculate the separation of both lineages as approximately about 723,000 years ago, during the Quaternary. Likely due to their bottom-dwelling body shape the Hypostomus species seems not a group of freshwater fishes prone to perform long river displacements (Weber, 2003). Genetic studies on other species of the genus Hypostomus as H. ancistroides (Sofia et al., 2008; Endo et al., 2012), H. margaritifer (Zawadzki et al., 2002), and H. regani (Zawadzki et al., 2008) have also found moderate structured populations as we found in the H. strigaticeps populations analyzed.
The hypothesis that H. strigaticeps is widely distributed in the Upper Parana River, is supported by the present data. Our phylogenetic results showed that all individuals analyzed from the four sub-basins investigated belong to H. strigaticeps. Genetic variability and the genetic distances found among individuals inhabiting all water systems analyzed indicated that all individuals belong to the same species: H. strigaticeps. Moreover, all these individuals form a monophyletic group highly supported, that were not intimately related to any of the sympatric Hypostomus species analyzed that occur in these area. Our results of the phylogeographic analysis confirm the presence of H. strigaticeps in other sub-basins of the upper Paraná River: Grande, Peixe and Paranapanema sub-basins.
The authors are grateful to Alessandro G. Bifi, Fábio F. Roxo, and Renato Devidé by helping to collect fishes in the fieldworks; to Cristiane Marcia Mileo and Edson Lourenço da Silva to image editing; to Bioedit(r) by helping with the English language; to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for financial support (process number: 07/58641-0). Nupélia provide logistic support.
Literature Cited
- Armbruster, J. W. 2004. Phylogenetic relaptionships of the sucker-mouch armored catfishes (Loricariidae) with particular emphasis on the Hypostominae and Ancistrinae. Zoological Journal of the Linnean Society, 141: 1-80.
- Artoni, R. F. & L. A. C. Bertollo. 2001. Trends in the karyotype evolution of Loricariidae fish (Siluriformes). Hereditas, 134: 201-210.
- Bermingham, E., S. S. Mccafferty & A. P. Martin. 1997. Fish biogeography and molecular clocks: perspectives from the Panamanian Isthmus. Pp. 113-128. In: Kocher, T., Stepien, C. A. (Ed.). Molecular Systematic of Fishes.
- Britto, M. R. 2003. Análise filogenética da ordem Siluriformes com ênfase nas relações da superfamília Loricarioidea (Teleostei: Ostariophysi). Unpublished Ph.D. Dissertation, Instituto de Biociências, Universidade Estadual de São Paulo, 512p.
- Burgess, W. E. 1989. An atlas of freshwater and marine catfishes a preliminary survey of the Siluriformes. T. F. H. Publications, Neptune, NJ.
- Cereali, S. S., E. Pomini, R. Rosa, C. H. Zawadzki, O. Froehlich & L. Giuliano-Caetano. 2008. Karyotype description of two species of Hypostomus (Siluriformes, Loricariidae) of the Planalto da Bodoquena, Brazil. Genetic and Molecular Research, 7: 583-591.
- Clement, M., D. Posada & K. A. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9: 1657-1659.
- Endo, K. S., E. R. M. Martinez, C. H. Zawadzki, L. R. S. Paiva & H. F. Júlio Jr. 2012. Karyotype description of possible new species of the Hypostomus ancistroides complex (Teleostei: Loricariidae) and other Hypostominae. Acta Scientiarum Biological Sciences, 34: 181-189.
- Ferraris, C. J. 1995. Catfishes and knifefishes. Pp. 106-112. In: Encyclopedia of fishes, J. R. Paxton & W. N. Eschmeyer (Eds.). Academic Press, San Diego.
- Hollanda Carvalho, P., F. C. T. Lima & C. H. Zawadzki. 2010. Two new species of the Hypostomus cochliodon group (Siluriformes: Loricariidae) from the rio Negro basin in Brazil. Neotropical Ichthyology, 8: 39-48.
- Huelsenbenck, J. P. & F. R. Ronquist. 2001. MrBayes: Bayesian inference of phylogeny, Bioinformatics, 17: 754-755.
- Montoya-Burgos, J. I. 2003. Historical biogeography of the catfish genus Hypostomus (Siluriformes: Loricariidae), with implications on the diversification of Neotropical ichthyofauna. Molecular Ecology, 12: 1855-1867.
- Panchal, M. 2007. The automation of Nested Clade Phylogeography Analysis. Bioinformatics Applications Note, 23: 509-510.
- Perdices, A., E. Bermingham, A. Montilla & I. Doadrio. 2002. Evolutionary history of genus Rhamdia (Teleostei: Pimelodidae) in Central America. Molecular Phylogenetic and Evolution, 25: 172-189.
- Posada, D. & K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics, 14: 817-818.
- Regan, C. T. 1908. Descriptions of new loricariid fishes from South America. Proceedings of the Zoological Society of London, 4: 795-800.
- Reis, R. E., S. O. Kullander & C. Ferraris (Eds.). 2003. Check List of the Freshwater Fishes of South and Central America, Porto Alegre, Edipucrs, 729p.
- Sambrook, J. & D. W. Russel. 2001.Molecular cloning. A laboratory manual. Cold New York: Spring Harbor Laboratory Press. Third Edition.
- Sofia, S. H., B. A. Galindo, F. M. Paula, L. M. K. Sodré & C. B. R. Martinez. 2008. Genetic diversity of Hypostomus ancistroides (Teleostei, Loricariidae) from an urban stream. Genetics and Molecular Biology, 31: 317-323.
- Templeton, A. R., K. A. Crandall & C. F. Sing. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics, 132: 619-633.
- Xia, X. & Z. Xie. 2001. DAMBE: Data analysis in molecular biology and evolution. Journal Heredity, 92: 371-373.
- Weber, C. 2003. Subfamily Hypostominae (Armored catfishes). Pp. 351-372. In: Check List of the freshwater fishes of South America. Reis, R. E., S. O. Kullander & C. J. Ferraris Jr. ( Eds.). Porto Alegre, Edipucrs, 729p.
- Zawadzki, C. H., C. Weber, C. S. Pavanelli & E. Renesto. 2002. Morphological and biochemical comparison of two allopatrid populations of Hypostomus margaritifer (Regan, 1907) (Osteichthyes, Loricariidae) from the upper Paraná River basin, Brazil. Acta Scientiarum, 24: 499-505.
- Zawadzki, C. H., E. Renesto, M. D. Peres & S. Paiva. 2008. Allozyme variation among three populations of the armored catfish Hypostomus regani (Ihering, 1905) (Siluriformes: Loricariidae) from the Paraná and Paraguay river basins, Brazil. Genetics and Molecular Biology, 31: 767-771.
- Zawadzki, C. H., C. Weber & C. S. Pavanelli. 2010. A new dark-saddled species of Hypostomus (Siluriformes: Loricariidae) from the upper Paraguay river basin. Neotropical Ichthyology, 8: 719-725.
-
Published March 31, 2013
Publication Dates
-
Publication in this collection
Jan-Mar 2013
History
-
Received
13 Dec 2011 -
Accepted
20 June 2012